Validation of a Liquid–Liquid Extraction Method to Study the Temporal Production of D-Series Resolvins by Head Kidney Cells from Atlantic Salmon (Salmon salar) Exposed to Docosahexaenoic Acid
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimal Concentration of Internal Standards
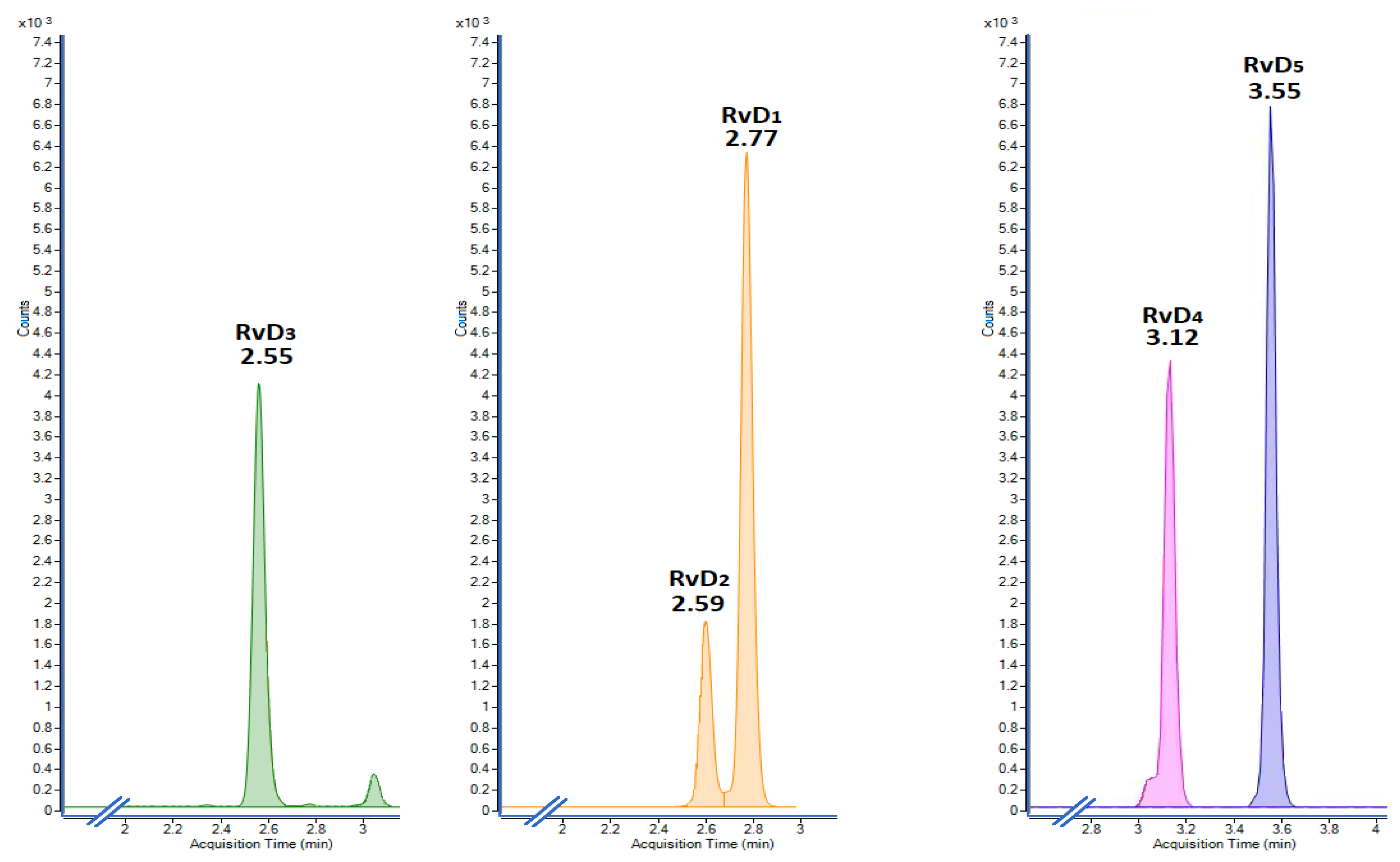
2.2. Analytical Validation
2.3. Analysis of Released Resolvins in L-15 Media by Head Kidney Cells
3. Materials and Methods
3.1. Reagents
3.2. Head Kidney Cells
3.3. Cell Cultures
3.4. Optimal Concentrations of the Internal Standards
3.5. Extraction Protocol
3.6. Analytical Performance
3.7. Liquid Chromatography Mass Spectrometry
3.8. Statistics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Serhan, C.N.; Dalli, J.; Colas, R.A.; Winkler, J.W.; Chiang, N. Protectins and maresins: New pro-resolving families of mediators in acute inflammation and resolution bioactive metabolome. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.L.; Kakazu, A.H.; He, J.; Nshimiyimana, R.; Petasis, N.A.; Jun, B.; Bazan, N.G.; Bazan, H.E.P. Elucidating the structure and functions of Resolvin D6 isomers on nerve regeneration with a distinctive trigeminal transcriptome. FASEB J. 2021, 35, e21775. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.; Serha, C.N. Specialized pro-resolving mediator network: An update on production and actions. Essays Biochem. 2020, 64, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.C.; Balas, L.; Bazan, N.G.; Brenna, J.T.; Chiang, N.; da Costa Souza, F.; Dalli, J.; Durand, T.; Galano, J.M.; Lein, P.J.; et al. Polyunsaturated fatty acids and fatty acid-derived lipid mediators: Recent advances in the understanding of their biosynthesis, structures, and functions. Prog. Lipid Res. 2022, 86, 101165. [Google Scholar] [CrossRef]
- Dalli, J.; Winkler, J.W.; Colas, R.A.; Arnardottir, H.; Cheng, C.Y.C.; Chiang, N.; Petasis, N.A.; Serhan, C.N. Resolvin D3 and Aspirin-Triggered Resolvin D3 Are Potent Immunoresolvents. Chem. Biol. 2013, 20, 188. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.W.; Orr, S.K.; Dalli, J.; Cheng, C.-Y.C.; Sanger, J.M.; Chiang, N.; Petasis, N.A.; Serhan, C.N. Resolvin D4 stereoassignment and its novel actions in host protection and bacterial clearance. Sci. Rep. 2015, 6, srep18972. [Google Scholar] [CrossRef]
- Kalyanaraman, C.; Tourdot, B.E.; Conrad, W.S.; Akinkugbe, O.; Freedman, J.C.; Holinstat, M.; Jacobson, M.P.; Holman, T.R.; Perry, S.C.; Kalyanaraman, C.; et al. 15-Lipoxygenase-1 biosynthesis of 7S,14S-diHDHA implicates 15-lipoxygenase-2 in biosynthesis of resolvin D5[S]. J. Lipid Res. 2020, 61, 1087–1103. [Google Scholar] [CrossRef]
- Pham, T.L.; Kakazu, A.H.; He, J.; Jun, B.; Bazan, N.G.; Bazan, H.E.P. Novel RvD6 stereoisomer induces corneal nerve regeneration and wound healing post-injury by modulating trigeminal transcriptomic signature. Sci. Rep. 2020, 10, 4582. [Google Scholar] [CrossRef]
- Serhan, C.N.; Krishnamoorthy, S.; Recchiuti, A.; Chiang, N. Novel Anti-Inflammatory-Pro-Resolving Mediators and Their Receptors. Curr. Top. Med. Chem. 2012, 11, 629–647. [Google Scholar] [CrossRef]
- Ruyter, B.; Bou, M.; Berge, G.M.; Mørkøre, T.; Sissener, N.H.; Sanden, M.; Lutfi, E.; Romarheim, O.H.; Krasnov, A.; Østbye, T.K.K. A dose-response study with omega-3 rich canola oil as a novel source of docosahexaenoic acid (DHA) in feed for Atlantic salmon (Salmo salar) in seawater; effects on performance, tissue fatty acid composition, and fillet quality. Aquaculture 2022, 561, 738733. [Google Scholar] [CrossRef]
- Shields, R.J. Larviculture of marine finfish in Europe. Aquaculture 2001, 200, 55–88. [Google Scholar] [CrossRef]
- Fedirko, V.; McKeown-Eyssen, G.; Serhan, C.N.; Barry, E.L.; Sandler, R.S.; Figueiredo, J.C.; Ahnen, D.J.; Bresalier, R.S.; Robertson, D.J.; Anderson, C.W.; et al. Plasma lipoxin A4 and resolvin D1 are not associated with reduced adenoma risk in a randomized trial of aspirin to prevent colon adenomas. Mol. Carcinog. 2017, 56, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Hong, S.; Gronert, K.; Colgan, S.P.; Devchand, P.R.; Mirick, G.; Moussignac, R.L. Resolvins: A Family of Bioactive Products of Omega-3 Fatty Acid Transformation Circuits Initiated by Aspirin Treatment that Counter Proinflammation Signals. J. Exp. Med. 2002, 196, 1025. [Google Scholar] [CrossRef]
- Lucena, E.; Yang, Y.; Mendez, C.; Holen, E.; Araujo, P. Extraction of Pro-and Anti-Inflammatory Biomarkers from fish Cells Exposed to Polyunsaturated Fatty Acids and Quantification by Liquid Chromatography Tandem Mass Spectrometry. Curr. Anal. Chem. 2018, 1, 1–9. [Google Scholar]
- Bordet, J.C.; Guichardant, M.; Lagarde, M. Arachidonic acid strongly stimulates prostaglandin I3 (PGI3) production from eicosapentaenoic acid in human endothelial cells. Biochem. Biophys. Res. Commun. 1986, 135, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D.; Schwedhelm, E.; Gutzki, F.M.; Frölich, J.C. Gas chromatographic-mass spectrometric discrimination between 8-iso-prostaglandin E2 and prostaglandin E2 through derivatization by O-(2,3,4,5,6-Pentafluorobenzyl)hydroxyl amine. Anal. Biochem. 1998, 261, 230–232. [Google Scholar] [CrossRef]
- Li, C.; Wu, X.; Liu, S.; Shen, D.; Zhu, J.; Liu, K. Role of Resolvins in the Inflammatory Resolution of Neurological Diseases. Front. Pharmacol. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fanti, F.; Oliva, E.; Tortolani, D.; Di Meo, C.; Fava, M.; Leuti, A.; Rapino, C.; Sergi, M.; Maccarrone, M.; Compagnone, D. μSPE followed by HPLC–MS/MS for the determination of series D and E resolvins in biological matrices. J. Pharm. Biomed. Anal. 2021, 203, 114181. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Eiroa, A.; Canle, M.; Leroy-Cancellieri, V.; Cerdà, V. Solid-phase extraction of organic compounds: A critical review. part ii. TrAC Trends Anal. Chem. 2016, 80, 655–667. [Google Scholar] [CrossRef]
- Dufour, D.; Khalil, A.; Nuyens, V.; Rousseau, A.; Delporte, C.; Noyon, C.; Cortese, M.; Reyé, F.; Pireaux, V.; Nève, J.; et al. Native and myeloperoxidase-oxidized low-density lipoproteins act in synergy to induce release of resolvin-D1 from endothelial cells. Atherosclerosis 2018, 272, 108–117. [Google Scholar] [CrossRef]
- Yang, Y.; Cruickshank, C.; Armstrong, M.; Mahaffey, S.; Reisdorph, R.; Reisdorph, N. New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. J. Chromatogr. A 2013, 1300, 217–226. [Google Scholar] [CrossRef]
- Masoodi, M.; Mir, A.A.; Petasis, N.A.; Serhan, C.N.; Nicolaou, A. Simultaneous lipidomic analysis of three families of bioactive lipid mediators leukotrienes, resolvins, protectins and related hydroxy-fatty acids by liquid chromatography/electrospray ionisation tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Kutzner, L.; Rund, K.M.; Ostermann, A.I.; Hartung, N.M.; Galano, J.M.; Balas, L.; Durand, T.; Balzer, M.S.; David, S.; Schebb, N.H. Development of an Optimized LC-MS Method for the Detection of Specialized Pro-Resolving Mediators in Biological Samples. Front. Pharmacol. 2019, 10, 169. [Google Scholar] [CrossRef]
- Norris, P.; Kapil, S.; Gorti, K. Targeted Profiling of Lipid Mediators. SCIEX Present 2017, 1–6. Available online: https://sciex.com/tech-notes/life-science-research/lipidomics/targeted-profiling-of-lipid-mediators (accessed on 10 June 2023).
- Agilent 6470B or Sciex 7500 Triple Quad—Chromatography Forum. Available online: https://www.chromforum.org/viewtopic.php?t=110834 (accessed on 19 May 2023).
- De Zavalía, N.; Fernandez, D.C.; Sande, P.H.; Keller Sarmiento, M.I.; Golombek, D.A.; Rosenstein, R.E.; Silberman, D.M. Circadian variations of prostaglandin E2 and F2α release in the golden hamster retina. J. Neurochem. 2010, 112, 972–979. [Google Scholar] [CrossRef]
- Nakahata, Y.; Akashi, M.; Trcka, D.; Yasuda, A.; Takumi, T. The in vitro real-time oscillation monitoring system identifies potential entrainment factors for circadian clocks. BMC Mol. Biol. 2006, 7, 5. [Google Scholar] [CrossRef]
- Araujo, P.; Janagap, S.; Holen, E. Application of Doehlert Uniform Shell Designs for Selecting Optimal Amounts of Internal Standards in the Analysis of Prostaglandins and Leukotrienes by Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. A 2012, 1260, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Analytical Method Committee. Is My Calibration Linear? Analyst 1994, 119, 2363–2366. [Google Scholar] [CrossRef]
- Stefanini-Oresic, L. Validation of analytical procedures: ICH guidelines Q2(R2). Farm. Glas. 2022, 2, 1–34. [Google Scholar]
- Araujo, P. Key aspects of analytical method validation and linearity evaluation. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 2224–2234. [Google Scholar] [CrossRef]


| Experiment | Resolvin [A] (ng/mL) | Internal Standard [IS] (ng/mL) |
|---|---|---|
| 1 | 15 | 15 |
| 2 | 30 | 15 |
| 3 | 45 | 15 |
| 4 | 15 | 30 |
| 5 | 30 | 30 |
| 6 | 45 | 30 |
| 7 | 15 | 45 |
| 8 | 30 | 45 |
| 9 | 45 | 45 |
| [IS] | RvD1-d5 | RvD2-d5 | RvD3-d5 | RvD1-d5 | RvD1-d5 | RvD1-d5 | RvD2-d5 | RvD3-d5 | RvD1-d5 | RvD1-d5 | RvD1-d5 | RvD2-d5 | RvD3-d5 | RvD1-d5 | RvD1-d5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 ng/mL | 30 ng/mL | 45/ng/mL | |||||||||||||
| [A] (ng/mL) | RvD1 | RvD2 | RvD3 | RvD4 | RvD5 | RvD1 | RvD2 | RvD3 | RvD4 | RvD5 | RvD1 | RvD2 | RvD3 | RvD4 | RvD5 |
| 15 | 2.70 | 12.41 | 33.74 | 0.79 | 1.00 | 2.85 | 12.81 | 34.63 | 0.83 | 0.95 | 3.32 | 16.68 | 39.22 | 1.00 | 1.16 |
| 15 | 2.57 | 13.78 | 31.24 | 0.79 | 0.92 | 2.77 | 14.39 | 32.76 | 0.87 | 0.93 | 3.35 | 16.73 | 38.37 | 0.99 | 1.15 |
| 15 | 3.04 | 15.80 | 36.96 | 0.93 | 1.10 | 2.85 | 14.05 | 34.03 | 0.87 | 0.96 | 3.27 | 16.00 | 39.58 | 1.01 | 1.22 |
| 30 | 3.06 | 15.07 | 36.22 | 0.85 | 1.07 | 3.03 | 14.97 | 32.95 | 0.89 | 1.03 | 3.12 | 15.41 | 35.83 | 0.95 | 1.25 |
| 30 | 2.94 | 14.01 | 32.97 | 0.80 | 1.08 | 3.24 | 16.35 | 37.58 | 0.98 | 1.14 | 3.23 | 15.43 | 35.06 | 0.91 | 1.26 |
| 30 | 2.98 | 15.17 | 33.63 | 0.86 | 1.26 | 3.09 | 15.24 | 32.57 | 0.95 | 1.00 | 3.18 | 16.22 | 38.42 | 0.94 | 1.32 |
| 45 | 2.92 | 14.63 | 34.31 | 0.84 | 1.11 | 2.82 | 14.53 | 32.44 | 0.82 | 1.01 | 2.92 | 14.15 | 34.87 | 0.83 | 1.16 |
| 45 | 2.97 | 14.58 | 33.71 | 0.84 | 1.11 | 2.87 | 14.33 | 34.57 | 0.87 | 0.98 | 2.92 | 14.86 | 32.89 | 0.90 | 1.09 |
| 45 | 2.96 | 14.80 | 36.16 | 0.82 | 1.24 | 2.60 | 13.35 | 31.01 | 0.80 | 1.05 | 2.97 | 14.57 | 32.29 | 0.82 | 1.14 |
| AVG | 2.90 ± 0.05 | 14.43 ± 0.33 | 34.10 ± 0.61 | 0.84 ± 0.01 | 1.08 ± 0.04 | 2.94 ± 0.06 | 14.58 ± 0.35 | 33.94 ± 0.63 | 0.89 ± 0.02 | 1.00 ± 0.02 | 3.17 ± 0.06 | 15.69 ± 0.31 | 36.78 ± 0.91 | 0.94 ± 0.02 | 1.20 ± 0.02 |
| %CV | 1.86 | 2.27 | 1.78 | 1.75 | 3.25 | 2.14 | 2.37 | 1.85 | 2.21 | 2.14 | 1.79 | 1.95 | 2.47 | 2.48 | 2.01 |
| Resolvin (0–50 ng/mL) | Slope (φ) | Intercept (β) | R2 | Fexperimental | LOD | LOQ | Recovery (%) |
|---|---|---|---|---|---|---|---|
| RvD1 | 0.197 | −0.063 | 0.997 | 0.526 | 0.042 | 0.127 | 98.2 ± 1.3 |
| RvD2 | 0.982 | −0.235 | 0.999 | 0.342 | 0.028 | 0.086 | 99.5 ± 1.1 |
| RvD3 | 2.304 | −0.318 | 0.998 | 0.142 | 0.032 | 0.097 | 99.5 ± 0.9 |
| RvD4 | 0.055 | 0.002 | 0.999 | 0.123 | 0.024 | 0.074 | 99.8 ± 1.2 |
| RvD5 | 0.077 | −0.048 | 0.994 | 0.624 | 0.059 | 0.180 | 96.9 ± 2.3 |
| Time | RvD1 | RvD2 | RvD3 | RvD4 | RvD5 | |
|---|---|---|---|---|---|---|
| Group | (hours) | Concentrations (ng/mL) | ||||
| Control | 6 | 0.127 ± 0.022 | 0.190 ± 0.109 | 0.186 ± 0.055 | 0.601 ± 0.090 | 0.055 ± 0.018 |
| 12 | 0.074 ± 0.009 | 0.238 ± 0.141 | 0.119 ± 0.049 | 0.382 ± 0.082 | 0.069 ± 0.009 | |
| 24 | 0.094 ± 0.014 | 0.154 ± 0.079 | 0.092 ± 0.018 | 0.419 ± 0.031 | 0.017 ± 0.003 | |
| DHA | 6 | 3.918 ± 1.798 | 4.412 ± 1.386 | 6.849 ± 3.156 | 10.231 ± 6.342 | 3.696 ± 2.022 |
| 12 | 1.388 ± 0.404 | 2.574 ± 1.143 | 5.212 ± 2.328 | 4.737 ± 1.710 | 1.395 ± 0.361 | |
| 24 | 4.391 ± 2.103 | 8.095 ± 4.129 | 9.583 ± 4.682 | 10.439 ± 5.121 | 4.256 ± 1.815 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araujo, P.; Iqbal, S.; Arnø, A.; Espe, M.; Holen, E. Validation of a Liquid–Liquid Extraction Method to Study the Temporal Production of D-Series Resolvins by Head Kidney Cells from Atlantic Salmon (Salmon salar) Exposed to Docosahexaenoic Acid. Molecules 2023, 28, 4728. https://doi.org/10.3390/molecules28124728
Araujo P, Iqbal S, Arnø A, Espe M, Holen E. Validation of a Liquid–Liquid Extraction Method to Study the Temporal Production of D-Series Resolvins by Head Kidney Cells from Atlantic Salmon (Salmon salar) Exposed to Docosahexaenoic Acid. Molecules. 2023; 28(12):4728. https://doi.org/10.3390/molecules28124728
Chicago/Turabian StyleAraujo, Pedro, Sarah Iqbal, Aleksander Arnø, Marit Espe, and Elisabeth Holen. 2023. "Validation of a Liquid–Liquid Extraction Method to Study the Temporal Production of D-Series Resolvins by Head Kidney Cells from Atlantic Salmon (Salmon salar) Exposed to Docosahexaenoic Acid" Molecules 28, no. 12: 4728. https://doi.org/10.3390/molecules28124728
APA StyleAraujo, P., Iqbal, S., Arnø, A., Espe, M., & Holen, E. (2023). Validation of a Liquid–Liquid Extraction Method to Study the Temporal Production of D-Series Resolvins by Head Kidney Cells from Atlantic Salmon (Salmon salar) Exposed to Docosahexaenoic Acid. Molecules, 28(12), 4728. https://doi.org/10.3390/molecules28124728






