Anticancer Activity and Mode of Action of Cu(II), Zn(II), and Mn(II) Complexes with 5-Chloro-2-N-(2-quinolylmethylene)aminophenol
Abstract
:1. Introduction
2. Results and Discussion
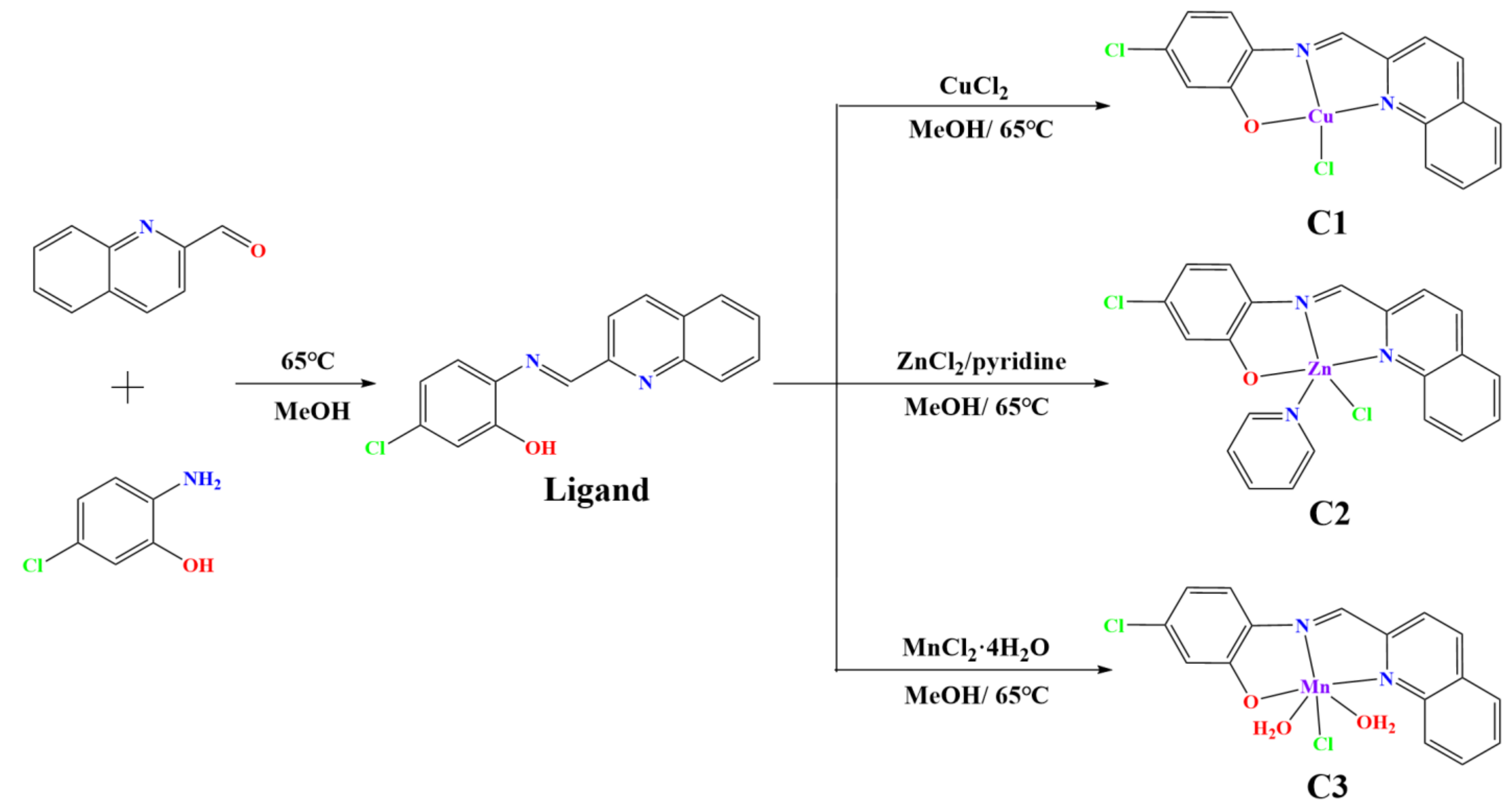
2.1. Synthesis and Characterization of Cu(II), Zn(II), and Mn(II) Complexes
2.2. The Solubility and Stability of Cu(II), Zn(II), and Mn(II) Complexes in Saline
2.3. The Cytotoxicity of Cu(II), Zn(II), and Mn(II) Complexes In Vitro
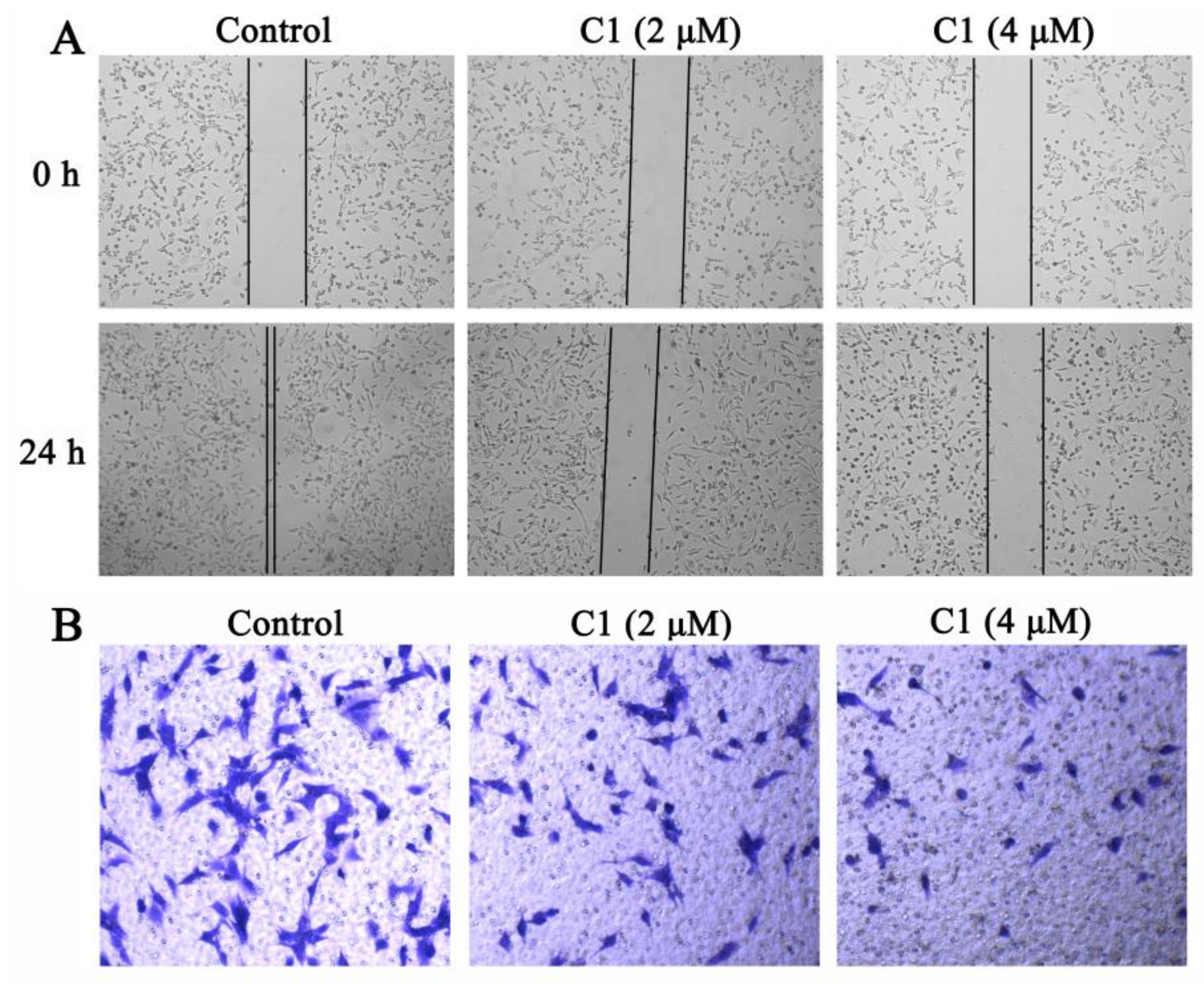
2.4. Inhibition of C1 on Migration and Invasion Activities
2.5. Mechanism of Action of C1
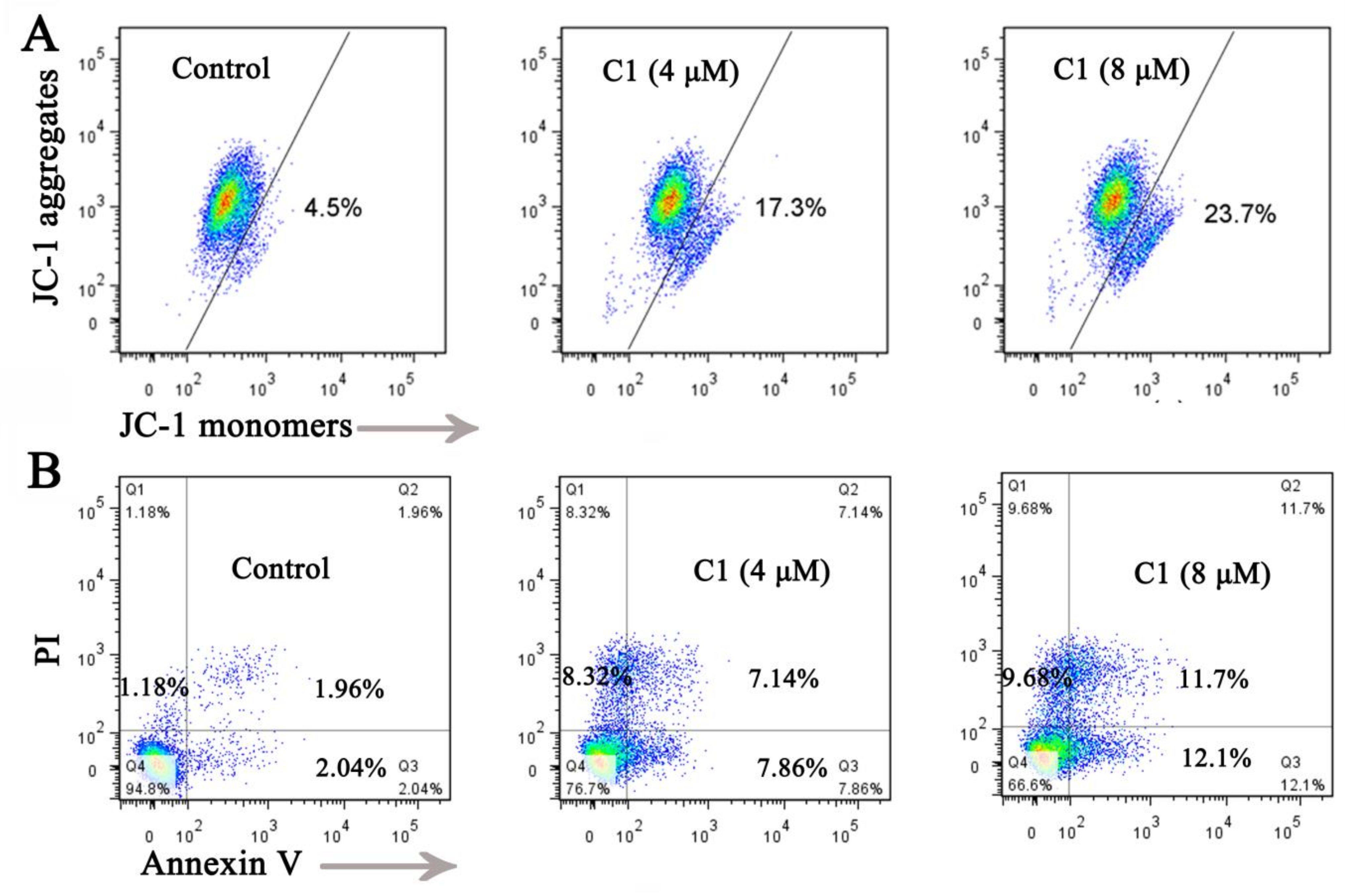
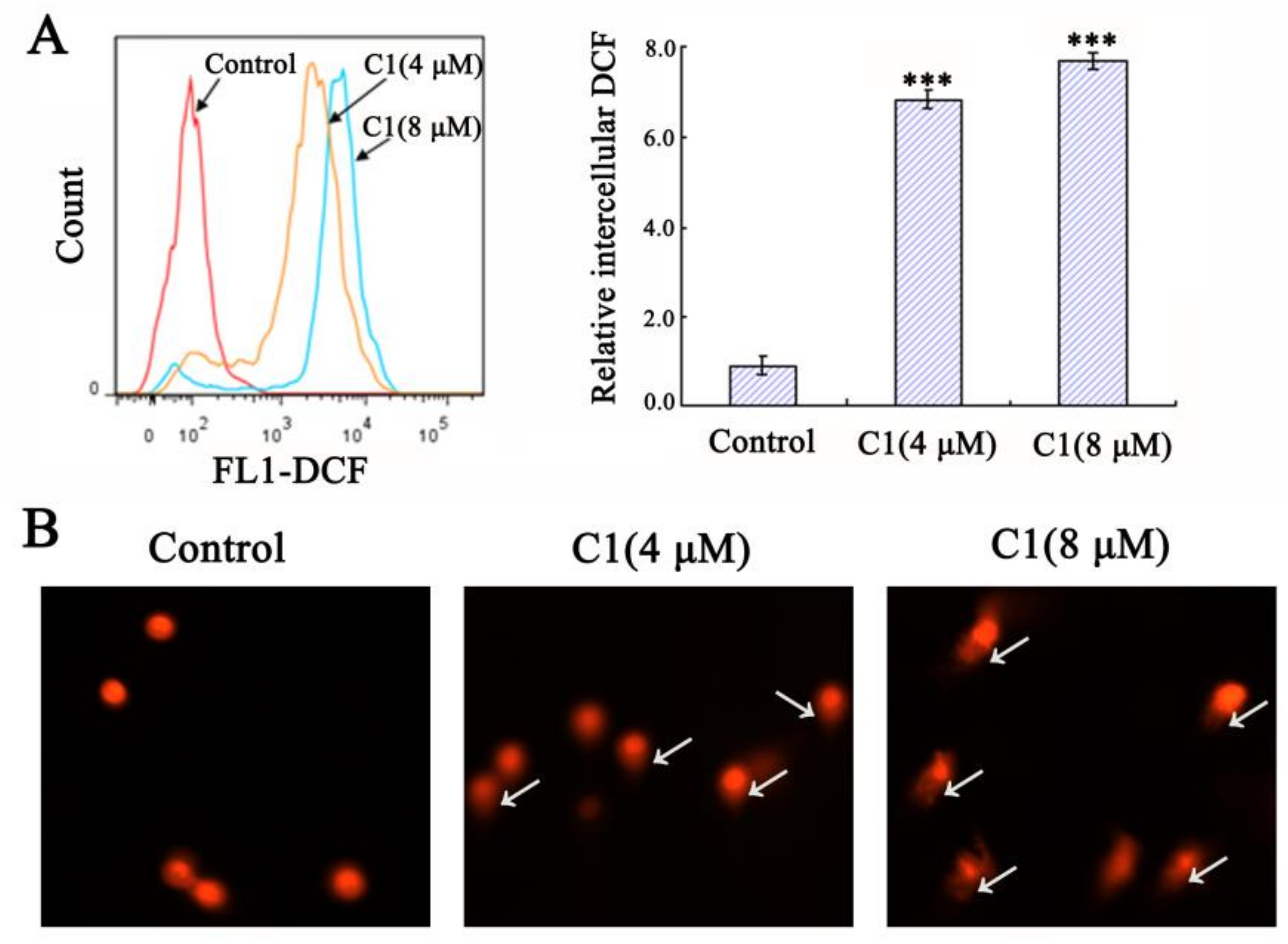
2.5.1. Inducing Mitochondrial-Mediated Apoptosis
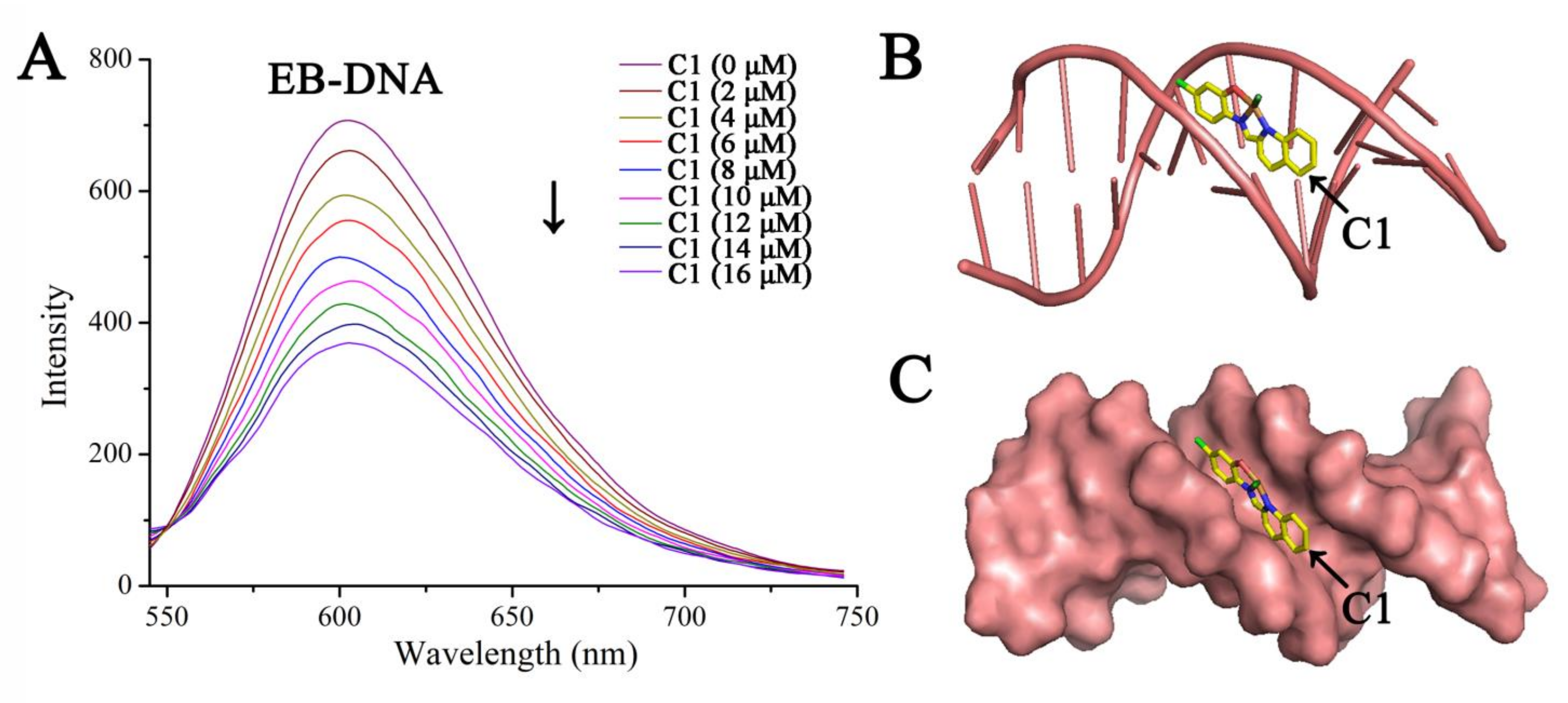
2.5.2. DNA Binding
2.5.3. Arresting Cell Cycle
2.5.4. Inducing A549 Cell Senescence
2.5.5. Inducing DNA Damage
2.6. In Vivo Antitumor Efficacy of C1
3. Materials and Methods
3.1. Reagents and Materials
3.2. Synthesis of Cu, Zn, and Mn Complexes
3.3. The Stability of Cu, Zn, and Mn Complexes
3.4. Wound-Healing Assay
3.5. Transwell Invasion Assay
3.6. Cell Apoptosis Analysis
3.7. Mitochondrial Membrane Potential (ΔΨm) Assay
3.8. Cell Cycle Analysis
3.9. β-Galactosidase Staining Assay
3.10. Measurement of Cellular ROS
3.11. Comet Assay
3.12. Fluorescence-Quenching Assay
3.13. Molecular Docking Study
3.14. In Vivo Antitumor Experiment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wang, X.; Li, Y.; Ren, X.; Zhou, Y.; Hu, W.; Zhou, C.; Jing, Q.; Yang, C.; Wang, L. DHA exhibits synergistic therapeutic efficacy with cisplatin to induce ferroptosis in pancreatic ductal adenocarcinoma via modulation of iron metabolism. Cell Death Dis. 2021, 12, 705. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Michels, J.; Brenner, C.; Szabadkai, G.; Harel-Bellan, A.; Castedo, M.; Kroemer, G. Systems biology of cisplatin resistance: Past, present and future. Cell Death Dis. 2014, 5, e1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romani, A.M. Cisplatin in cancer treatment. Biochem. Pharmacol. 2022, 206, 115323. [Google Scholar] [CrossRef]
- Callejo, A.; Sedó-Cabezón, L.; Domenech Juan, I.; Llorens, J. Cisplatin-induced ototoxicity: Effects, mechanisms and protection strategies. Toxics 2015, 3, 268–293. [Google Scholar] [CrossRef] [Green Version]
- Sancho-Garnier, H.; Colonna, M. Breast cancer epidemiology. Presse Med. 2019, 48, 1076–1084. [Google Scholar] [CrossRef]
- Li, N.; Deng, Y.; Zhou, L.; Tian, T.; Yang, S.; Wu, Y.; Zheng, Y.; Zhai, Z.; Hao, Q.; Song, D. Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: Results from the Global Burden of Disease Study 2017. J. Hematol. Oncol. 2019, 12, 140. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Zhu, Y.; Cheng, J.; Shen, J.; Huang, H.; Liu, M.; Chen, Z.; Liu, Y. Nitric oxide-releasing platinum (IV) prodrug efficiently inhibits proliferation and metastasis of cancer cells. Chem. Commun. 2020, 56, 14051–14054. [Google Scholar] [CrossRef]
- Ayipo, Y.O.; Osunniran, W.A.; Mordi, M.N. Metal complexes of β-carboline: Advances in anticancer therapeutics. Coord. Chem. Rev. 2021, 432, 213746. [Google Scholar] [CrossRef]
- Gourdon, L.; Cariou, K.; Gasser, G. Phototherapeutic anticancer strategies with first-row transition metal complexes: A critical review. Chem. Soc. Rev. 2022, 51, 1167–1195. [Google Scholar] [CrossRef] [PubMed]
- Heras, B.L.; Amesty, Á.; Estévez-Braun, A.; Hortelano, S. Metal complexes of natural product like-compounds with antitumor activity. Anti-Cancer Agents Med. Chem. 2019, 19, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.; Huang, G.; Li, J.; Yang, F.; Liang, H. Versatile delivery systems for non-platinum metal-based anticancer therapeutic agents. Coord. Chem. Rev. 2021, 441, 213975. [Google Scholar] [CrossRef]
- Paprocka, R.; Wiese-Szadkowska, M.; Janciauskiene, S.; Kosmalski, T.; Kulik, M.; Helmin-Basa, A. Latest developments in metal complexes as anticancer agents. Coord. Chem. Rev. 2022, 452, 214307. [Google Scholar] [CrossRef]
- Silva, C.S.; Moutinho, C.; Ferreira da Vinha, A.; Matos, C. Trace minerals in human health: Iron, zinc, copper, manganese and fluorine. Int. J. Sci. Res. Methodol. 2019, 13, 57–80. [Google Scholar]
- Zabłocka-Słowińska, K.; Płaczkowska, S.; Prescha, A.; Pawełczyk, K.; Porębska, I.; Kosacka, M.; Pawlik-Sobecka, L.; Grajeta, H. Serum and whole blood Zn, Cu and Mn profiles and their relation to redox status in lung cancer patients. J. Trace Elem. Med. Biol. 2018, 45, 78–84. [Google Scholar] [CrossRef]
- Bocca, B.; Ruggieri, F.; Pino, A.; Rovira, J.; Calamandrei, G.; Martínez, M.Á.; Domingo, J.L.; Alimonti, A.; Schuhmacher, M. Human biomonitoring to evaluate exposure to toxic and essential trace elements during pregnancy. Part A. concentrations in maternal blood, urine and cord blood. Environ. Res. 2019, 177, 108599. [Google Scholar] [CrossRef]
- Pellei, M.; Del Bello, F.; Porchia, M.; Santini, C. Zinc coordination complexes as anticancer agents. Coord. Chem. Rev. 2021, 445, 214088. [Google Scholar] [CrossRef]
- Kovala-Demertzi, D.; Staninska, M.; Garcia-Santos, I.; Castineiras, A.; Demertzis, M.A. Synthesis, crystal structures and spectroscopy of meclofenamic acid and its metal complexes with manganese (II), copper (II), zinc (II) and cadmium (II). Antiproliferative and superoxide dismutase activity. J. Inorg. Biochem. 2011, 105, 1187–1195. [Google Scholar] [CrossRef]
- Parekh, N.M.; Mistry, B.M.; Pandurangan, M.; Shinde, S.K.; Patel, R.V. Investigation of anticancer potencies of newly generated Schiff base imidazolylphenylheterocyclic-2-ylmethylenethiazole-2-amines. Chin. Chem. Lett. 2017, 28, 602–606. [Google Scholar] [CrossRef]
- Hameed, A.; Al-Rashida, M.; Uroos, M.; Abid Ali, S.; Khan, K.M. Schiff bases in medicinal chemistry: A patent review (2010–2015). Expert Opin. Ther. Pat. 2017, 27, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Ali, M.; Latif, A.; Rehman, N.U.; Saher, S.; Khan, A.; Ullah, S.; Ullah, O.; Halim, S.A.; Sani, F. Novel Bis-Schiff’s base derivatives of 4-nitroacetophenone as potent α-glucosidase agents: Design, synthesis and in silico approach. Bioorg. Chem. 2022, 128, 106058. [Google Scholar] [CrossRef] [PubMed]
- Agbektas, T.; Zontul, C.; Ozturk, A.; Huseynzada, A.; Ganbarova, R.; Hasanova, U.; Cinar, G.; Tas, A.; Kaya, S.; Chtita, S. Effect of azomethine group containing compounds on gene profiles in Wnt and MAPK signal patterns in lung cancer cell line: In Silico and In Vitro analyses. J. Mol. Struct. 2023, 1275, 134619. [Google Scholar] [CrossRef]
- Tan, Y.-X.; Zhang, Z.-J.; Liu, Y.; Yu, J.-X.; Zhu, X.-M.; Kuang, D.-Z.; Jiang, W.-J. Synthesis, crystal structure and biological activity of the Schiff base organotin(IV) complexes based on salicylaldehyde-o-aminophenol. J. Mol. Struct. 2017, 1149, 874–881. [Google Scholar] [CrossRef]
- Wazalwar, S.S.; Banpurkar, A.R.; Perdih, F. Synthesis, Characterization, Molecular Docking Studies and Anticancer Activity of Schiff Bases Derived from 3-(Substituted phenyl)-1-phenyl-1H-pyrazole-4-carbaldehyde and 2-Aminophenol. J. Chem. Crystallogr. 2018, 48, 185–199. [Google Scholar] [CrossRef]
- Liu, X.; Hamon, J.-R. Recent developments in penta-, hexa-and heptadentate Schiff base ligands and their metal complexes. Coord. Chem. Rev. 2019, 389, 94–118. [Google Scholar] [CrossRef]
- Alfonso-Herrera, L.A.; Rosete-Luna, S.; Hernández-Romero, D.; Rivera-Villanueva, J.M.; Olivares-Romero, J.L.; Cruz-Navarro, J.A.; Soto-Contreras, A.; Arenaza-Corona, A.; Morales-Morales, D.; Colorado-Peralta, R. Transition Metal Complexes with Tridentate Schiff Bases (O N O and O N N) Derived from Salicylaldehyde: An Analysis of Their Potential Anticancer Activity. Chemmedchem 2022, 17, e202200367. [Google Scholar] [CrossRef] [PubMed]
- Jana, N.C.; Patra, M.; Brandão, P.; Panja, A. Synthesis, structure and diverse coordination chemistry of cobalt (III) complexes derived from a Schiff base ligand and their biomimetic catalytic oxidation of o-aminophenols. Polyhedron 2019, 164, 23–34. [Google Scholar] [CrossRef]
- Panja, A.; Shyamal, M.; Saha, A.; Mandal, T.K. Methylene bridge regulated geometrical preferences of ligands in cobalt (III) coordination chemistry and phenoxazinone synthase mimicking activity. Dalton Trans. 2014, 43, 5443–5452. [Google Scholar] [CrossRef]
- Abd El-Halim, H.F.; Mohamed, G.G.; Anwar, M.N. Antimicrobial and anticancer activities of Schiff base ligand and its transition metal mixed ligand complexes with heterocyclic base. Appl. Organomet. Chem. 2018, 32, e3899. [Google Scholar] [CrossRef]
- Gurgul, I.; Janczy-Cempa, E.; Mazuryk, O.; Lekka, M.; Łomzik, M.; Suzenet, F.; Gros, P.C.; Brindell, M. Inhibition of Metastasis by Polypyridyl Ru (II) Complexes through Modification of Cancer Cell Adhesion–In Vitro Functional and Molecular Studies. J. Med. Chem. 2022, 65, 10459–10470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, F.; Chen, W.; Miao, H.; Liang, H.; Liao, Z.; Zhang, Z.; Zhang, B. The role of RNA m5C modification in cancer metastasis. Int. J. Biol. Sci. 2021, 17, 3369. [Google Scholar] [CrossRef]
- Duff, D.; Long, A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal. 2017, 35, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Lyden, D.; Ghajar, C.M.; Correia, A.L.; Aguirre-Ghiso, J.A.; Cai, S.; Rescigno, M.; Zhang, P.; Hu, G.; Fendt, S.-M.; Boire, A. Metastasis. Cancer Cell 2022, 40, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Brindell, M.; Gurgul, I.; Janczy-Cempa, E.; Gajda-Morszewski, P.; Mazuryk, O. Moving Ru polypyridyl complexes beyond cytotoxic activity towards metastasis inhibition. J. Inorg. Biochem. 2022, 226, 111652. [Google Scholar] [CrossRef] [PubMed]
- Al-Aamri, H.M.; Irving, H.R.; Bradley, C.; Meehan-Andrews, T. Intrinsic and extrinsic apoptosis responses in leukaemia cells following daunorubicin treatment. BMC Cancer 2021, 21, 438. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.B.; Wang, F.Y.; Tang, X.M.; Feng, H.W.; Chen, Z.F.; Liu, Y.C.; Liu, Y.N.; Liang, H. Organometallic gold (III) complexes similar to tetrahydroisoquinoline induce ER-stress-mediated apoptosis and pro-death autophagy in A549 cancer cells. J. Med. Chem. 2018, 61, 3478–3490. [Google Scholar] [CrossRef]
- Das, A.; Mohammed, T.P.; Kumar, R.; Bhunia, S.; Sankaralingam, M. Carbazole appended trans-dicationic pyridinium porphyrin finds supremacy in DNA binding/photocleavage over a non-carbazolyl analogue. Dalton Trans. 2022, 51, 12453–12466. [Google Scholar] [CrossRef]
- Rafique, B.; Kalsoom, S.; Sajini, A.A.; Ismail, H.; Iqbal, M. Synthesis, Characterization, Biological Evaluation and DNA Interaction Studies of 4-Aminophenol Derivatives: Theoretical and Experimental Approach. Molecules 2022, 27, 1352. [Google Scholar] [CrossRef]
- Gałczyńska, K.; Drulis-Kawa, Z.; Arabski, M. Antitumor activity of Pt (II), Ru (III) and Cu (II) complexes. Molecules 2020, 25, 3492. [Google Scholar] [CrossRef]
- Park, C.W.; Bak, Y.; Kim, M.J.; Srinivasrao, G.; Hwang, J.; Sung, N.K.; Kim, B.Y.; Yu, J.-H.; Hong, J.T.; Yoon, D.-Y. The Novel Small Molecule STK899704 Promotes Senescence of the Human A549 NSCLC Cells by Inducing DNA Damage Responses and Cell Cycle Arrest. Front. Pharmacol. 2018, 9, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, H.U.; Haj-Yehia, A.; Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000, 5, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2018, 25, 101084. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Yang, D.L.; Qin, H.X.; He, L.J.; Huang, J.H.; Tang, D.Y.; Xu, Z.G.; Chen, Z.Z.; Li, Y. DMAPT-D6 induces death-receptor-mediated apoptosis to inhibit glioblastoma cell oncogenesis via induction of DNA damage through accumulation of intracellular ROS. Oncol. Rep. 2021, 45, 1261–1272. [Google Scholar] [CrossRef]
- Gai, S.S.; Jiang, M. Biological properties of Cu(II) complex with human serum albumin. Huaxue Tongbao 2018, 81, 253–257. [Google Scholar]




| Compound | A549 | MCF-7 | Hela | A549cisR | WI-38 | SI a |
|---|---|---|---|---|---|---|
| L | >40 | >40 | >40 | >40 | >40 | - |
| C1 | 3.93 ± 0.28 | 5.30 ± 0.35 | 6.31 ± 0.32 | 6.79 ± 0.38 | 10.22 ± 0.43 | 2.60 |
| C2 | 18.26 ± 0.52 | 23.61 ± 0.98 | 22.75 ± 0.80 | 29.33 ± 0.85 | 33.96 ± 1.06 | 1.86 |
| C3 | 33.61 ± 0.98 | >40 | >40 | >40 | >40 | - |
| Cisplatin | 12.54 ± 0.45 | 14.53 ± 0.48 | 15.33 ± 0.51 | >40 | 9.61 ± 0.58 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gai, S.; He, L.; He, M.; Zhong, X.; Jiang, C.; Qin, Y.; Jiang, M. Anticancer Activity and Mode of Action of Cu(II), Zn(II), and Mn(II) Complexes with 5-Chloro-2-N-(2-quinolylmethylene)aminophenol. Molecules 2023, 28, 4876. https://doi.org/10.3390/molecules28124876
Gai S, He L, He M, Zhong X, Jiang C, Qin Y, Jiang M. Anticancer Activity and Mode of Action of Cu(II), Zn(II), and Mn(II) Complexes with 5-Chloro-2-N-(2-quinolylmethylene)aminophenol. Molecules. 2023; 28(12):4876. https://doi.org/10.3390/molecules28124876
Chicago/Turabian StyleGai, Shuangshuang, Liqin He, Mingxian He, Xuwei Zhong, Caiyun Jiang, Yiming Qin, and Ming Jiang. 2023. "Anticancer Activity and Mode of Action of Cu(II), Zn(II), and Mn(II) Complexes with 5-Chloro-2-N-(2-quinolylmethylene)aminophenol" Molecules 28, no. 12: 4876. https://doi.org/10.3390/molecules28124876
APA StyleGai, S., He, L., He, M., Zhong, X., Jiang, C., Qin, Y., & Jiang, M. (2023). Anticancer Activity and Mode of Action of Cu(II), Zn(II), and Mn(II) Complexes with 5-Chloro-2-N-(2-quinolylmethylene)aminophenol. Molecules, 28(12), 4876. https://doi.org/10.3390/molecules28124876






