Computational Tool to Design Small Synthetic Inhibitors Selective for XIAP-BIR3 Domain
Abstract
1. Introduction
2. Results and Discussion
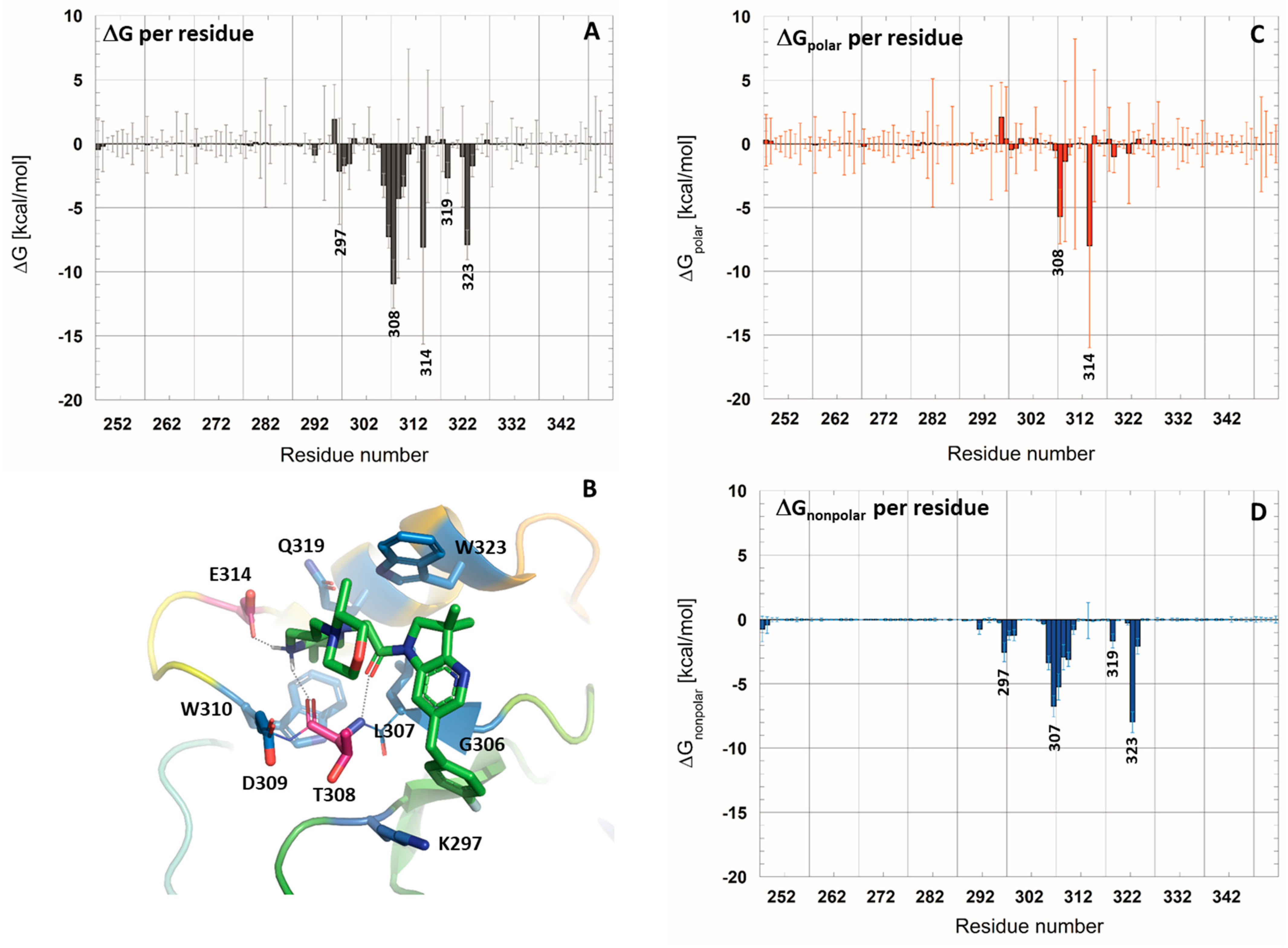
2.1. MM-PBSA Binding Free-Energy Prediction
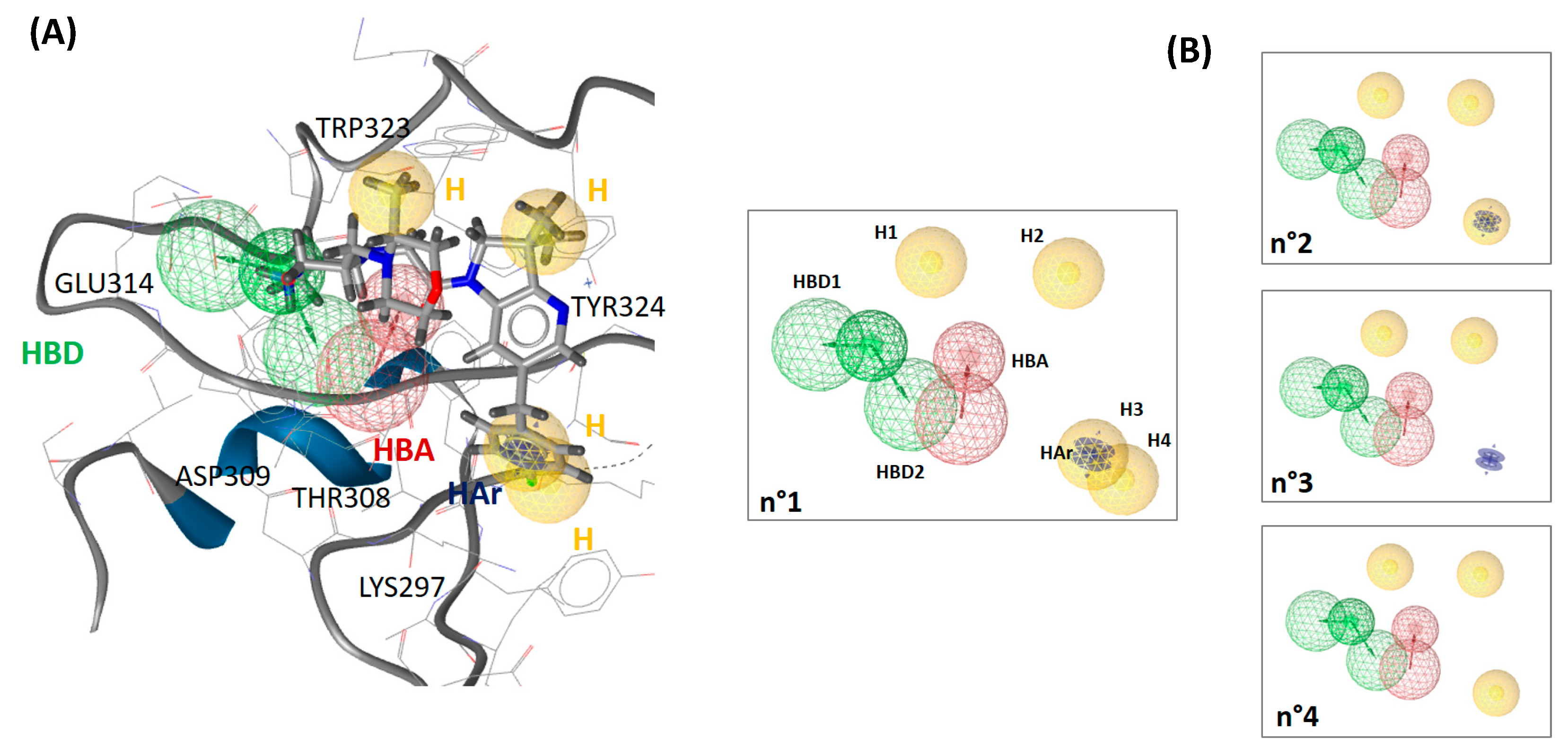
2.2. Construction and Validation of a 3D Pharmacophore of Synthetic XIAP-BIR3 Inhibitors
3. Conclusions
4. Materials and Methods
4.1. Structure Preparation
4.2. Molecular Dynamics Simulations
4.3. MM-PBSA Prediction
4.4. 3D Pharmacophore Building
4.5. Testing Chemical Library
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Takahashi, R.; Salvesen, G.S.; Reed, J.C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 1997, 388, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Eckelman, B.P.; Salvesen, G.S.; Scott, F.L. Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Rep. 2006, 7, 988–994. [Google Scholar] [CrossRef]
- Hussain, A.R.; Siraj, A.K.; Ahmed, M.; Bu, R.; Pratheeshkumar, P.; Alrashed, A.M.; Qadri, Z.; Ajarim, D.; Al-Dayel, F.; Beg, S.; et al. XIAP over-expression is an independent poor prognostic marker in Middle Eastern breast cancer and can be targeted to induce efficient apoptosis. BMC Cancer 2017, 17, 640. [Google Scholar] [CrossRef] [PubMed]
- Mace, P.D.; Shirley, S.; Day, C.L. Assembling the building blocks: Structure and function of inhibitor of apoptosis proteins. Cell Death Differ. 2010, 17, 46–53. [Google Scholar] [CrossRef]
- Chai, J.; Wu, Q.; Shiozaki, E.; Srinivasula, S.M.; Alnemri, E.S.; Shi, Y. Crystal Structure of a Procaspase-7 Zymogen: Mechanisms of Activation and Substrate Binding. Cell 2001, 107, 399–407. [Google Scholar] [CrossRef]
- Huang, Y.; Park, Y.C.; Rich, R.L.; Segal, D.; Myszka, D.G.; Wu, H. Structural basis of caspase inhibition by XIAP: Differential roles of the linker versus the BIR domain. Cell 2001, 104, 781–790. [Google Scholar] [CrossRef]
- Riedl, S.J.; Renatus, M.; Schwarzenbacher, R.; Zhou, Q.; Sun, C.; Fesik, S.W.; Liddington, R.C.; Salvesen, G.S. Structural basis for the inhibition of caspase-3 by XIAP. Cell 2001, 104, 791–800. [Google Scholar] [CrossRef]
- Chai, J.; Shiozaki, E.; Srinivasula, S.M.; Wu, Q.; Dataa, P.; Alnemri, E.S.; Shi, Y. Structural Basis of Caspase-7 Inhibition by XIAP. Cell 2001, 104, 769–780. [Google Scholar] [CrossRef]
- Sun, C.; Cai, M.; Gunasekera, A.H.; Meadows, R.P.; Wang, H.; Chen, J.; Zhang, H.; Wu, W.; Xu, N.; Ng, S.-C.; et al. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 1999, 401, 818–822. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, C.; Olejniczak, E.T.; Meadows, R.P.; Betz, S.F.; Oost, T.; Herrmann, J.; Wu, J.C.; Fesik, S.W. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 2000, 408, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Chai, J.; Suber, T.L.; Wu, J.-W.; Du, C.; Wang, X.; Shi, Y. Structural basis of IAP recognition by Smac/DIABLO. Nature 2000, 408, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, C.; Belunis, C.; Crowther, R.; Danho, W.; Gao, L.; Goggin, B.; Janson, C.A.; Li, S.; Remiszewski, S.; Schutt, A.; et al. The structure of XIAP BIR2: Understanding the selectivity of the BIR domains. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Cong, H.; Xu, L.; Wu, Y.; Qu, Z.; Bian, T.; Zhang, W.; Xing, C.; Zhuang, C. Inhibitor of Apoptosis Protein (IAP) Antagonists in Anticancer Agent Discovery: Current Status and Perspectives. J. Med. Chem. 2019, 62, 5750–5772. [Google Scholar] [CrossRef]
- Mannhold, R.; Fulda, S.; Carosati, E. IAP antagonists: Promising candidates for cancer therapy. Drug Discov. Today 2010, 15, 210–219. [Google Scholar] [CrossRef]
- Fulda, S.; Vucic, D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012, 11, 109–124. [Google Scholar] [CrossRef]
- Fulda, S. Smac mimetics as IAP antagonists. Semin. Cell Dev. Biol. 2015, 39, 132–138. [Google Scholar] [CrossRef]
- Infante, J.R.; Dees, E.C.; Olszanski, A.J.; Dhuria, S.V.; Sen, S.; Cameron, S.; Cohen, R.B. Phase I dose-escalation study of LCL161, an oral inhibitor of apoptosis proteins inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 3103–3110. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Chessari, G.; Buck, I.M.; Day, J.E.; Day, P.J.; Iqbal, A.; Johnson, C.N.; Lewis, E.J.; Martins, V.; Miller, D.; Reader, M.; et al. Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP. J. Med. Chem. 2015, 58, 6574–6588. [Google Scholar] [CrossRef]
- Tamanini, E.; Buck, I.M.; Chessari, G.; Chiarparin, E.; Day, J.E.H.; Frederickson, M.; Griffiths-Jones, C.M.; Hearn, K.; Heightman, T.D.; Iqbal, A.; et al. Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP). J. Med. Chem. 2017, 60, 4611–4625. [Google Scholar] [CrossRef] [PubMed]
- Ward, G.A.; Lewis, E.J.; Ahn, J.S.; Johnson, C.N.; Lyons, J.F.; Martins, V.; Munck, J.M.; Rich, S.J.; Smyth, T.; Thompson, N.T.; et al. ASTX660, a Novel Non-peptidomimetic Antagonist of cIAP1/2 and XIAP, Potently Induces TNF alpha-Dependent Apoptosis in Cancer Cell Lines and Inhibits Tumor Growth. Mol. Cancer Ther. 2018, 17, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, C.W.; Le Grand, S.; Walker, R.C.; Roitberg, A.E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.M.; MacKerell, A.D.; Reuter, N. Interactions between Methylated Ammonium Groups and Tryptophan in the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2019, 15, 7–12. [Google Scholar] [CrossRef]
- Mouawad, L.; Perahia, D. All-atom normal mode calculations of large molecular systems using iterative methods. In Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems; Chapman and Hall/CRC: Boca Raton, FL, USA, 2005; pp. 17–39. [Google Scholar]
- Duan, L.; Liu, X.; Zhang, J.Z.H. Interaction entropy: A new paradigm for highly efficient and reliable computation of protein–ligand binding free energy. J. Am. Chem. Soc. 2016, 138, 5722–5728. [Google Scholar] [CrossRef]
- Cong, Y.; Li, M.; Feng, G.; Li, Y.; Wang, X.; Duan, L. Trypsin-Ligand binding affinities calculated using an effective interaction entropy method under polarized force field. Sci. Rep. 2017, 7, 17708. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Lingands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Roy, K.; Kar, S.; Das, R.N. Chapter 10—Other Related Techniques. In Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment; Roy, K., Kar, S., Das, R.N., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 357–425. [Google Scholar] [CrossRef]
- Opo, F.A.D.M.; Rahman, M.M.; Ahammad, F.; Ahmed, I.; Bhuiyan, M.A.; Asiri, A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021, 11, 4049. [Google Scholar] [CrossRef]
- Schrödinger. Schrödinger Release 2023-1; LLC: New York, NY, USA, 2021. [Google Scholar]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treat- ment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Jenson, C.J. Temperature Dependence of TIP3P, SPC, and TIP4P Water from NPT Monte Carlo Simulations: Seeking Temperatures of Maximum Density. J. Comput. Chem. 1998, 19, 1179–1186. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks III, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular simulation Program. J. Comp. Chem. 2009, 30, 1545–1615. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Srinivasan, J.; Miller, J.; Kollman, P.A.; Case, D.A. Continuum solvent studies of the stability of RNA hairpin loops and helices. J. Biomol. Struct. Dyn. 1998, 16, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Kirchmair, J.; Markt, P.; Distinto, S.; Wolber, G.; Langer, T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection--what can we learn from earlier mistakes? J. Comput. -Aided Mol. Des. 2008, 22, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
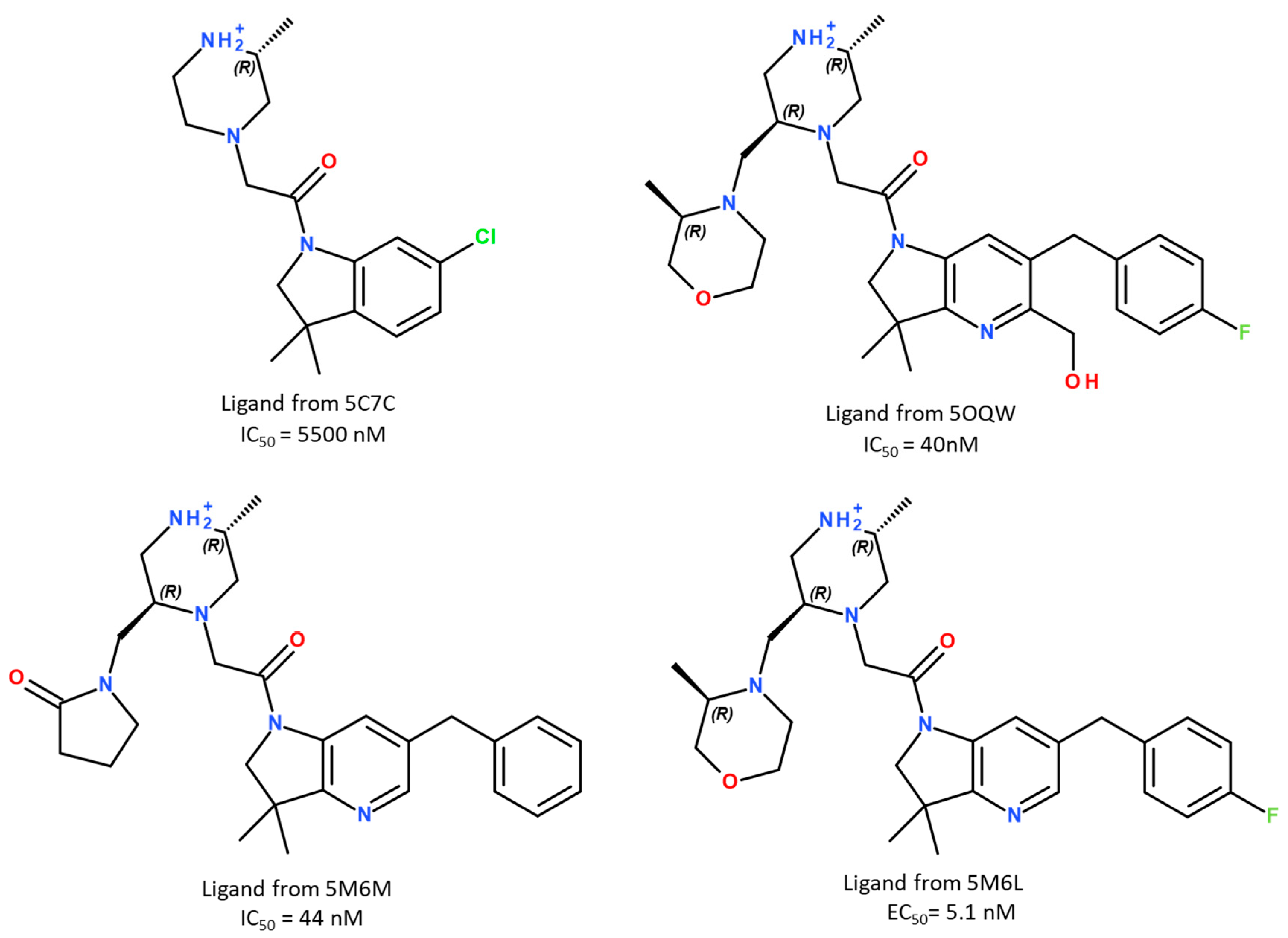
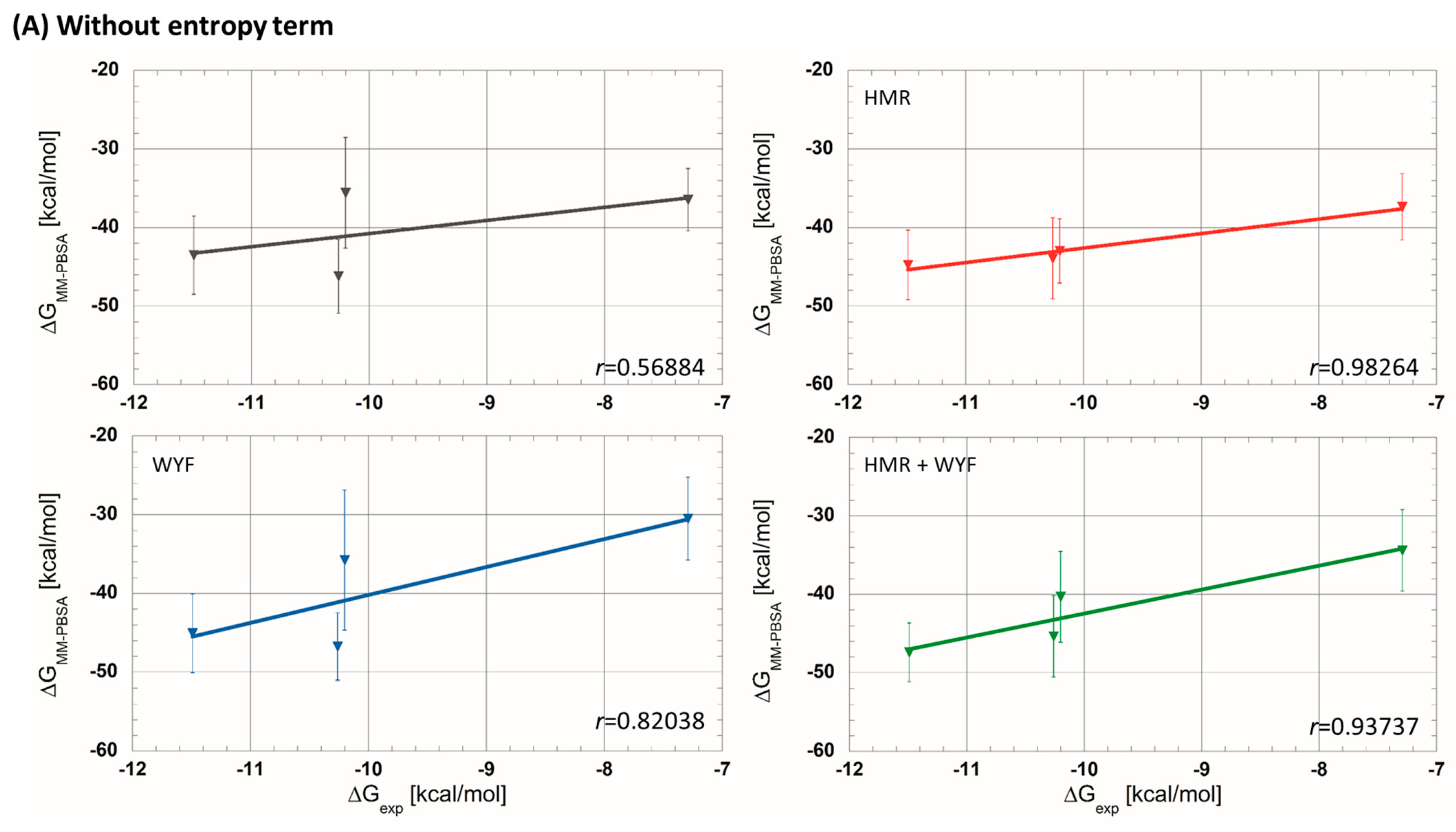
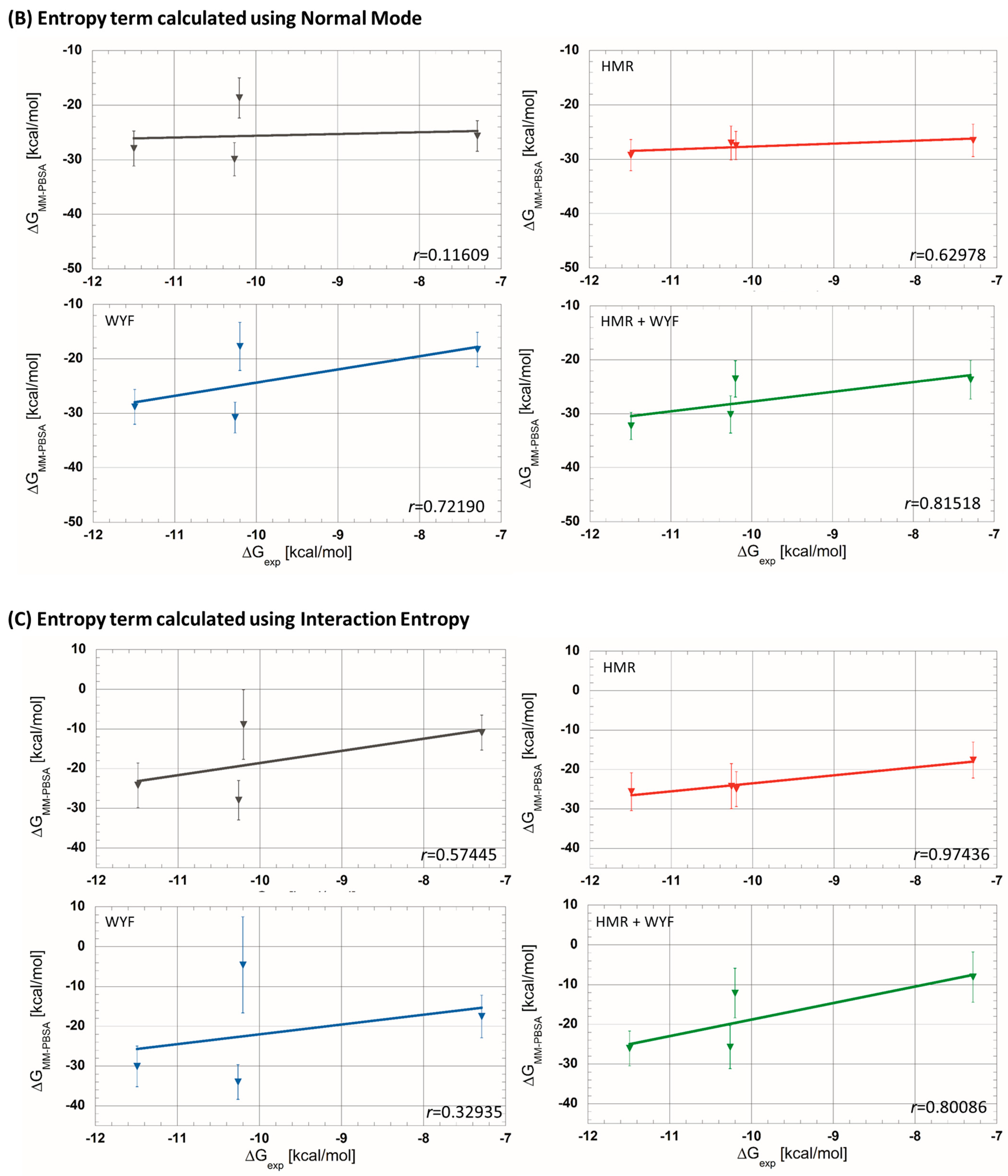

| (a) Without entropic term. | |||||||||
| ΔGexp (kcal/mol) | ΔGMM-PBSA (kcal/mol) | ||||||||
| Without Additional Parametrization | HMR | WYF | HMR + WYF | ||||||
| 5C7C | −7.29 | −36.5 ± 4.0 | −37.4 ± 4.2 | −30.5 ± 5.3 | −34.4 ± 5.2 | ||||
| 5M6M | −10.20 | −35.6 ± 7.0 | −43.0 ± 4.1 | −35.8 ± 8.9 | −40.3 ± 5.8 | ||||
| 5OQW | −10.26 | −46.2 ± 4.7 | −43.9 ± 5.1 | −46.8 ± 4.3 | −45.4 ± 5.2 | ||||
| 5M6L | −11.49 | −43.5 ± 5.0 | −44.7 ± 4.4 | −45.1 ± 5.0 | −47.4 ± 3.7 | ||||
| AVPI | −8.91 | −46.2 ± 4.9 | −39.5 ± 6.6 | −47.5 ± 9.2 | −38.2 ± 5.9 | ||||
| (b) With entropic term calculated using normal mode analysis of harmonic frequencies (NM) or using the interaction entropy method (IE). | |||||||||
| ΔGexp (kcal/mol) | ΔGMM-PBSA (kcal/mol) | ||||||||
| Without Additional Parameterization | HMR | WYF | HMR + WYF | ||||||
| NM | IE | NM | IE | NM | IE | NM | IE | ||
| 5C7C | −7.29 | −25.6 ± 2.8 | −10.9 ± 4.4 | −26.5 ± 3.0 | −17.6 ± 4.6 | −18.3 ± 3.2 | −17.5 ± 5.4 | −23.7 ± 3.6 | −8.1 ± 6.3 |
| 5M6M | −10.20 | −18.7 ± 3.7 | − 8.9 ± 8.8 | −27.5 ± 2.6 | −24.9 ± 4.4 | −17.7 ± 4.4 | −4.6 ± 12.1 | −23.5 ± 3.4 | −12.1 ± 6.3 |
| 5OQW | −10.26 | −29.9 ± 3.0 | −28.0 ± 5.0 | −27.0 ± 3.1 | −24.2 ± 5.7 | −30.8 ± 2.8 | −34.0 ± 4.4 | −30.1 ± 3.5 | −25.8 ± 5.4 |
| 5M6L | −11.49 | −27.9 ± 3.2 | −24.2 ± 5.6 | −29.2 ± 2.9 | −25.6 ± 4.8 | −28.8 ± 3.2 | −30.0 ± 5.1 | −32.3 ± 2.5 | −26.1 ± 4.4 |
| AVPI | −8.91 | −28.7 ± 3.0 | −33.2 ± 5.0 | −23.0 ± 3.8 | −22.8 ± 7.0 | −32.2 ± 6.2 | −31.7 ± 9.9 | −21.0 ± 3.2 | −19.5 ± 6.2 |
| Active/inactive cutoff in pIC50 | 6 |
| Total number of ligands | 5590 |
| Number of active molecules | 173 |
| Number of inactive molecules | 5417 |
| Molecules with 6 > pIC50 > 4.5 not retained | 96 |
| Active/inactive ratio | 1/31 |
| Number of ligands retained in the testing library | 5494 |
| Pharmacophore | Number of Omitted Features during Ligand Alignments | Sensitivity | Specificity | Enrichment Factor | AUC at 1.5% | AUC at 100% |
|---|---|---|---|---|---|---|
| n°1 | 0 | TP = 5 2.9% | TN = 5417 100% | 32.3 | 1 | 0.51 |
| 1 | TP = 53 30.6% | TN = 5413 99.9% | 30.0 | 1 | 0.65 | |
| n°2 | 0 | TP = 7 4.0% | TN = 5417 100% | 32.3 | 1 | 0.52 |
| 1 | TP = 97 56.1% | TN = 5411 99.9% | 30.4 | 1 | 0.78 | |
| n°3 | 0 | TP = 7 4.0% | TN = 5417 100% | 32.3 | 1 | 0.52 |
| 1 | TP = 96 55.5% | TN = 5155 95.2% | 8.7 | 1 | 0.76 | |
| n°4 | 0 | TP = 17 9.8% | TN = 5416 100% | 30.5 | 1 | 0.55 |
| 1 | TP = 134 77.5% | TN = 5056 93.3% | 8.7 | 1 | 0.87 | |
| n°5 | 0 | TP = 136 78.1% | TN = 5338 98.5% | 20.4 | 1 | 0.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farag, M.; Kieffer, C.; Guedeney, N.; Voisin-Chiret, A.S.; Sopkova-de Oliveira Santos, J. Computational Tool to Design Small Synthetic Inhibitors Selective for XIAP-BIR3 Domain. Molecules 2023, 28, 5155. https://doi.org/10.3390/molecules28135155
Farag M, Kieffer C, Guedeney N, Voisin-Chiret AS, Sopkova-de Oliveira Santos J. Computational Tool to Design Small Synthetic Inhibitors Selective for XIAP-BIR3 Domain. Molecules. 2023; 28(13):5155. https://doi.org/10.3390/molecules28135155
Chicago/Turabian StyleFarag, Marc, Charline Kieffer, Nicolas Guedeney, Anne Sophie Voisin-Chiret, and Jana Sopkova-de Oliveira Santos. 2023. "Computational Tool to Design Small Synthetic Inhibitors Selective for XIAP-BIR3 Domain" Molecules 28, no. 13: 5155. https://doi.org/10.3390/molecules28135155
APA StyleFarag, M., Kieffer, C., Guedeney, N., Voisin-Chiret, A. S., & Sopkova-de Oliveira Santos, J. (2023). Computational Tool to Design Small Synthetic Inhibitors Selective for XIAP-BIR3 Domain. Molecules, 28(13), 5155. https://doi.org/10.3390/molecules28135155








