Bipyridine Ruthenium(II) Complexes with Halogen-Substituted Salicylates: Synthesis, Crystal Structure, and Biological Activity
Abstract
1. Introduction
2. Results and Discussion
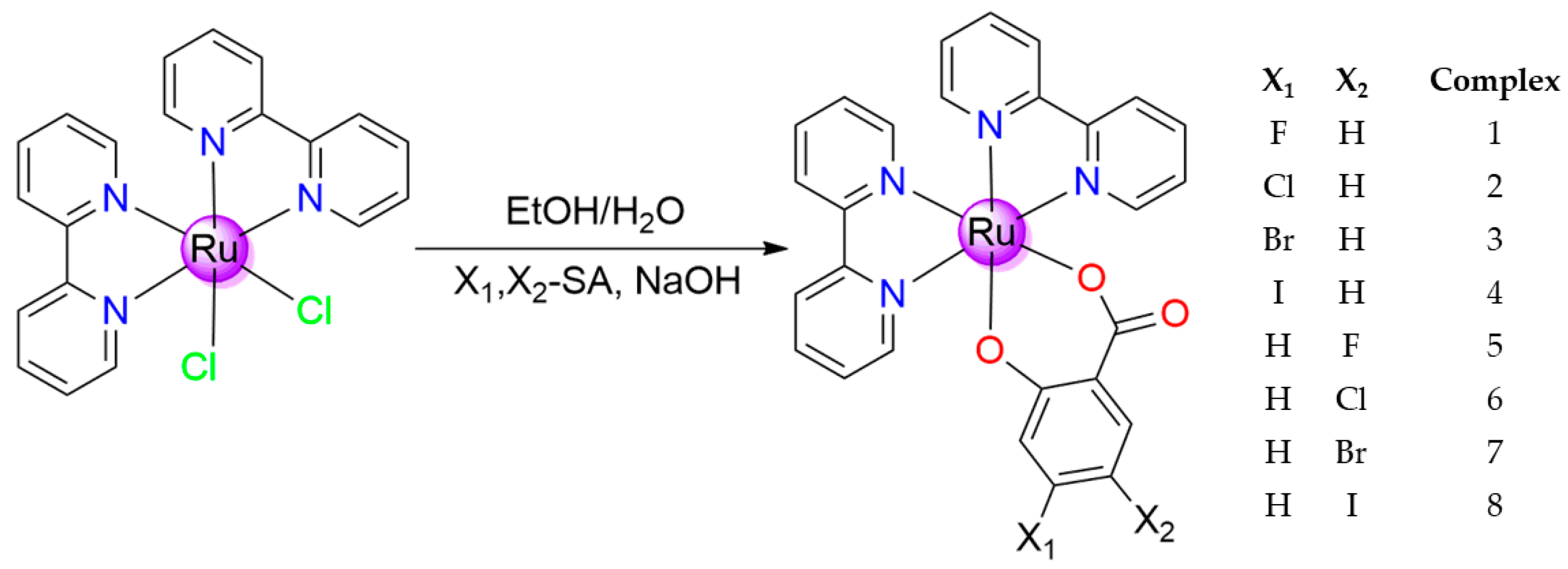
2.1. Synthesis, Stability and Spectroscopical Study of Prepared Complexes
2.1.1. Infrared Spectroscopy
2.1.2. 1H-NMR Spectra of Prepared Complexes
2.1.3. Electronic Spectra, ESI-MS and Study of Solvatochromism
2.2. Crystal and Molecular Structures of Prepared Complexes
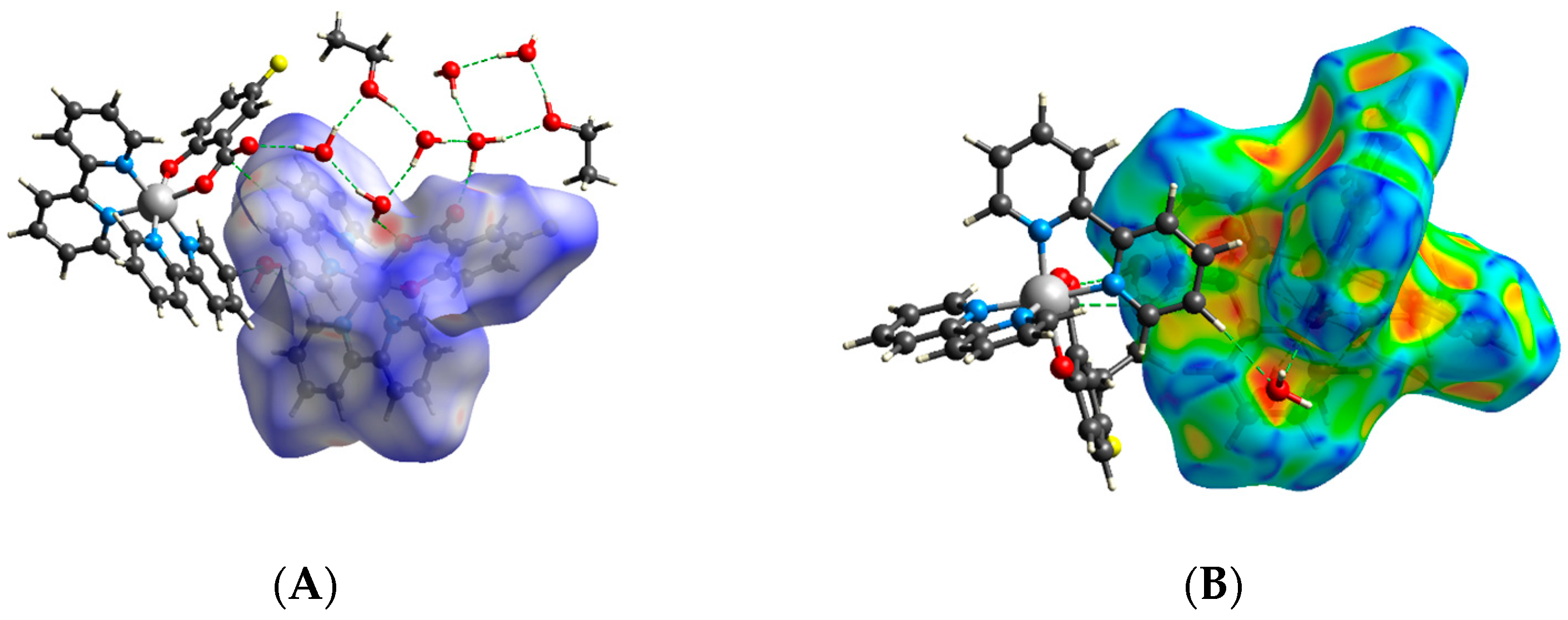
2.3. Hirshfeld Surfaces Analysis
2.4. Interaction with Bovine Serum Albumin
2.5. Interaction with DNA
2.6. In Vitro Study of Anticancer Activity
3. Materials and Methods
3.1. Synthesis and Chemicals
3.2. X-ray Crystallography
3.3. Hirshfeld Surface Analysis
3.4. Molecular Spectroscopy
3.5. Interaction with Bovine Serum Albumin (BSA)
3.6. Interaction with ct-DNA
3.6.1. Absorption Titrations
3.6.2. Quenching of the Fluorescence of EB-DNA Adduct
3.7. Study of Anticancer Activity
3.7.1. Cell Culture
3.7.2. Cytotoxic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Cashman, J.N. The Mechanisms of Action of NSAIDs in Analgesia. Drugs 1996, 52, 13–23. [Google Scholar] [CrossRef]
- Vane, J.R.; Botting, R.M. The mechanism of action of aspirin. Tromb. Res. 2003, 5–6, 225–258. [Google Scholar] [CrossRef] [PubMed]
- Kirkby, N.S.; Sampaio, W.; Etelvino, G.; Alves, D.T.; Anders, K.L.; Temponi, R.; Shala, F.; Nair, A.S.; Ahmetaj-Shala, B.; Jiao, J.; et al. Cyclooxygenase-2 Selectively Controls Renal Blood Flow Through a Novel PPARβ/δ-Dependent Vasodilator Pathway. Hypertension 2018, 71, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Peskar, B.M.; Maricic, N.; Gretzer, B.; Schuligoi, R.; Schmassmann, A. Role of cyclooxygenase-2 in gastric mucosal defense. Life Sci. 2001, 25–26, 2993–3003. [Google Scholar] [CrossRef]
- Vane, J.R.; Botting, R.M. Mechanism of action of antiinflammatory drugs. Int. J. Tissue React. 1998, 20, 3–15. [Google Scholar] [PubMed]
- Srivastava, P.; Mishra, R.; Verma, M.; Sivakumar, S.; Patra, A.K. Cytotoxic ruthenium(II) polypyridyl complexes with naproxen as NSAID: Synthesis, biological interactions and antioxidant activity. Polyhedron 2019, 172, 132–140. [Google Scholar] [CrossRef]
- Lipsky, P.E.; Brooks, P.; Crofford, L.J.; DuBois, R.; Graham, D.; Simon, L.S.; van de Putte, L.B.; Abrsamson, S.B. Unresolved issues in the role of cyclooxygenase-2 in normal physiologic processes and disease. Arch. Intern. Med. 2000, 160, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharan, N.V.; Dai, H.; Roos, K.L.T.; Evanson, N.K.; Tomsik, J.; Elton, T.S.; Simmons, D.L. COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: Cloning, structure, and expression. Proc. Natl. Acad. Sci. USA 2002, 99, 13926–13931. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.S.Y. Role of Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) in Cancer Prevention and Cancer Promotion. Adv. Pharmacol. Sci. 2019, 2019, 3418975. [Google Scholar] [CrossRef]
- Rayburn, E.R.; Ezell, S.J.; Zhang, R. Anti-Inflammatory Agents for Cancer Therapy. Mol. Cell. Pharmacol. 2009, 1, 29–43. [Google Scholar] [CrossRef]
- Wang, H.; Li, M.; Rinehart, J.J.; Zhang, R. Pretreatment with dexamethasone increases antitumor activity of carboplatin and gemcitabine in mice bearing human cancer xenografts: In vivo activity, pharmacokinetics, and clinical implications for cancer chemotherapy. Clin. Cancer Res. 2004, 10, 1633–1644. [Google Scholar] [CrossRef]
- Banti, C.N.; Hadjikakou, K. Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) in Metal Complexes and Their Effect at the Cellular Level. Eur. J. Inorg. Chem. 2016, 2016, 3048–3071. [Google Scholar] [CrossRef]
- Psomas, G. Copper(II) and zinc(II) coordination compounds of non-steroidal anti-inflammatory drugs: Structural deatures and antioxidant octivity. Coord. Chem. Rev. 2020, 412, 213259. [Google Scholar] [CrossRef]
- Khan, H.Y.; Parveen, S.; Yousuf, I.; Tabassum, S.; Arjmand, F. Metal complexes of NSAIDs as potent anti-tumor chemotherapeutics: Mechanistic insighhts into cytotoxic activity via multiple pathways primarily by inhibition of COX-1 and COX-2 enzymes. Coord. Chem. Rev. 2022, 453, 214316. [Google Scholar] [CrossRef]
- Jayamani, A.; Sengottuvelan, N.; Chakkaravarthi, G. Synthesis, structural, electrochemical, DNA interaction, antimicrobial and molecular docking studies on dimeric copper(II) complexes involving some potential bidentate ligands. Polyhedron 2014, 81, 764–776. [Google Scholar] [CrossRef]
- Kucková, L.; Jomová, K.; Švorcová, A.; Valko, M.; Segľa, P.; Moncoľ, J.; Kožíšek, J. Synthesis, Crystal Structure, Spectroscopic Properties and Potential Biological Activities of Salicylate–Neocuproine Ternary Copper(II) Complexes. Molecules 2015, 20, 2115–2137. [Google Scholar] [CrossRef] [PubMed]
- Kostova, I. Ruthenium Complexes as Anticancer Agents. Curr. Med. Chem. 2006, 13, 1085–1107. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.; Nam, T.-G. Ruthenium Complexes as Anticancer Agents: A Brief History and Perspective. Drug Des. Dev. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef]
- Bouma, M.; Nuijen, B.; Jansen, M.T.; Sava, G.; Flaibani, A.; Bult, A.; Beijnen, J.H. A kinetic study of the chemical stability of the antimetastatic ruthenium complex NAMI-A. Int. J. Pharm. 2002, 248, 239–246. [Google Scholar] [CrossRef]
- Wernitznig, D.; Kiakos, K.; Favero, G.D.; Harrer, N.; Machat, H.; Osswald, A.; Jakupec, M.A.; Wernitznig, A.; Sommergruber, W.; Keppler, B.K. First-in-class ruthenium anticancer drug (KP1339/IT-139) induces an immunogenic cell death signature in colorectal spheroids in vitro. Metallomics 2019, 6, 1044–1048. [Google Scholar] [CrossRef]
- Rausch, M.; Dyson, P.J.; Nowak-Sliwinska, P. Recent Considerations in the Application of RAPTA-C for Cancer Treatment and Perspectives for Its Combination with Immunotherapies. Adv. Ther. 2019, 2, 1900042. [Google Scholar] [CrossRef]
- Monro, S.; Colón, K.L.; Yin, H.; Roque III, J.; Konda, P.; Gujar, S.; Thummel, R.P.; Lilge, L.; Cameron, C.G.; McFarland, S.A. Transition Metal Complexes and Photodynamic Therapy from a Tumor-Centered Approach: Challenges, Opportunities, and Highlights from the Development of TLD1433. Chem. Rev. 2019, 119, 797–828. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-C.; Zhang, Y.; Jie, X.-M.; She, J.; Dongye, G.-Z.; Zhong, Y.; Deng, Y.-Y.; Wang, J.; Guo, B.-Y.; Chen, L.-M. Ruthenium(II) salicylate complexes inducing ROS-mediated apoptosis by targeting thioredoxin reductase. J. Inorg. Biochem. 2019, 193, 112–123. [Google Scholar] [CrossRef]
- Chen, J.-C.; Wang, J.; Deng, Y.-Y.; Wang, T.; Miao, T.-F.; Li, C.-P.; Cai, X.-H.; Liu, Y.; Henri, J.; Chen, L.-M. Ruthenium(II) Complexes Bearing O,O-Chelating Ligands Induced Apoptosis in A549 Cells through the Mitochondrial Apoptotic Pathway. Bioinorg. Chem. Appl. 2020, 2020, 8890950. [Google Scholar] [CrossRef] [PubMed]
- Paunescu, E.; McArthur, S.; Soudani, M.; Scopelliti, R.; Dyson, P.J. Nonsteroidal Anti-inflammatory—Organometallic Anticancer Compounds. Inorg. Chem. 2016, 55, 1788–1808. [Google Scholar] [CrossRef] [PubMed]
- School, M.T.; Rudbari, H.A.; Gil-Antón, T.; Cuevas-Vicario, J.V.; García, B.; Busto, N.; Moini, N.; Blacque, O. The effect of halogenation of salicylaldehyde on the antiproliferative activities of {Δ/Λ-[Ru(bpy)2(X,Y-sal)]BF4} complexes. Dalton Trans. 2022, 51, 7658–7672. [Google Scholar] [CrossRef]
- Nakamoto, I. Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B, 6th ed.; Wiley: New York, NY, USA, 2009. [Google Scholar]
- Reichardt, C.; Erlton, T. Solvents and Solvent Effects in Organic Chemistry; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2010. [Google Scholar]
- Ji, Z.; Huang, S.D.; Guadalupe, A.R. Synthesis, X-ray structures, spectroscopic and electrochemical properties of ruthenium(II) complexes containing 2,2′-bipyrimidine. Inorg. Chim. Acta 2000, 305, 127–134. [Google Scholar] [CrossRef]
- Ghosh, A.; Mandoli, A.; Damdoaran, K.K.; Yadav, N.S.; Ghosh, T.; Jha, B.; Thomas, J.A.; Das, A. DNA binding and cleavage properties of a newly synthesised Ru(II)-polypyridyl complex. Dalton Trans. 2009, 42, 9312–9321. [Google Scholar] [CrossRef]
- Constantino, V.R.L.; Toma, H.E.; de Oliveira, L.F.C.; Rein, F.N.; Rocha, R.C.; de Oliveira Silva, D. Structure, spectroscopy and electrochemistry of the bis(2,2’-bipyridine)(salicylato)ruthenium(II) complex. J. Chem. Soc. Dalton Trans. 1999, 11, 1735–1740. [Google Scholar] [CrossRef]
- Topala, T.; Bodocki, A.; Opean, L.; Opean, R. Bovine Serum Albumin Interactions with Metal Complexes. Clujul. Med. 2014, 87, 215–219. [Google Scholar] [CrossRef]
- Jozefíková, F.; Perontsis, S.; Koňáriková, K.; Švorc, Ľ.; Mazúr, M.; Psomas, G.; Moncoľ, J. In vitro biological activity of copper(II) complexes with NSAIDs and nicotinamide: Characterization, DNA- and BSA-interaction study and anticancer activity. J. Inorg. Biochem. 2022, 228, 111696. [Google Scholar] [CrossRef] [PubMed]
- Topala, T.; Bodoki, A.; Oprean, L.; Oprean, R. Experimental techniques in the study of metal complex-DNA-interactions. Farmacia 2014, 62, 1049–1061. [Google Scholar]
- Lay, P.A.; Sargeson, A.M.; Taube, H.; Chou, M.H.; Creutz, C. Cis-Bis(2,2′-Bipyridine-N,N′) Complexes of Ruthenium(III)/(II) and Osmium(III)/(II). Inorg. Synth. 1986, 24, 291–299. [Google Scholar] [CrossRef]
- Koziskova, J.; Hahn, F.; Richter, J.; Kožíšek, J. Comparison of different absorption corrections on the model structure of tetrakis(μ2-acetato)-diaqua-di-copper(II). Acta Chim. Slovaca 2016, 9, 136–140. [Google Scholar] [CrossRef]
- CrysAlisPRO; Oxford Diffraction/Agilent Technologies UK Ltd.: Yarnton, UK, 2009.
- Sheldrick, G.M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayalitaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Vobded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayalitaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Parkin, A.; Barr, G.; Dong, W.; Gilmore, C.J.; Jayalitaka, D.; McKinnon, J.J.; Spackman, M.A.; Wilson, C.C. Comparing entire crystal structures: Structural genetic fingerprinting. CrystEngComm 2007, 9, 648–652. [Google Scholar] [CrossRef]
- Malis, G.; Geromichalou, E.; Geromichalos, G.D.; Hatzidimitriou, A.G.; Psomas, G. Copper(II) complexes with non-steroidal anti-inflammatory drugs: Structural characterization, in vitro and in silico biological profile. J. Inorg. Biochem. 2021, 224, 111563. [Google Scholar] [CrossRef] [PubMed]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-ligand complexes of ruthenium(II)—Factors governing binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Jozefíková, F.; Perontsis, S.; Šimunková, M.; Barbieriková, Z.; Švorc, Ľ.; Valko, M.; Psomas, G.; Moncol, J. Novel copper(II) complexes with fenamates and isonicotinamide: Structure and properties, and interactions with DNA and serum albumin. New J. Chem. 2020, 44, 12827–12842. [Google Scholar] [CrossRef]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936–942. [Google Scholar] [PubMed]
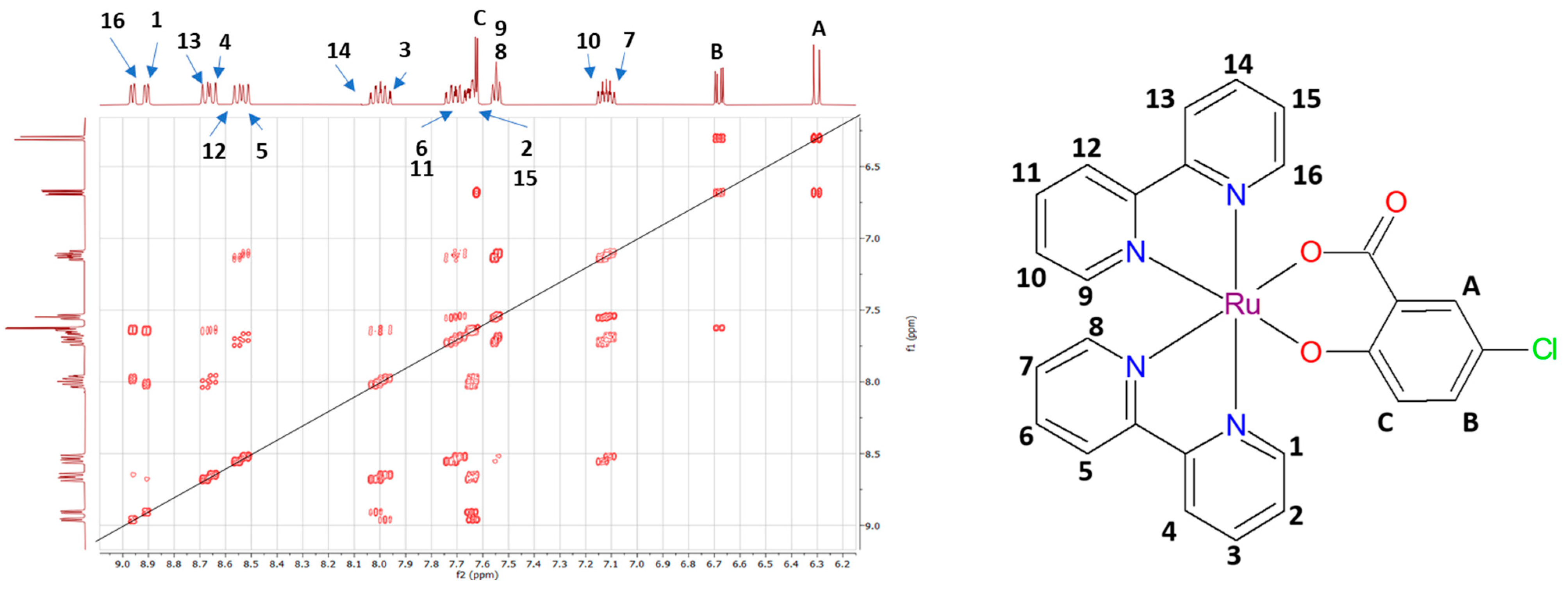












| Complex | ν(O-H)/cm−1 | ν(C-H)ar./cm−1 | ν(C=N)/cm−1 | νas(COO)/cm−1 | νs(COO)/cm−1 | δ/cm−1 | ν(C-O)/cm−1 | ν(Ru-N)/cm−1 |
|---|---|---|---|---|---|---|---|---|
| 1 | 3234 (br, m) | 3068 (w) | 1609 (m) | 1556 (s) | 1416 (s) | 140 | 1253 (s) | 410 (w) |
| 2 | 3356 (br, m) | 3101, 3068 (w) | 1583 (m) | 1542 (s) | 1415 (s) | 127 | 1237 (s) | 421 (w) |
| 3 | 3354 (br, m) | 3101, 3068 (w) | 1578 (m) | 1540 (s) | 1416 (s) | 124 | 1234 (s) | 420 (w) |
| 4 | 3294 (br, m) | 3064 (w) | 1571 (s) | 1542 (s) | 1413 (s) | 129 | 1245 (s) | 421 (w) |
| 5 | 3383 (br, m) | 3072 (w) | 1603 (w) | 1547 (s) | 1412 (s) | 135 | 1233 (s) | 424 (w) |
| 6 | 3384 (br, m) | 3066 (w) | 1589 (s) | 1557 (s) | 1402 (s) | 155 | 1241 (m) | 420 (w) |
| 7 | 3320 (br, m) | 3064 (w) | 1584 (m) | 1545 (s) | 1400 (s) | 145 | 1243 (s) | 421 (w) |
| 8 | 3359 (br, m) | 3067 (w) | 1571 (s) | 1556 (s) | 1417 (m) | 139 | 1250 (s) | 421 (w) |
| Complex | ILCT I | ILCT II | 1MLCT I | 1MLCT II | MLCT I | MLCT II |
|---|---|---|---|---|---|---|
| P1 | 245 | 293 | 349 | 497 | 363 | 544 (583, 641) |
| 1 | 245 | 295 | 363 | 524 | 369, 396 | 575 (688) |
| 2 | 244 | 295 | 370 | 524 | 368, 394 | 571, 604 (708) |
| 3 | 244 | 295 | 370 | 524 | 364, 393 | 575 (716) |
| 4 | 264 | 295 | 367 | 524 | 360, 393 | 573 (711) |
| 5 | 245 | 295 | 368 | 526 | 395 | 573 (725) |
| 6 | 244 | 295 | 371 | 525 | 389, 503 | 602 (719) |
| 7 | 244 | 295 | 370 | 524 | 362 | 601 |
| 8 | 242 | 295 | 366 | 525 | 391, 496 | 595 (742) |
| Compound | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Ru1-O1 | 2.066 (5) | 2.0587 (18) | 2.070 (6) | 2.081 (5) |
| Ru1-O3 | 2.067 (5) | 2.0649 (18) | 2.065 (6) | 2.066 (5) |
| Ru1-N1 | 2.035 (6) | 2.027 (2) | 2.031 (7) | 2.047 (6) |
| Ru1-N2 | 2.057 (7) | 2.052 (2) | 2.052 (8) | 2.044 (6) |
| Ru1-N3 | 2.020 (7) | 2.043 (2) | 2.049 (8) | 2.052 (5) |
| Ru1-N4 | 2.023 (6) | 2.028 (2) | 2.029 (7) | 2.027 (6) |
| Ru2-O4 | 2.082 (5) | - | - | 2.073 (5) |
| Ru2-O6 | 2.073 (5) | - | - | 2.081 (5) |
| Ru2-N5 | 2.032 (6) | - | - | 2.020 (6) |
| Ru2-N6 | 2.050 (7) | - | - | 2.081 (5) |
| Ru2-N7 | 2.046 (7) | - | - | 2.046 (5) |
| Ru2-N8 | 2.031 (6) | - | - | 2.020 (6) |
| Compound | 5 | 7A | 7B | 8 |
|---|---|---|---|---|
| Ru1-O1 | 2.061 (7) | 2.062 (4) | 2.067 (3) | 2.091 (4) |
| Ru1-O3 | 2.053 (6) | 2.064 (4) | 2.070 (3) | 2.071 (4) |
| Ru1-N1 | 2.050 (8) | 2.020 (4) | 2.029 (3) | 2.032 (5) |
| Ru1-N2 | 2.009 (8) | 2.053 (4) | 2.053 (3) | 2.055 (5) |
| Ru1-N3 | 2.037 (8) | 2.049 (4) | 2.044 (3) | 2.051 (5) |
| Ru1-N4 | 2.000 (8) | 2.016 (4) | 2.024 (3) | 2.022 (5) |
| Compound | ΔI/I0 (%) | KSV (M−1) × 105 | kq (M−1s−1) × 1013 | K × 104 | n |
|---|---|---|---|---|---|
| 1 | 69.7 | 1.21 (±0.05) | 1.21 (±0.02) | 7.32 (±0.34) | 1.20 |
| 2 | 78.0 | 1.89 (±0.05) | 1.89 (±0.02) | 12.82 (±0.61) | 1.12 |
| 3 | 75.1 | 1.61 (±0.03) | 1.61 (±0.01) | 14.80 (±0.41) | 1.02 |
| 4 | 66.9 | 1.07 (±0.02) | 1.07 (±0.01) | 16.75 (±0.940) | 0.86 |
| 5 | 73.0 | 1.45 (±0.06) | 1.45 (±0.02) | 7.23 (±0.250) | 1.28 |
| 6 | 74.9 | 1.56 (±0.03) | 1.56 (±0.01) | 18.99 (±0.93) | 0.93 |
| 7 | 79.1 | 1.99 (±0.03) | 1.99 (±0.01) | 22.42 (±0.27) | 0.97 |
| 8 | 75.7 | 1.57 (±0.11) | 1.57 (±0.05) | 36.52 (±2.42) | 0.87 |
| Compound | λ (nm) | ΔA/A0 (%) | Δλ (nm) | Kb (M−1) |
|---|---|---|---|---|
| 1 | 245 | 40.5 | +3 | 9.72 (±2.43) × 103 |
| 2 | 244 | 20.0 | +1 | 6.33 (±1.67) × 105 |
| 3 | 244 | 18.8 | +1 | 4.09 (±1.33) × 105 |
| 4 | 264 | 20.3 | −16 | 1.26 (±0.13) × 105 |
| 5 | 245 | 11.9 | +2 | 2.05 (±0.48) × 104 |
| 6 | 244 | 20.6 | +3 | 3.97 (±0.44) × 103 |
| 7 | 244 | 14.8 | +3 | 4.63 (±0.93) × 103 |
| 8 | 242 | 24.2 | +3 | 8.790 (±1.26) × 103 |
| Compound | ΔI/I0 (%) | KSV (M−1) × 104 | kq (M−1s−1) × 1012 |
|---|---|---|---|
| 1 | 85.2 | 3.15 (±0.23) | 1.37 (±0.10) |
| 2 | 94.7 | 10.70 (±0.98) | 4.65 (±0.43) |
| 3 | 91.1 | 6.41 (±0.55) | 2.79 (±0.23) |
| 4 | 83.5 | 4.76 (±0.41) | 2.07 (±0.17) |
| 5 | 92.0 | 7.10 (±0.58) | 3.09 (±0.25) |
| 6 | 79.4 | 6.68 (±0.83) | 2.90 (±0.36) |
| 7 | 94.4 | 9.39 (±1.16) | 4.08 (±0.50) |
| 8 | 86.2 | 5.22 (±0.51) | 2.27 (±0.22) |
| Compound | U-118MG | MCF-7 | ||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| 1 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | 5.76 × 10−6 | 4.75 × 10−6 | 3.30 × 10−6 |
| 2 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | 4.97 × 10−6 | 5.75 × 10−6 | 4.23 × 10−6 |
| 3 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | 6.78 × 10−6 | 5.69 × 10−6 |
| 4 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | 4.92 × 10−6 | 6.18 × 10−6 | 6.65 × 10−6 |
| 5 | 3.56 × 10−6 | 4.72 × 10−6 | 3.49 × 10−6 | <2 × 10−6 | <2 × 10−6 | <2 × 10−6 |
| 6 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | >10 × 10−6 | 6.20 × 10−6 | 3.79 × 10−6 |
| 7 | 5.35 × 10−6 | 3.95 × 10−6 | 8.38 × 10−6 | 4.23 × 10−6 | 4.92 × 10−6 | <2 × 10−6 |
| 8 | 4.08 × 10−6 | 2.65 × 10−6 | >10 × 10−6 | <2 × 10−6 | <2 × 10−6 | <2 × 10−6 |
| Compound | 1·3H2O·EtOH | 2·2.6H2O·2EtOH | 3·6H2O | 4·3H2O | 5·1.55H2O·EtOH | 7A·1.75H2O | 7B·H2O·EtOH | 8·4H2O |
|---|---|---|---|---|---|---|---|---|
| Chemical formula | C27H21FN4O4Ru | C31H33ClN4O6Ru | C27H21BrN4O4Ru | C27H21.5IN4O4.25Ru | C29H31FN4O7Ru | C27H21BrN4O4Ru | C29H27BrN4O5Ru | C27H27IN4O7Ru |
| Mr | 585.55 | 694.13 | 646.46 | 697.95 | 667.65 | 646.46 | 692.52 | 747.49 |
| Crystal system | Monoclinic | Trigonal | Trigonal | Triclinic | Monoclinic | Triclinic | Triclinic | Monoclinic |
| Space group | P21/c | R | R | P21/c | P21/c | |||
| T/K | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| a/Å | 15.5799 (6) | 25.4969 (2) | 25.7119 (7) | 11.3038 (2) | 10.2179 (2) | 9.4768 (6) | 9.79637 (20) | 9.9270 (5) |
| b/Å | 24.1583 (9) | 25.4969 (2) | 25.7119 (7) | 16.1443 (4) | 26.3315 (6) | 9.9798 (6) | 10.27595 (16) | 27.7411 (12) |
| c/Å | 14.9993 (5) | 24.4547 (2) | 24.1522 (6) | 16.2832 (3) | 10.8839 (2) | 14.9267 (5) | 14.58477 (17) | 11.0554 (6) |
| α/° | 90 | 90 | 90 | 79.887 (2) | 90 | 99.343 (4) | 73.1374 (12) | 90 |
| β/° | 101.057 (3) | 90 | 90 | 71.323 (2) | 106.823 (2) | 90.086 (4) | 79.6472 (15) | 112.761 (6) |
| γ/° | 90 | 120 | 120 | 74.945 (2) | 90 | 112.648 (6) | 87.7349 (15) | 90 |
| V/Å3 | 5540.7 (4) | 13767.9 (2) | 13827.9 (8) | 2704.55 (11) | 2803.02 (10) | 1282.39 (13) | 1382.08 (4) | 2807.4 (3) |
| Z | 8 | 18 | 18 | 2 | 4 | 2 | 2 | 4 |
| μ/mm−1 | 4.959 | 5.369 | 5.942 | 13.995 | 5.058 | 7.119 | 6.676 | 13.602 |
| Crystal size/mm | 0.12 × 0.15 × 0.24 | 0.13 × 0.22 × 0.22 | 0.15 × 0.18 × 0.23 | 0.05 × 0.08 × 0.12 | 0.01 × 0.25 × 0.38 | 0.18 × 0.21 × 0.24 | 0.24 × 0.33 × 0.33 | 0.13 × 0.14 × 0.18 |
| ρcalc/g·cm−3 | 1.404 | 1.507 | 1.397 | 1.714 | 1.582 | 1.674 | 1.664 | 1.769 |
| S | 1.046 | 1.033 | 1.068 | 1.047 | 1.185 | 1.060 | 1.070 | 1.023 |
| R1 [I > 2σ(I)] | 0.0763 | 0.0391 | 0.0549 | 0.0648 | 0.0869 | 0.0478 | 0.0437 | 0.0511 |
| wR2 [All data] | 0.2088 | 0.1076 | 0.1473 | 0.1800 | 0.2456 | 0.1296 | 0.1172 | 0.1358 |
| Largest diff. peak/hole e/Å−3 | 1.82/−1.44 | 0.70/−0.83 | 2.38/−1.60 | 2.98/−2.97 | 2.93/−1.62 | 1.27/−1.87 | 1.20/−1.38 | 1.04/−1.14 |
| CCDC | 2259382 | 2259383 | 2259384 | 2259385 | 2259386 | 2259387 | 2259388 | 2259389 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schoeller, M.; Piroš, M.; Litecká, M.; Koňariková, K.; Jozefíková, F.; Šagátová, A.; Zahradníková, E.; Valentová, J.; Moncol, J. Bipyridine Ruthenium(II) Complexes with Halogen-Substituted Salicylates: Synthesis, Crystal Structure, and Biological Activity. Molecules 2023, 28, 4609. https://doi.org/10.3390/molecules28124609
Schoeller M, Piroš M, Litecká M, Koňariková K, Jozefíková F, Šagátová A, Zahradníková E, Valentová J, Moncol J. Bipyridine Ruthenium(II) Complexes with Halogen-Substituted Salicylates: Synthesis, Crystal Structure, and Biological Activity. Molecules. 2023; 28(12):4609. https://doi.org/10.3390/molecules28124609
Chicago/Turabian StyleSchoeller, Martin, Milan Piroš, Miroslava Litecká, Katarína Koňariková, Flóra Jozefíková, Alexandra Šagátová, Eva Zahradníková, Jindra Valentová, and Ján Moncol. 2023. "Bipyridine Ruthenium(II) Complexes with Halogen-Substituted Salicylates: Synthesis, Crystal Structure, and Biological Activity" Molecules 28, no. 12: 4609. https://doi.org/10.3390/molecules28124609
APA StyleSchoeller, M., Piroš, M., Litecká, M., Koňariková, K., Jozefíková, F., Šagátová, A., Zahradníková, E., Valentová, J., & Moncol, J. (2023). Bipyridine Ruthenium(II) Complexes with Halogen-Substituted Salicylates: Synthesis, Crystal Structure, and Biological Activity. Molecules, 28(12), 4609. https://doi.org/10.3390/molecules28124609






