Structural and Functional Characterization of Drosophila melanogaster α-Amylase
Abstract
1. Introduction
2. Results
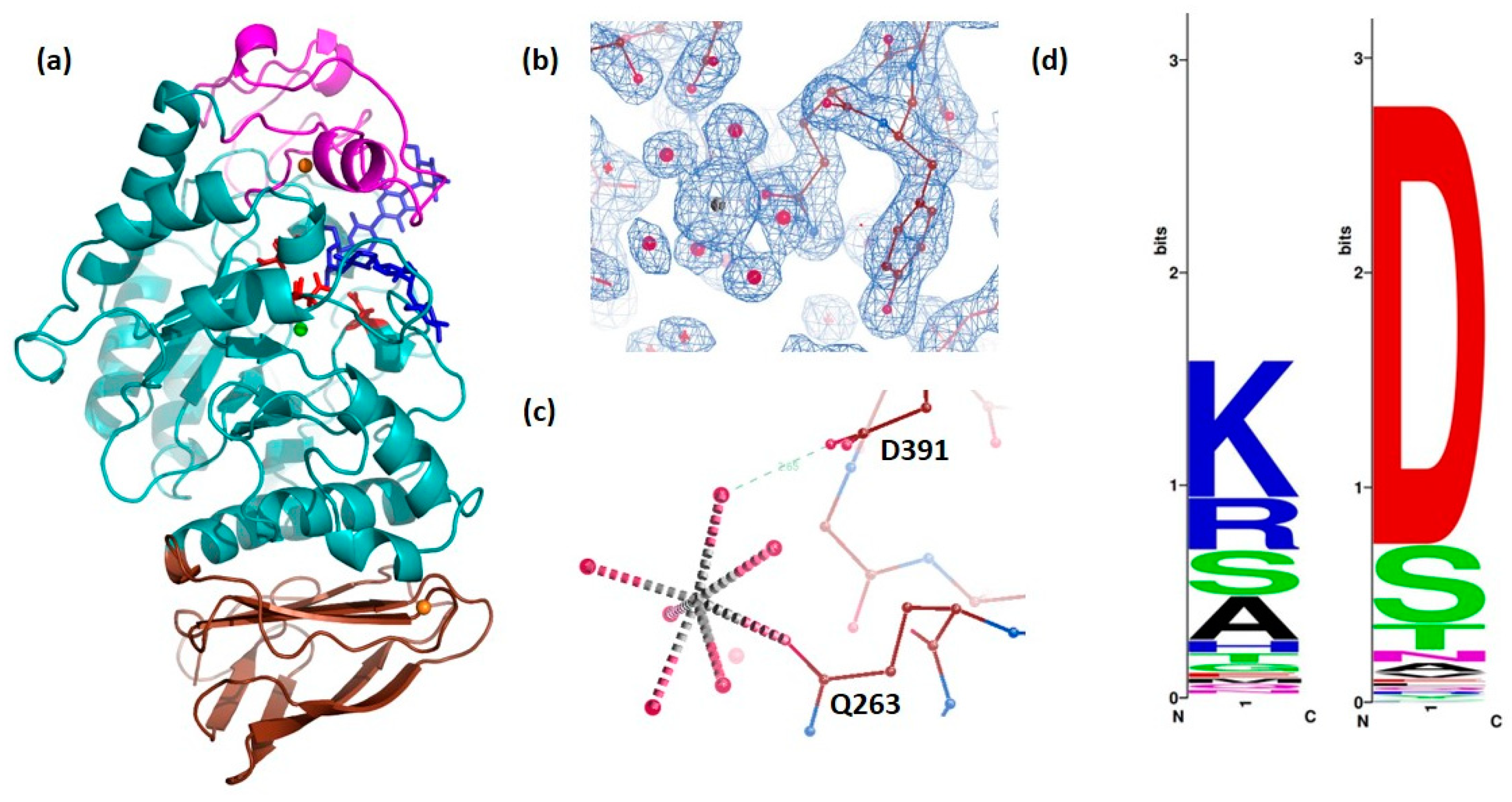
2.1. Three-Dimensional Structure of DMA
2.2. Three-Dimensional Structure of DMA in Complex with Acarbose
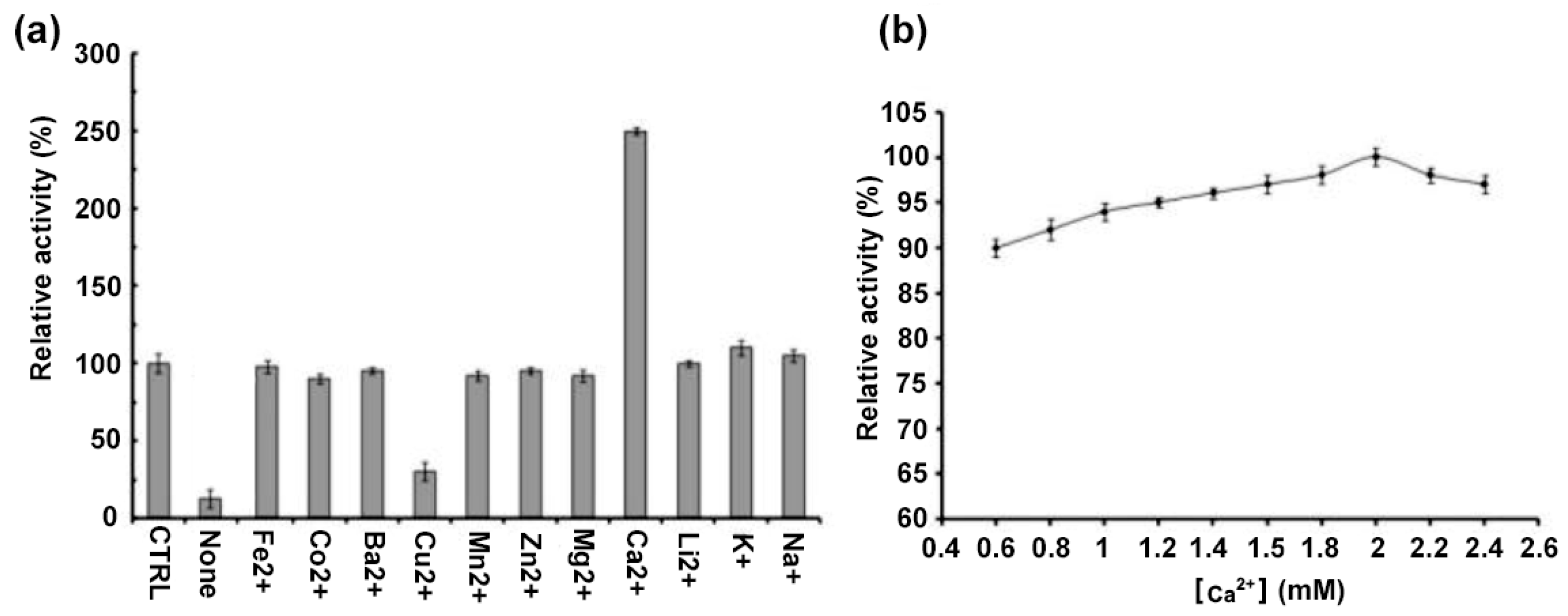
2.3. Biochemical Properties
2.4. Thermal and pH Stability
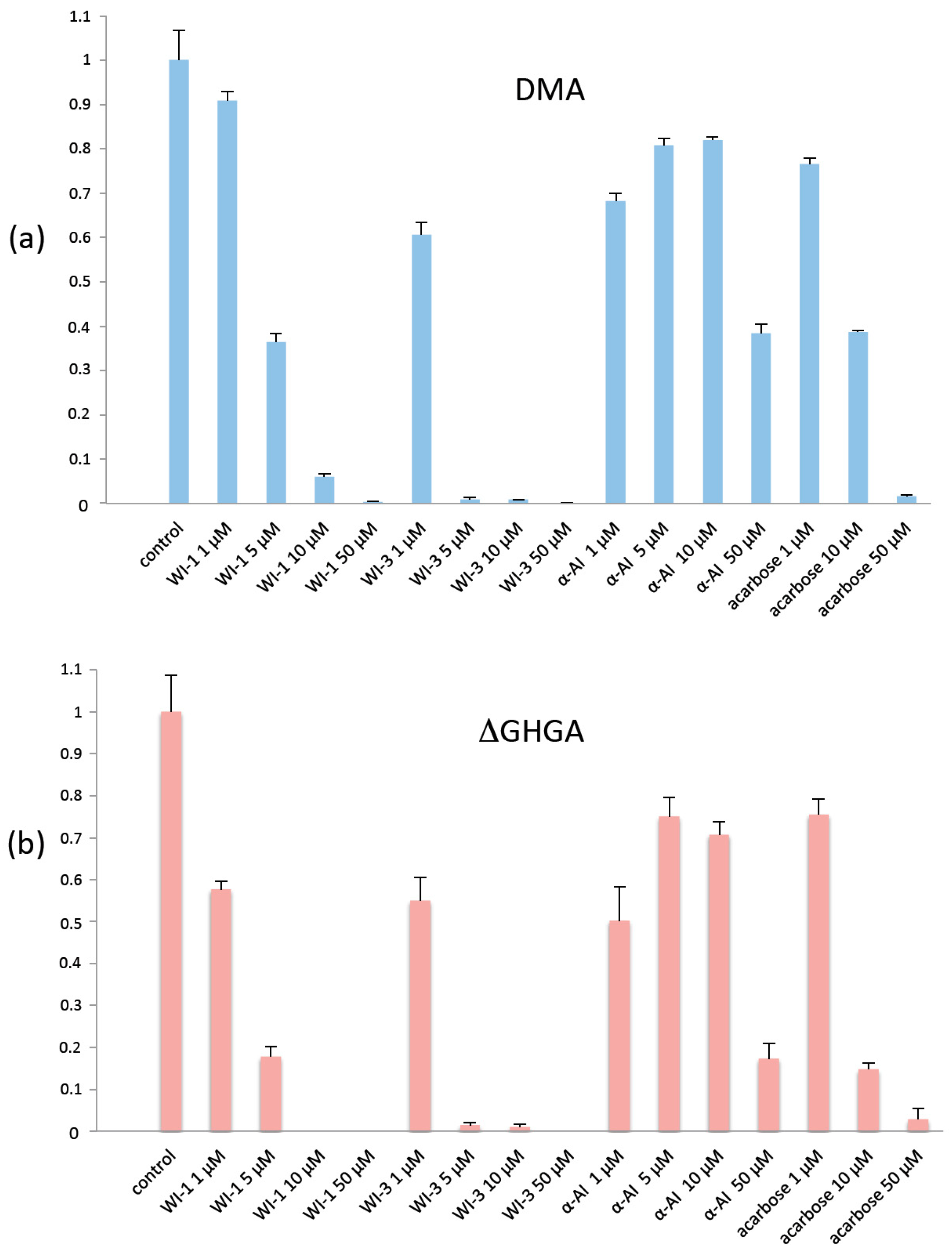
2.5. Role of the Flexible Loop
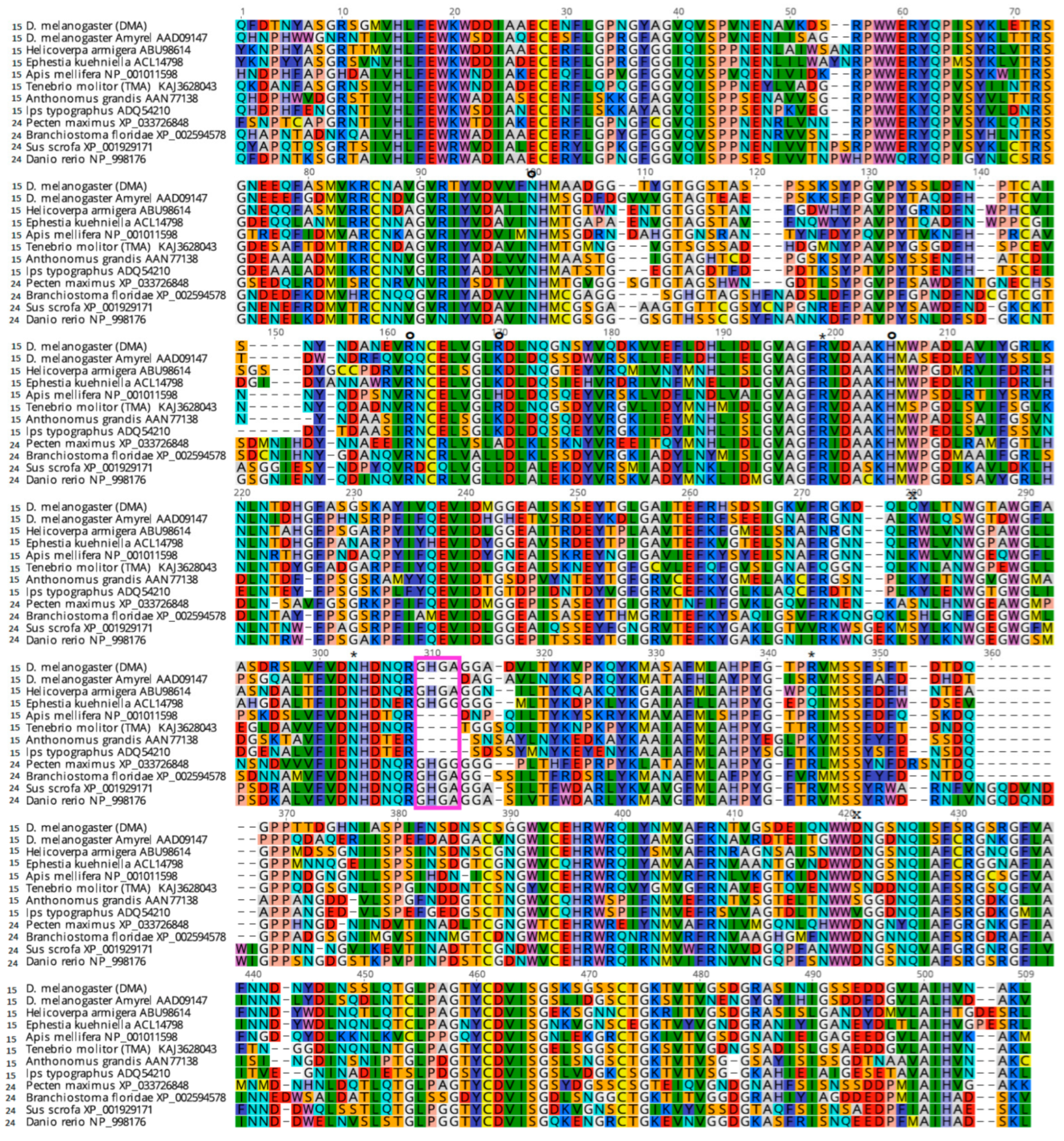
2.6. Comparative Studies with Other α-Amylases
2.7. Docking Studies of DMA and Insect Inhibitors
2.8. Phylogenetic Studies
2.9. Enzyme Variability in Drosophila melanogaster
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media, Plasmids and Growth Conditions
4.2. Over-Expression and Purification of Drosophila melanogaster α-Amylase
4.3. Crystallization
4.4. Data Collection
4.5. Structure Determination and Refinement
4.6. Bioinformatics and Figure Rendering
4.7. Activity Measurements
4.7.1. Temperature, pH and Stability Profiles
4.7.2. Effect of Metal Ions
4.7.3. Determination of Kinetic Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Dadd, R. Nutrition: Organisms, Comprehensive Insect Physiology, Biochemistry and Pharmacology; Kerkut, G.A., Gilbert, L.I., Eds.; National Academy Press: Oxford, UK; Washington, DC, USA, 1985; Volume 4. [Google Scholar]
- Dojnov, B.; Bozić, N.; Nenadović, V.; Ivanović, J.; Vujcić, Z. Purification and Properties of Midgut α-Amylase Isolated from Morimus Funereus (Coleoptera: Cerambycidae) Larvae. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 149, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Terra, W.R.; Ferreira, C. Insect Digestive Enzymes: Properties, Compartmentalization and Function. Comp. Biochem. Physiol.-Part B Biochem. Mol. Biol. 1994, 109, 1–62. [Google Scholar] [CrossRef]
- Da Lage, J.-L. The Amylases of Insects. Int. J. Insect Sci. 2018, 10, 1179543318804783. [Google Scholar] [CrossRef] [PubMed]
- Da Lage, J.; Maczkowiak, F.; Cariou, M. Molecular Characterization and Evolution of the Amylase Multigene Family of Drosophila Ananassae. J. Mol. Evol. 2000, 51, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L.; Rigden, D.J.; Melo, F.R.; Grossi-De-Sá, M.F. Plant α-Amylase Inhibitors and Their Interaction with Insect α-Amylases. Eur. J. Biochem. 2002, 269, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Ferry, N.; Edwards, M.G.; Gatehouse, J.A.; Gatehouse, A.M. Plant–Insect Interactions: Molecular Approaches to Insect Resistance. Curr. Opin. Biotechnol. 2004, 15, 155–161. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- MacGregor, E.A.; Janecek, S.; Svensson, B. Relationship of Sequence and Structure to Specificity in the α-Amylase Family of Enzymes. Biochim. Biophys. Acta 2001, 1546, 1–20. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. A Remote but Significant Sequence Homology between Glycoside Hydrolase Clan GH-H and Family GH31. FEBS Lett. 2007, 581, 1261–1268. [Google Scholar] [CrossRef]
- Da Lage, J.-L.; Van Wormhoudt, A.; Cariou, M.-L. Diversity and Evolution of the α-Amylase Genes in Animals. Biol. Bratisl. 2002, 57, 181–189. [Google Scholar]
- Grossman, G.L.; James, A.A. The Salivary Glands of the Vector Mosquito, Aedes Aegypti, Express a Novel Member of the Amylase Gene Family. Insect Mol. Biol. 1993, 1, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Grossman, G.L.; Campos, Y.; Severson, D.W.; James, A.A. Evidence for Two Distinct Members of the Amylase Gene Family in the Yellow Fever Mosquito, Aedes Aegypti. Insect Biochem. Mol. Biol. 1997, 27, 769–781. [Google Scholar] [CrossRef]
- Claisse, G.; Feller, G.; Bonneau, M.; Da Lage, J.-L. A Single Amino-Acid Substitution Toggles Chloride Dependence of the α-Amylase Paralog Amyrel in Drosophila Melanogaster and Drosophila Virilis Species. Insect Biochem. Mol. Biol. 2016, 75, 70–77. [Google Scholar] [CrossRef]
- Feller, G.; Bonneau, M.; Da Lage, J.-L. Amyrel, a Novel Glucose-Forming α-Amylase from Drosophila with 4-α-Glucanotransferase Activity by Disproportionation and Hydrolysis of Maltooligosaccharides. Glycobiology 2021, 31, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Ramasubbu, N.; Paloth, V.; Luo, Y.; Brayer, G.D.; Levine, M.J. Structure of Human Salivary α-Amylase at 1.6 Å Resolution: Implications for Its Role in the Oral Cavity. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Strobl, S.; Maskos, K.; Wiegand, G.; Huber, R.; Gomis-Rüth, F.X.; Glockshuber, R. A Novel Strategy for Inhibition of α-Amylases: Yellow Meal Worm α-Amylase in Complex with the Ragi Bifunctional Inhibitor at 2.5 Å Resolution. Structure 1998, 6, 911–921. [Google Scholar] [CrossRef]
- Kikkawa, H. Biochemical Genetics of Bombyx Mori (Silkworm). In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 1953; Volume 5, pp. 107–140. ISBN 978-0-12-017605-2. [Google Scholar]
- Doane, W.W. Amylase Variants in Drosophila Melanogaster: Linkage Studies and Characterization of Enzyme Extracts. J. Exp. Zool. 1969, 171, 31–41. [Google Scholar] [CrossRef]
- Hoorn, A.J.W.; Scharloo, W. Functional Significance of Amylase Polymorphism in Drosophila Melanogaster. III. Ontogeny of Amylase and Some α-Glucosidases. Biochem. Genet. 1980, 18, 51–63. [Google Scholar] [CrossRef]
- Hickey, D.A. Selection on Amylase Allozymes in Drosophila Melanogaster: Selection Experiments Using Several Independently Derived Pairs of Chromosomes. Evolution 1979, 33, 1128. [Google Scholar] [CrossRef]
- Qian, M.; Haser, R.; Payan, F. Structure and Molecular Model Refinement of Pig Pancreatic α-Amylase at 2.1 Å Resolution. J. Mol. Biol. 1993, 231, 785–799. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Davies, G.J. Structure of the Aspergillus Oryzae α-Amylase Complexed with the Inhibitor Acarbose at 2.0 Å Resolution. Biochemistry 1997, 36, 10837–10845. [Google Scholar] [CrossRef] [PubMed]
- Kadziola, A.; Søgaard, M.; Svensson, B.; Haser, R. Molecular Structure of a Barley α-Amylase-Inhibitor Complex: Implications for Starch Binding and Catalysis. J. Mol. Biol. 1998, 278, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the Psychrophilic Alteromonas Haloplanctis α-Amylase Give Insights into Cold Adaptation at a Molecular Level. Structure 1998, 6, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.J.; Lozanov, V.; Patthy, A.; Huber, R.; Bode, W.; Pongor, S.; Strobl, S. Specific Inhibition of Insect α-Amylases: Yellow Meal Worm α-Amylase in Complex with the Amaranth α-Amylase Inhibitor at 2.0 Å Resolution. Structure 1999, 7, 1079–1088. [Google Scholar] [CrossRef]
- Aghajari, N.; Roth, M.; Haser, R. Crystallographic Evidence of a Transglycosylation Reaction: Ternary Complexes of a Psychrophilic α-Amylase. Biochemistry 2002, 41, 4273–4280. [Google Scholar] [CrossRef]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structural Basis of α-Amylase Activation by Chloride. Protein Sci. 2002, 11, 1435–1441. [Google Scholar] [CrossRef]
- Maurus, R.; Begum, A.; Williams, L.K.; Fredriksen, J.R.; Zhang, R.; Withers, S.G.; Brayer, G.D. Alternative Catalytic Anions Differentially Modulate Human α-Amylase Activity and Specificity. Biochemistry 2008, 47, 3332–3344. [Google Scholar] [CrossRef]
- Robert, X.; Haser, R.; Mori, H.; Svensson, B.; Aghajari, N. Oligosaccharide Binding to Barley α-Amylase 1. J. Biol. Chem. 2005, 280, 32968–32978. [Google Scholar] [CrossRef]
- Strobl, S.; Maskos, K.; Betz, M.; Wiegand, G.; Huber, R.; Gomis-Rüth, F.X.; Glockshuber, R. Crystal Structure of Yellow Meal Worm α-Amylase at 1.64 Å Resolution. J. Mol. Biol. 1998, 278, 617–628. [Google Scholar] [CrossRef]
- Brünger, A.T. Free R Value: A Novel Statistical Quantity for Assessing the Accuracy of Crystal Structures. Nature 1992, 355, 472–475. [Google Scholar] [CrossRef]
- Brayer, G.D.; Luo, Y.; Withers, S.G. The structure of Human Pancreatic α-Amylase at 1.8 Å Resolution and Comparisons with Related Enzymes. Protein Sci. 1995, 4, 1730–1742. [Google Scholar] [CrossRef] [PubMed]
- Strobl, S.; Gomis-Rüth, F.X.; Maskos, K.; Frank, G.; Huber, R.; Glockshuber, R. The α-Amylase from the Yellow Meal Worm: Complete Primary Structure, Crystallization and Preliminary X-Ray Analysis. FEBS Lett. 1997, 409, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Kadziola, A.; Abe, J.; Svensson, B.; Haser, R. Crystal and Molecular Structure of Barley α-Amylase. J. Mol. Biol. 1994, 239, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Haser, R.; Gottschalk, T.E.; Ratajczak, F.; Driguez, H.; Svensson, B.; Aghajari, N. The Structure of Barley α-Amylase Isozyme 1 Reveals a Novel Role of Domain C in Substrate Recognition and Binding: A Pair of Sugar Tongs. Structure 2003, 11, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Kusunoki, M.; Harada, W.; Tanaka, N.; Iga, Y.; Yasuoka, N.; Toda, H.; Narita, K.; Kakudo, M. Molecular Structure of Taka-Amylase A. I. Backbone Chain Folding at 3 Å Resolution. J. Biochem. 1980, 87, 1555–1558. [Google Scholar] [CrossRef]
- Boel, E.; Brady, L.; Brzozowski, A.M.; Derewenda, Z.; Dodson, G.G.; Jensen, V.J.; Petersen, S.B.; Swift, H.; Thim, L.; Woldike, H.F. Calcium Binding in α-Amylases: An X-Ray Diffraction Study at 2.1-Å Resolution of Two Enzymes from Aspergillus. Biochemistry 1990, 29, 6244–6249. [Google Scholar] [CrossRef]
- Linden, A.; Wilmanns, M. Adaptation of Class-13 α-Amylases to Diverse Living Conditions. Chembiochem 2004, 5, 231–239. [Google Scholar] [CrossRef]
- Machius, M.; Wiegand, G.; Huber, R. Crystal Structure of Calcium-Depleted Bacillus Licheniformis α-Amylase at 2.2 Å Resolution. J. Mol. Biol. 1995, 246, 545–559. [Google Scholar] [CrossRef]
- Hwang, K.Y.; Song, H.K.; Chang, C.; Lee, J.; Lee, S.Y.; Kim, K.K.; Choe, S.; Sweet, R.M.; Suh, S.W. Crystal Structure of Thermostable α-Amylase from Bacillus Licheniformis Refined at 1.7 Å Resolution. Mol. Cells 1997, 7, 251–258. [Google Scholar]
- Shirai, T.; Hung, V.S.; Morinaka, K.; Kobayashi, T.; Ito, S. Crystal Structure of GH13 α-Glucosidase GSJ from One of the Deepest Sea Bacteria. Proteins 2008, 73, 126–133. [Google Scholar] [CrossRef]
- Tan, T.-C.; Mijts, B.N.; Swaminathan, K.; Patel, B.K.C.; Divne, C. Crystal Structure of the Polyextremophilic α-Amylase AmyB from Halothermothrix Orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch. J. Mol. Biol. 2008, 378, 852–870. [Google Scholar] [CrossRef] [PubMed]
- Alikhajeh, J.; Khajeh, K.; Ranjbar, B.; Naderi-Manesh, H.; Lin, Y.H.; Liu, E.; Guan, H.H.; Hsieh, Y.C.; Chuankhayan, P.; Huang, Y.C.; et al. Structure of Bacillus Amyloliquefaciens α-Amylase at High Resolution: Implications for Thermal Stability. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Maczkowiak, F.; Da Lage, J.-L. Origin and Evolution of the Amyrel Gene in the α-Amylase Multigene Family of Diptera. Genetica 2006, 128, 145–158. [Google Scholar] [CrossRef]
- Feller, G.; Bussy, O.; Houssier, C.; Gerday, C. Structural and Functional Aspects of Chloride Binding to Alteromonas Haloplanctis α-Amylase. J. Biol. Chem. 1996, 271, 23836–23841. [Google Scholar] [CrossRef]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Crystal Structures of the Psychrophilic α-Amylase from Alteromonas Haloplanctis in Its Native Form and Complexed with an Inhibitor. Protein Sci. 1998, 7, 564–572. [Google Scholar] [CrossRef]
- Brayer, G.D.; Sidhu, G.; Maurus, R.; Rydberg, E.H.; Braun, C.; Wang, Y.; Nguyen, N.T.; Overall, C.M.; Withers, S.G. Subsite Mapping of the Human Pancreatic α-Amylase Active Site through Structural, Kinetic, and Mutagenesis Techniques. Biochemistry 2000, 39, 4778–4791. [Google Scholar] [CrossRef]
- Roth, C.; Moroz, O.V.; Turkenburg, J.P.; Blagova, E.; Waterman, J.; Ariza, A.; Ming, L.; Tianqi, S.; Andersen, C.; Davies, G.J.; et al. Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance. Int. J. Mol. Sci. 2019, 20, 4902. [Google Scholar] [CrossRef]
- Ramasubbu, N.; Ragunath, C.; Mishra, P.J.; Thomas, L.M.; Gyémánt, G.; Kandra, L. Human Salivary α-Amylase Trp58 Situated at Subsite -2 Is Critical for Enzyme Activity. Eur. J. Biochem. 2004, 271, 2517–2529. [Google Scholar] [CrossRef]
- Davies, G.J.; Brzozowski, A.M.; Dauter, Z.; Rasmussen, M.D.; Borchert, T.V.; Wilson, K.S. Structure of a Bacillus Halmapalus Family 13 α-Amylase, BHA, in Complex with an Acarbose-Derived Nonasaccharide at 2.1 Å Resolution. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 190–193. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Lawson, D.M.; Turkenburg, J.P.; Bisgaard-Frantzen, H.; Svendsen, A.; Borchert, T.V.; Dauter, Z.; Wilson, K.S.; Davies, G.J. Structural Analysis of a Chimeric Bacterial α-Amylase. High-Resolution Analysis of Native and Ligand Complexes. Biochemistry 2000, 39, 9099–9107. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Haser, R.; Buisson, G.; Duée, E.; Payan, F. The Active Center of a Mammalian α-Amylase. Structure of the Complex of a Pancreatic α-Amylase with a Carbohydrate Inhibitor Refined to 2.2-Å Resolution. Biochemistry 1994, 33, 6284–6294. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, M.; Fujimoto, Z.; Momma, M.; Takase, K.; Mizuno, H. Crystal Structure of Bacillus Subtilis α-Amylase in Complex with Acarbose. J. Bacteriol. 2003, 185, 6981–6984. [Google Scholar] [CrossRef] [PubMed]
- Ramasubbu, N.; Ragunath, C.; Mishra, P.J. Probing the Role of a Mobile Loop in Substrate Binding and Enzyme Activity of Human Salivary Amylase. J. Mol. Biol. 2003, 325, 1061–1076. [Google Scholar] [CrossRef]
- Yardley, D.G.; Moussatos, V.V.; Martin, M.P. The Amylase System of Drosophila-II. Insect Biochem. 1983, 13, 543–548. [Google Scholar] [CrossRef]
- Prigent, S.; Matoub, M.; Rouland, C.; Cariou, M.L. Metabolic Evolution in α-Amylases from Drosophila Virilis and D. Repleta, Two Species with Different Ecological Niches. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1998, 119, 407–412. [Google Scholar] [CrossRef]
- Najafi, M.F.; Deobagkar, D.; Deobagkar, D. Purification and Characterization of an Extracellular α-Amylase from Bacillus Subtilis AX20. Protein Expr. Purif. 2005, 41, 349–354. [Google Scholar] [CrossRef]
- Bernfeld, P. [17] Amylases, α and β. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1955; Volume 1, pp. 149–158. ISBN 978-0-12-181801-2. [Google Scholar]
- Pytelková, J.; Hubert, J.; Lepšík, M.; Šobotník, J.; Šindelka, R.; Křížková, I.; Horn, M.; Mareš, M. Digestive α-Amylases of the Flour Moth Ephestia Kuehniella- Adaptation to Alkaline Environment and Plant Inhibitors: Alkaline Digestive α-Amylases of Lepidoptera. FEBS J. 2009, 276, 3531–3546. [Google Scholar] [CrossRef]
- Ferruz, N.; Schmidt, S.; Höcker, B. ProteinTools: A Toolkit to Analyze Protein Structures. Nucleic Acids Res. 2021, 49, W559–W566. [Google Scholar] [CrossRef]
- Nahoum, V.; Farisei, F.; Le-Berre-Anton, V.; Egloff, M.P.; Rougé, P.; Poerio, E.; Payan, F. A Plant-Seed Inhibitor of Two Classes of α-Amylases: X-Ray Analysis of Tenebrio Molitor Larvae α-Amylase in Complex with the Bean Phaseolus Vulgaris Inhibitor. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 360–362. [Google Scholar] [CrossRef]
- Shibata, H.; Yamazaki, T. A Comparative Study of the Enzymological Features of α-Amylase in the Drosophila Melanogaster Species Subgroup. Jpn. J. Genet. 1994, 69, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Commin, C.; Aumont-Nicaise, M.; Claisse, G.; Feller, G.; Da Lage, J.-L. Enzymatic Characterization of Recombinant α-Amylase in the Drosophila Melanogaster Species Subgroup: Is There an Effect of Specialization on Digestive Enzyme? Genes. Genet. Syst. 2013, 88, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Inomata, N.; Shibata, H.; Okuyama, E.; Yamazaki, T. Evolutionary Relationships and Sequence Variation of α-Amylase Variants Encoded by Duplicated Genes in the Amy Locus of Drosophila Melanogaster. Genetics 1995, 141, 237–244. [Google Scholar] [CrossRef]
- Araki, H.; Inomata, N.; Yamazaki, T. Molecular Evolution of Duplicated Amylase Gene Regions in Drosophila Melanogaster: Evidence of Positive Selection in the Coding Regions and Selective Constraints in the Cis-Regulatory Regions. Genetics 2001, 157, 667–677. [Google Scholar] [CrossRef]
- Uitdehaag, J.C.; Mosi, R.; Kalk, K.H.; van der Veen, B.A.; Dijkhuizen, L.; Withers, S.G.; Dijkstra, B.W. X-ray Structures along the Reaction Pathway of Cyclodextrin Glycosyltransferase Elucidate Catalysis in the α-Amylase Family. Nat. Struct. Biol. 1999, 6, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Rydberg, E.H.; Li, C.; Maurus, R.; Overall, C.M.; Brayer, G.D.; Withers, S.G. Mechanistic Analyses of Catalysis in Human Pancreatic α-Amylase: Detailed Kinetic and Structural Studies of Mutants of Three Conserved Carboxylic Acids. Biochemistry 2002, 41, 4492–4502. [Google Scholar] [CrossRef]
- Vallée, F.; Kadziola, A.; Bourne, Y.; Juy, M.; Rodenburg, K.W.; Svensson, B.; Haser, R. Barley α-Amylase Bound to Its Endogenous Protein Inhibitor BASI: Crystal Structure of the Complex at 1.9 Å Resolution. Structure 1998, 6, 649–659. [Google Scholar] [CrossRef]
- Machius, M.; Declerck, N.; Huber, R.; Wiegand, G. Activation of Bacillus Licheniformis α-Amylase through a Disorder→order Transition of the Substrate-Binding Site Mediated by a Calcium–Sodium–Calcium Metal Triad. Structure 1998, 6, 281–292. [Google Scholar] [CrossRef]
- Linden, A.; Mayans, O.; Meyer-Klaucke, W.; Antranikian, G.; Wilmanns, M. Differential Regulation of a Hyperthermophilic α-Amylase with a Novel (Ca,Zn) Two-Metal Center by Zinc. J. Biol. Chem. 2003, 278, 9875–9884. [Google Scholar] [CrossRef]
- D’Amico, S.; Gerday, C.; Feller, G. Structural Similarities and Evolutionary Relationships in Chloride-Dependent α-Amylases. Gene 2000, 253, 95–105. [Google Scholar] [CrossRef]
- Maurus, R.; Begum, A.; Kuo, H.-H.; Racaza, A.; Numao, S.; Andersen, C.; Tams, J.W.; Vind, J.; Overall, C.M.; Withers, S.G.; et al. Structural and Mechanistic Studies of Chloride Induced Activation of Human Pancreatic α-Amylase. Protein Sci. 2005, 14, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Klarenberg, A.J.; Vermeulen, J.W.C.; Jacobs, P.J.M.; Scharloo, W. Genetic and Dietary Regulation of Tissue-Specific Expression Patterns of α-Amylase in Larvae of Drosophila Melanogaster. Comp. Biochem. Physiol. Part B Comp. Biochem. 1988, 89, 143–146. [Google Scholar] [CrossRef]
- Overend, G.; Luo, Y.; Henderson, L.; Douglas, A.E.; Davies, S.A.; Dow, J.A.T. Molecular Mechanism and Functional Significance of Acid Generation in the Drosophila Midgut. Sci. Rep. 2016, 6, 27242. [Google Scholar] [CrossRef] [PubMed]
- Prigent, S.; Renard, E.; Cariou, M.-L. Electrophoretic Mobility of Amylase in Drosophilids Indicates Adaptation to Ecological Diversity. Genetica 2003, 119, 133–145. [Google Scholar] [CrossRef]
- Daïnou, O.; Cariou, M.; Goux, J.; David, J. Amylase Polymorphism in Drosophila Melanogaster: Haplotype Frequencies in Tropical African and American Populations. Genet. Sel. Evol. 1993, 25, 133. [Google Scholar] [CrossRef]
- Araki, H.; Yoshizumi, S.; Inomata, N.; Yamazaki, T. Genetic Coadaptation of the Amylase Gene System in Drosophila Melanogaster: Evidence for the Selective Advantage of the Lowest AMY Activity and of Its Epistatic Genetic Background. J. Hered. 2005, 96, 388–395. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.-W.; Read, R.J.; Adams, P.D. Iterative Model Building, Structure Refinement and Density Modification with the PHENIX AutoBuild Wizard. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 61–69. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Vagin, A.A.; Steiner, R.A.; Lebedev, A.A.; Potterton, L.; McNicholas, S.; Long, F.; Murshudov, G.N. REFMAC5 Dictionary: Organization of Prior Chemical Knowledge and Guidelines for Its Use. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards Automated Crystallographic Structure Refinement with Phenix.Refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Kleywegt, G.J.; Jones, T.A. Detection, Delineation, Measurement and Display of Cavities in Macromolecular Structures. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 178–185. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Hooft, R.W.; Vriend, G.; Sander, C.; Abola, E.E. Errors in Protein Structures. Nature 1996, 381, 272. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis Testing Using Phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef]
- Pupko, T.; Pe, I.; Shamir, R.; Graur, D. A Fast Algorithm for Joint Reconstruction of Ancestral Amino Acid Sequences. Mol. Biol. Evol. 2000, 17, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Kumar, S.; Nei, M. A New Method of Inference of Ancestral Nucleotide and Amino Acid Sequences. Genetics 1995, 141, 1641–1650. [Google Scholar] [CrossRef]
- Nielsen, R. Mapping Mutations on Phylogenies. Syst. Biol. 2002, 51, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]



| Native DMA | DMA/Acarbose | |
|---|---|---|
| PDB-ID Data collection | 8OR6 | 8ORP |
| Beamline | ID23-2 | ID14-1 |
| Temperature (K) | 100 | 100 |
| Wavelength (Å) | 0.8726 | 0.9334 |
| Resolution range (Å) | 44.8–2.5 | 30.0–2.0 |
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | a = 80.4, b = 73.1, c = 99.3 | a = 80.1, b = 72.5, c = 98.2 |
| α, β, γ (°) | β = 98.3 | β = 98.4 |
| n° reflections | 148,467 | 280,188 |
| n° unique reflections | 39,454 | 75,240 |
| Rmerge (%) | 21.6 | 13.5 |
| CC1/2 | 0.967 (0.641) | 0.993 (0.778) |
| I/σ(I) | 6.4 (3.0) | 9.0 (2.3) |
| Multiplicity | 3.8 | 3.7 |
| Completeness (%) | 99.2 (93.9) | 99.8 (99.7) |
| n° molecules/asymm. unit | 2 | 2 |
| Refinement | ||
| R/Rfree 1 (%) | 19.0/27.6 | 17.6/21.2 |
| n° atoms | ||
| protein | 7312 | 7318 |
| water | 493 | 874 |
| ligands | - | 304 3 |
| n° ions per molecule | ||
| strontium 2 | 2 | 2 |
| chloride | 1 | 1 |
| Average B-factor (Å2) | ||
| protein | 18.7 | 17.6 |
| water | 19.7 | 27.8 |
| RMSD | ||
| Bond lengths (Å) | 0.002 | 0.003 |
| Bond angles (°) | 0.47 | 0.64 |
| Ramachandran | ||
| Favored (%) | 97.2 | 97.5 |
| Allowed (%) | 2.6 | 2.3 |
| Outliers (%) | 0.2 | 0.2 |
| Substrate | Km (g L−1) | kcat (s−1) | kcat/Km (s−1/g L−1) |
| starch | 0.68 ± 0.06 | 571 ± 1 | 840 ± 17 |
| Substrate | Km (µM) | kcat (s−1) | kcat/Km (µM−1/s−1) |
| maltotriose | 94 ± 5 | 194 ± 6 | 2.1 ± 0.1 |
| maltotetraose | 70 ± 1 | 223 ± 3 | 3.2 ± 0.9 |
| maltopentaose | 68 ± 8 | 278 ± 2 | 4.1 ± 0.7 |
| maltohexaose | 61 ± 6 | 370 ± 4 | 6.1 ± 0.6 |
| maltoheptaose | 52 ± 4 | 493 ± 1 | 9.5 ± 0.4 |
| Temperatures (°C) | Half-Life (t1/2, h) | pH | Half-Life (t1/2, h) |
|---|---|---|---|
| 4 | 44 ± 0.1 | 3.0 | 16 ± 0.1 |
| 10 | 46 ± 0.3 | 3.5 | 19 ± 0.2 |
| 15 | 47 ± 0.2 | 4.0 | 32 ± 0.1 |
| 20 | 48 ± 0.6 | 4.5 | 47 ± 0.3 |
| 25 | 47 ± 0.2 | 5.0 | 49 ± 0.6 |
| 30 | 46 ± 0.4 | 5.5 | 48 ± 0.4 |
| 35 | 40 ± 0.2 | 6.0 | 48 ± 0.2 |
| 40 | 38 ± 0.2 | 6.5 | 47 ± 0.5 |
| 45 | 29 ± 0.2 | 7.0 | 48 ± 0.7 |
| 50 | 6.0 ± 0.1 | 7.5 | 47 ± 0.3 |
| 55 | 4.0 ± 0.2 | 8.0 | 46 ± 0.2 |
| 60 | 0.9 ± 0.3 | 8.5 | 44 ± 0.1 |
| 9.0 | 38 ± 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rhimi, M.; Da Lage, J.-L.; Haser, R.; Feller, G.; Aghajari, N. Structural and Functional Characterization of Drosophila melanogaster α-Amylase. Molecules 2023, 28, 5327. https://doi.org/10.3390/molecules28145327
Rhimi M, Da Lage J-L, Haser R, Feller G, Aghajari N. Structural and Functional Characterization of Drosophila melanogaster α-Amylase. Molecules. 2023; 28(14):5327. https://doi.org/10.3390/molecules28145327
Chicago/Turabian StyleRhimi, Moez, Jean-Luc Da Lage, Richard Haser, Georges Feller, and Nushin Aghajari. 2023. "Structural and Functional Characterization of Drosophila melanogaster α-Amylase" Molecules 28, no. 14: 5327. https://doi.org/10.3390/molecules28145327
APA StyleRhimi, M., Da Lage, J.-L., Haser, R., Feller, G., & Aghajari, N. (2023). Structural and Functional Characterization of Drosophila melanogaster α-Amylase. Molecules, 28(14), 5327. https://doi.org/10.3390/molecules28145327






