New Crystal Form of Human Neuropilin-1 b1 Fragment with Six Electrostatic Mutations Complexed with KDKPPR Peptide Ligand
Abstract
:1. Introduction
2. Results and Discussion
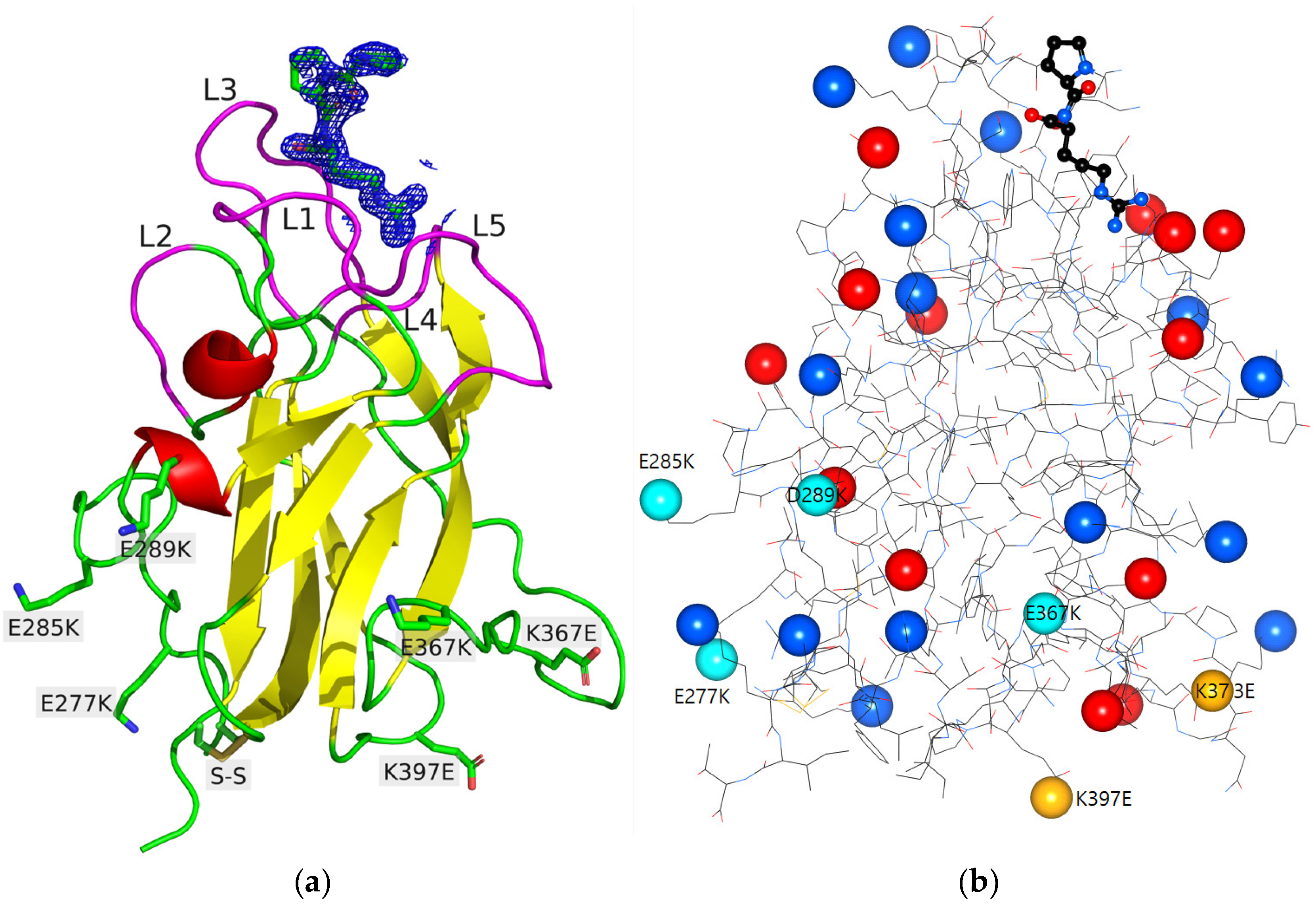
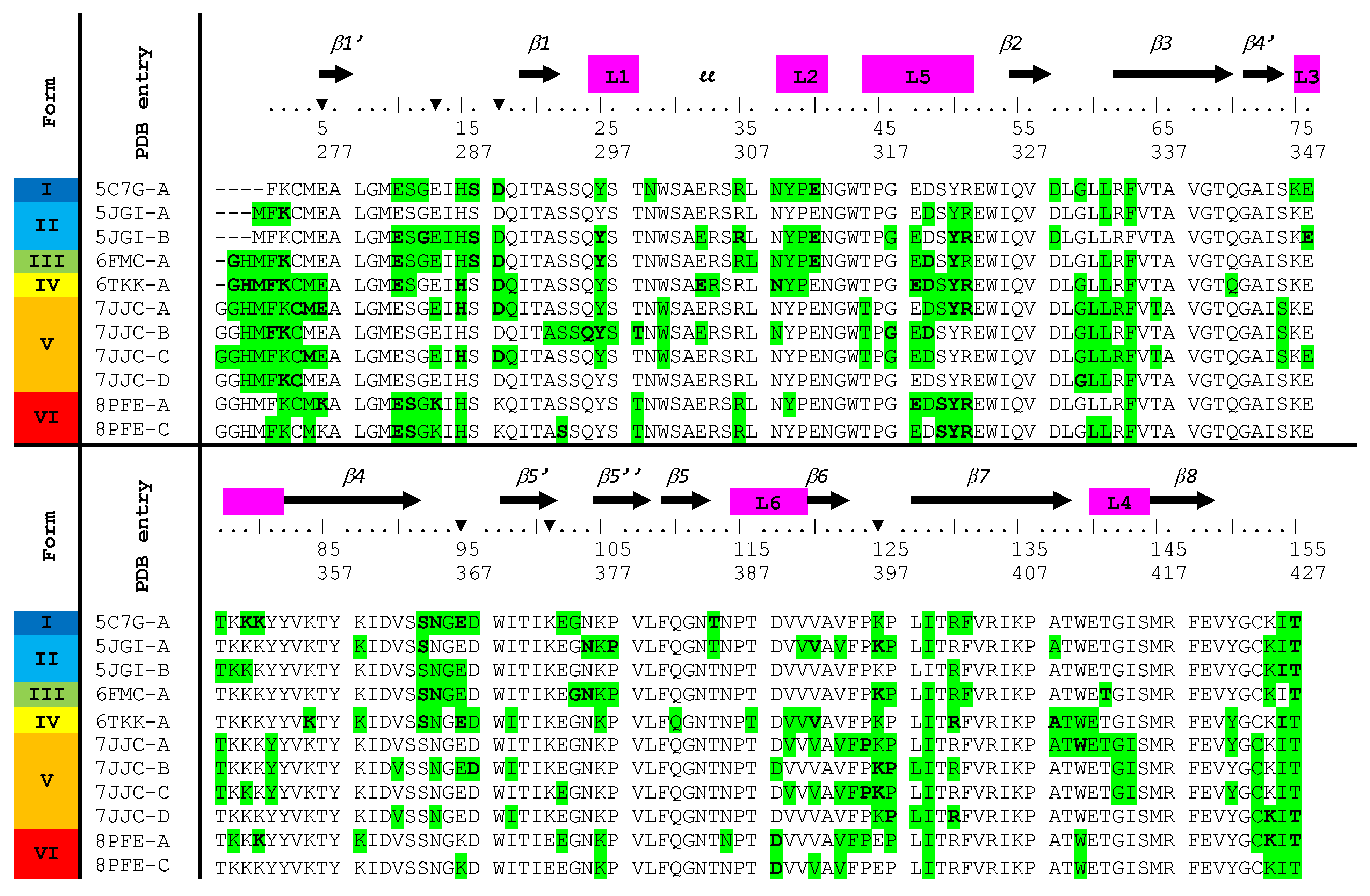
2.1. Design of the NRP1-b1 Hexavariant
2.2. Crystal Structure of the NRP1-b1 Hexavariant
2.2.1. Description of the Structure
2.2.2. Crystal Packing and Intermolecular Contacts
2.3. Protein Ligand Interaction
2.3.1. Description of Ligand Binding
2.3.2. Electrostatic Influence of the Protein on the Peptide
2.3.3. Hirshfeld Surface and Contacts Enrichment Ratio
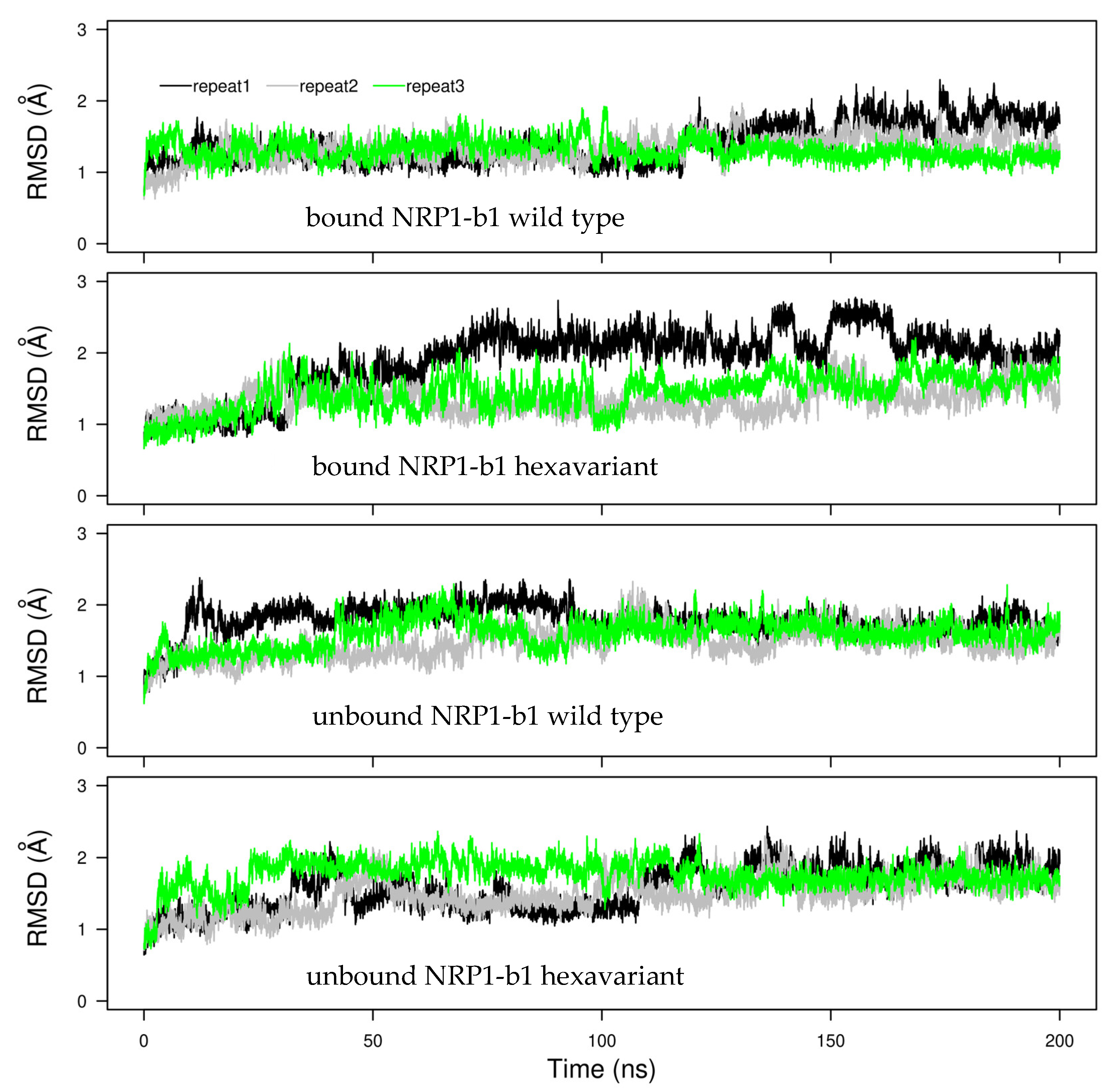
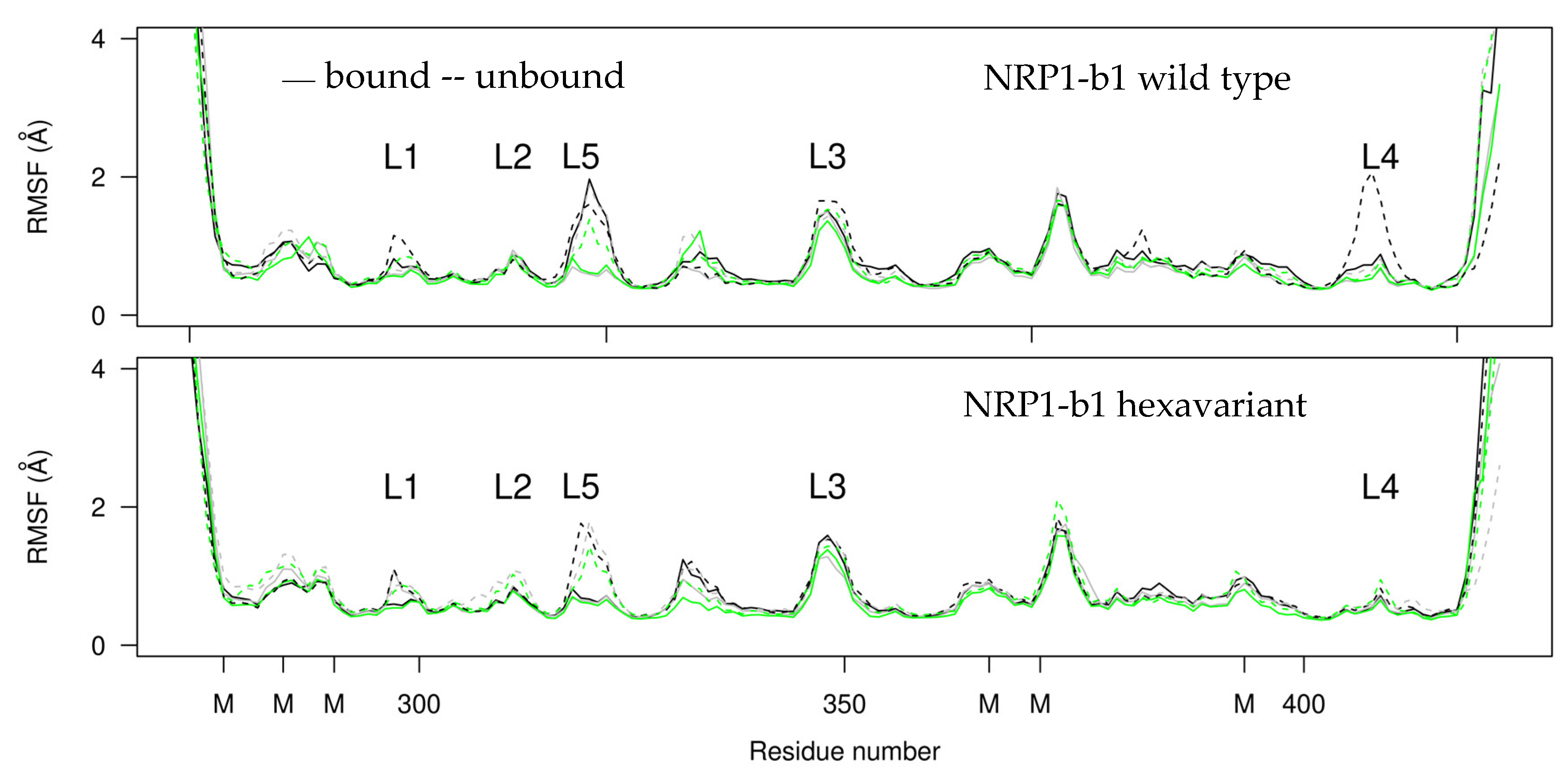
2.4. Molecular Dynamics
3. Materials and Methods
3.1. Protein Production and Purification
3.2. KDKPPR Synthesis on Solid Phase
3.3. Crystallization
3.4. X-ray Diffraction Data Collection and Crystal Structure Determination
3.5. Nucleophilic Influence Zones
3.6. Hirshfeld Analysis
3.7. Molecular Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lu, D.; Shang, G.; He, X.; Bai, X.C.; Zhang, X. Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex. Nat. Commun. 2021, 12, 3172. [Google Scholar] [CrossRef] [PubMed]
- Soker, S.; Miao, H.-Q.; Nomi, M.; Takashima, S.; Klagsbrun, M. VEGF165 mediates formation of complexes containing VEGFR-2 and neuropilin-1 that enhance VEGF165-receptor binding. J. Cell. Biochem. 2002, 85, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 Is Expressed by Endothelial and Tumor Cells as an Isoform-Specific Receptor for Vascular Endothelial Growth Factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, B.; Khaled, Y.S.; Ammori, B.J.; Elkord, E. Neuropilin 1: Function and therapeutic potential in cancer. Cancer Immunol. Immunother. 2014, 63, 81–99. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Lerouge, L.; Gries, M.; Chateau, A.; Daouk, J.; Lux, F.; Rocchi, P.; Cedervall, J.; Olsson, A.K.; Tillement, O.; Frochot, C.; et al. Targeting Glioblastoma-Associated Macrophages for Photodynamic Therapy Using AGuIX((R))-Design Nanoparticles. Pharmaceutics 2023, 15, 997. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.D.; Zhong, L.P.; He, J.; Zhao, Y.X. Targeting neuropilin-1 interactions is a promising anti-tumor strategy. Chin. Med. J. 2020, 134, 508–517. [Google Scholar] [CrossRef]
- Pellet-Many, C.; Frankel, P.; Jia, H.; Zachary, I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008, 411, 211–226. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.C.; Kreusch, A.; McMullan, D.; Ng, K.; Spraggon, G. Crystal Structure of the Human Neuropilin-1 b1 Domain. Structure 2003, 11, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Powell, J.; Mota, F.; Steadman, D.; Soudy, C.; Miyauchi, J.T.; Crosby, S.; Jarvis, A.; Reisinger, T.; Winfield, N.; Evans, G.; et al. Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGFbeta) Production in Regulatory T-Cells. J. Med. Chem. 2018, 61, 4135–4154. [Google Scholar] [CrossRef]
- Mota, F.; Fotinou, C.; Rana, R.R.; Chan, A.W.E.; Yelland, T.; Arooz, M.T.; O’Leary, A.P.; Hutton, J.; Frankel, P.; Zachary, I.; et al. Architecture and hydration of the arginine-binding site of neuropilin-1. FEBS J. 2018, 285, 1290–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, M.W.; Xu, P.; Li, X.; Vander Kooi, C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012, 287, 11082–11089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvis, A.; Allerston, C.K.; Jia, H.; Herzog, B.; Garza-Garcia, A.; Winfield, N.; Ellard, K.; Aqil, R.; Lynch, R.; Chapman, C.; et al. Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J. Med. Chem. 2010, 53, 2215–2226. [Google Scholar] [CrossRef]
- Kamarulzaman, E.E.; Vanderesse, R.; Gazzali, A.M.; Barberi-Heyob, M.; Boura, C.; Frochot, C.; Shawkataly, O.; Aubry, A.; Wahab, H.A. Molecular modelling, synthesis and biological evaluation of peptide inhibitors as anti-angiogenic agent targeting neuropilin-1 for anticancer application. J. Biomol. Struct. Dyn. 2017, 35, 26–45. [Google Scholar] [CrossRef] [Green Version]
- Kamarulzaman, E.E.; Gazzali, A.M.; Acherar, S.; Frochot, C.; Barberi-Heyob, M.; Boura, C.; Chaimbault, P.; Sibille, E.; Wahab, H.A.; Vanderesse, R. New Peptide-Conjugated Chlorin-Type Photosensitizer Targeting Neuropilin-1 for Anti-Vascular Targeted Photodynamic Therapy. Int. J. Mol. Sci. 2015, 16, 24059–24080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechet, D.; Mordon, S.R.; Guillemin, F.; Barberi-Heyob, M.A. Photodynamic therapy of malignant brain tumours: A complementary approach to conventional therapies. Cancer Treat. Rev. 2014, 40, 229–241. [Google Scholar] [CrossRef]
- Gries, M.; Thomas, N.; Daouk, J.; Rocchi, P.; Choulier, L.; Jubreaux, J.; Pierson, J.; Reinhard, A.; Jouan-Hureaux, V.; Chateau, A.; et al. Multiscale Selectivity and in vivo Biodistribution of NRP-1-Targeted Theranostic AGuIX Nanoparticles for PDT of Glioblastoma. Int. J. Nanomed. 2020, 15, 8739–8758. [Google Scholar] [CrossRef]
- Thomas, E.; Colombeau, L.; Gries, M.; Peterlini, T.; Mathieu, C.; Thomas, N.; Boura, C.; Frochot, C.; Vanderesse, R.; Lux, F.; et al. Ultrasmall AGuIX theranostic nanoparticles for vascular-targeted interstitial photodynamic therapy of glioblastoma. Int. J. Nanomed. 2017, 12, 7075–7088. [Google Scholar] [CrossRef] [Green Version]
- Richard, M.; Chateau, A.; Jelsch, C.; Didierjean, C.; Manival, X.; Charron, C.; Maigret, B.; Barberi-Heyob, M.; Chapleur, Y.; Boura, C.; et al. Carbohydrate-based peptidomimetics targeting neuropilin-1: Synthesis, molecular docking study and in vitro biological activities. Bioorgan. Med. Chem. 2016, 24, 5315–5325. [Google Scholar] [CrossRef]
- Jelsch, C.; Longhi, S.; Cambillau, C. Packing forces in nine crystal forms of cutinase. Proteins Struct. Funct. Genet. 1998, 31, 320–333. [Google Scholar] [CrossRef]
- Domagala, S.; Fournier, B.; Liebschner, D.; Guillot, B.; Jelsch, C. An improved experimental databank of transferable multipolar atom models—ELMAM2. Construction details and applications. Acta Crystallogr. A 2012, 68, 337–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- wwPDB consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mata, I.; Molins, E.; Espinosa, E. Zero-Flux Surfaces of the Electrostatic Potential: The Border of Influence Zones of Nucleophilic and Electrophilic Sites in Crystalline Environment. J. Phys. Chem. A 2007, 111, 9859–9870. [Google Scholar] [CrossRef] [PubMed]
- Vuković, V.; Leduc, T.; Jelić-Matošević, Z.; Didierjean, C.; Favier, F.; Guillot, B.; Jelsch, C. A rush to explore protein–ligand electrostatic interaction energy with Charger. Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 1292–1304. [Google Scholar] [CrossRef] [PubMed]
- Wade, R.C.; Gabdoulline, R.R.; Lüdemann, S.K.; Lounnas, V. Electrostatic steering and ionic tethering in enzyme–ligand binding: Insights from simulations. Proc. Natl. Acad. Sci. USA 1998, 95, 5942–5949. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Guillot, B.; Enrique, E.; Huder, L.; Jelsch, C. MoProViewer: A tool to study proteins from a charge density science perspective. Acta Crystallogr. Sect. A 2014, 70, C279. [Google Scholar] [CrossRef]
- Jelsch, C.; Ejsmont, K.; Huder, L. The enrichment ratio of atomic contacts in crystals, an indicator derived from the Hirshfeld surface analysis. IUCrJ 2014, 1, 119–128. [Google Scholar] [CrossRef]
- Jelsch, C.; Bibila Mayaya Bisseyou, Y. Atom interaction propensities of oxygenated chemical functions in crystal packings. IUCrJ 2017, 4, 158–174. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of B-Factors in Protein Science: Interpreting Rigidity, Flexibility, and Internal Motion and Engineering Thermostability. Chem. Rev. 2019, 119, 1626–1665. [Google Scholar] [CrossRef]
- Ahlstrom, L.S.; Miyashita, O. Packing interface energetics in different crystal forms of the λ Cro dimer. Proteins Struct. Funct. Bioinform. 2014, 82, 1128–1141. [Google Scholar] [CrossRef] [Green Version]
- Ahlstrom, L.S.; Vorontsov, I.I.; Shi, J.; Miyashita, O. Effect of the Crystal Environment on Side-Chain Conformational Dynamics in Cyanovirin-N Investigated through Crystal and Solution Molecular Dynamics Simulations. PLoS ONE 2017, 12, e0170337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janowski, P.A.; Liu, C.; Deckman, J.; Case, D.A. Molecular dynamics simulation of triclinic lysozyme in a crystal lattice. Protein Sci. 2016, 25, 87–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmanic, A.; Pannu, N.S.; Zagrovic, B. X-ray refinement significantly underestimates the level of microscopic heterogeneity in biomolecular crystals. Nat. Commun. 2014, 5, 3220. [Google Scholar] [CrossRef] [PubMed]
- Alshawaf, E.; Hammad, M.M.; Marafie, S.K.; Ali, H.; Al-Mulla, F.; Abubaker, J.; Mohammad, A. Discovery of natural products to block SARS-CoV-2 S-protein interaction with Neuropilin-1 receptor: A molecular dynamics simulation approach. Microb. Pathog. 2022, 170, 105701. [Google Scholar] [CrossRef]
- Appleton, B.A.; Wu, P.; Maloney, J.; Yin, J.; Liang, W.-C.; Stawicki, S.; Mortara, K.; Bowman, K.K.; Elliott, J.M.; Desmarais, W.; et al. Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. EMBO J. 2007, 26, 4902–4912. [Google Scholar] [CrossRef] [Green Version]
- Janssen, B.J.C.; Malinauskas, T.; Weir, G.A.; Cader, M.Z.; Siebold, C.; Jones, E.Y. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat. Struct. Mol. Biol. 2012, 19, 1293–1299. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Smart, O.S.; Womack, T.O.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. Sect. D 2012, 68, 368–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Voss, N.R.; Gerstein, M. 3V: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010, 38, W555–W562. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D., Jr. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]

| Atom Type | C | Hc | N | Ho/n | O | W # |
|---|---|---|---|---|---|---|
| surface_peptide | 8.8 | 42.7 | 5.5 | 22.9 | 20.1 | 0 |
| surface_protein | 21.4 | 30.0 | 1.2 | 15.6 | 15.9 | 16.0 |
| C | 1.4 | 21.3 | 1.3 | 3.4 | 0.4 | 1.0 |
| Hc | 10.3 | 3.5 | 7.2 | 15.3 | 4.8 | |
| N | 0 | 0.5 | 0.2 | 1.3 | ||
| Ho/n | CXY | (%) | 1.6 | 17.4 | 6.9 | |
| O | 0.3 | 2.0 | ||||
| C | 0.75 | 1.80 | 1.02 | 0.54 | 0.08 | 0.73 |
| Hc | 0.81 | 1.62 | 0.53 | 1.20 | 0.70 | |
| N | 0 | 0.40 | 0.17 | 1.44 | ||
| Ho/n | EXY | 0.44 | 2.58 | 1.88 | ||
| O | 0.09 | 0.64 | ||||
| Hphob | Hphil | Hphob * Hphil | ||||
| surface % | peptide | 57.1 | 43.0 | |||
| surface % | protein | 52.6 | 47.4 | |||
| contacts | % | 37.8 | 28.2 | 34.0 | ||
| enrichment | 1.26 | 1.38 | 0.69 |
| Data Collection | |
|---|---|
| Diffraction source | ESRF FIP2-BM07 |
| Wavelength (Å) | 0.9795 |
| Space group | P322 |
| a, b, c (Å) | 59.77, 59.77, 174.60 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution range (Å) | 44.53–1.35 (1.37–1.35) 1 |
| Total number of measured intensities | 696,189 (16,486) 1 |
| Number of unique reflections | 80,230 (3895) 1 |
| Average redundancy | 8.7 (4.2) 1 |
| Mean I/sig(I) | 28.7 (1.9) 1 |
| Completeness (%) | 99.7 (95.7) 1 |
| Rmerge 2; Rmeas 3 | 0.031 (0.681) 1; 0.033 (0.779) 1 |
| CC1/2 4 | 1.00 (0.69) 1 |
| Wilson B-factor (Å2) | 17.9 (Aimless)/21.14 (Buster) |
| Refinement and structure | |
| Resolution range (Å) | 19.57–1.35 (1.36–1.35) 1 |
| Number of reflections | 80,203 (1605) 1 |
| Rwork/Rfree 5 | 0.1942/0.2100 (0.2826/0.2760) 1 |
| Correlation Fo − Fc/Fo − Fcfree | 0.966/0.963 |
| Total number of atoms | 2910 |
| Average B factor (Å2) | 25.55 |
| Model quality | |
| RMSZ bond lengths 6 | 1.28 |
| RMSZ bond angles 6 | 1.14 |
| Ramachandran favored (%) | 97.5 |
| Ramachandran allowed (%) | 2.4 |
| Rotamer outliers (%) | 1.8 |
| Clash-score 7 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goudiaby, I.; Malliavin, T.E.; Mocchetti, E.; Mathiot, S.; Acherar, S.; Frochot, C.; Barberi-Heyob, M.; Guillot, B.; Favier, F.; Didierjean, C.; et al. New Crystal Form of Human Neuropilin-1 b1 Fragment with Six Electrostatic Mutations Complexed with KDKPPR Peptide Ligand. Molecules 2023, 28, 5603. https://doi.org/10.3390/molecules28145603
Goudiaby I, Malliavin TE, Mocchetti E, Mathiot S, Acherar S, Frochot C, Barberi-Heyob M, Guillot B, Favier F, Didierjean C, et al. New Crystal Form of Human Neuropilin-1 b1 Fragment with Six Electrostatic Mutations Complexed with KDKPPR Peptide Ligand. Molecules. 2023; 28(14):5603. https://doi.org/10.3390/molecules28145603
Chicago/Turabian StyleGoudiaby, Ibrahima, Thérèse E. Malliavin, Eva Mocchetti, Sandrine Mathiot, Samir Acherar, Céline Frochot, Muriel Barberi-Heyob, Benoît Guillot, Frédérique Favier, Claude Didierjean, and et al. 2023. "New Crystal Form of Human Neuropilin-1 b1 Fragment with Six Electrostatic Mutations Complexed with KDKPPR Peptide Ligand" Molecules 28, no. 14: 5603. https://doi.org/10.3390/molecules28145603
APA StyleGoudiaby, I., Malliavin, T. E., Mocchetti, E., Mathiot, S., Acherar, S., Frochot, C., Barberi-Heyob, M., Guillot, B., Favier, F., Didierjean, C., & Jelsch, C. (2023). New Crystal Form of Human Neuropilin-1 b1 Fragment with Six Electrostatic Mutations Complexed with KDKPPR Peptide Ligand. Molecules, 28(14), 5603. https://doi.org/10.3390/molecules28145603









