Actinomycins from Soil-Inhabiting Streptomyces as Sources of Antibacterial Pigments for Silk Dyeing
Abstract
1. Introduction
2. Results
2.1. Isolation of Actinobacteria
2.2. Screening for Pigment Production and Antibacterial Activity
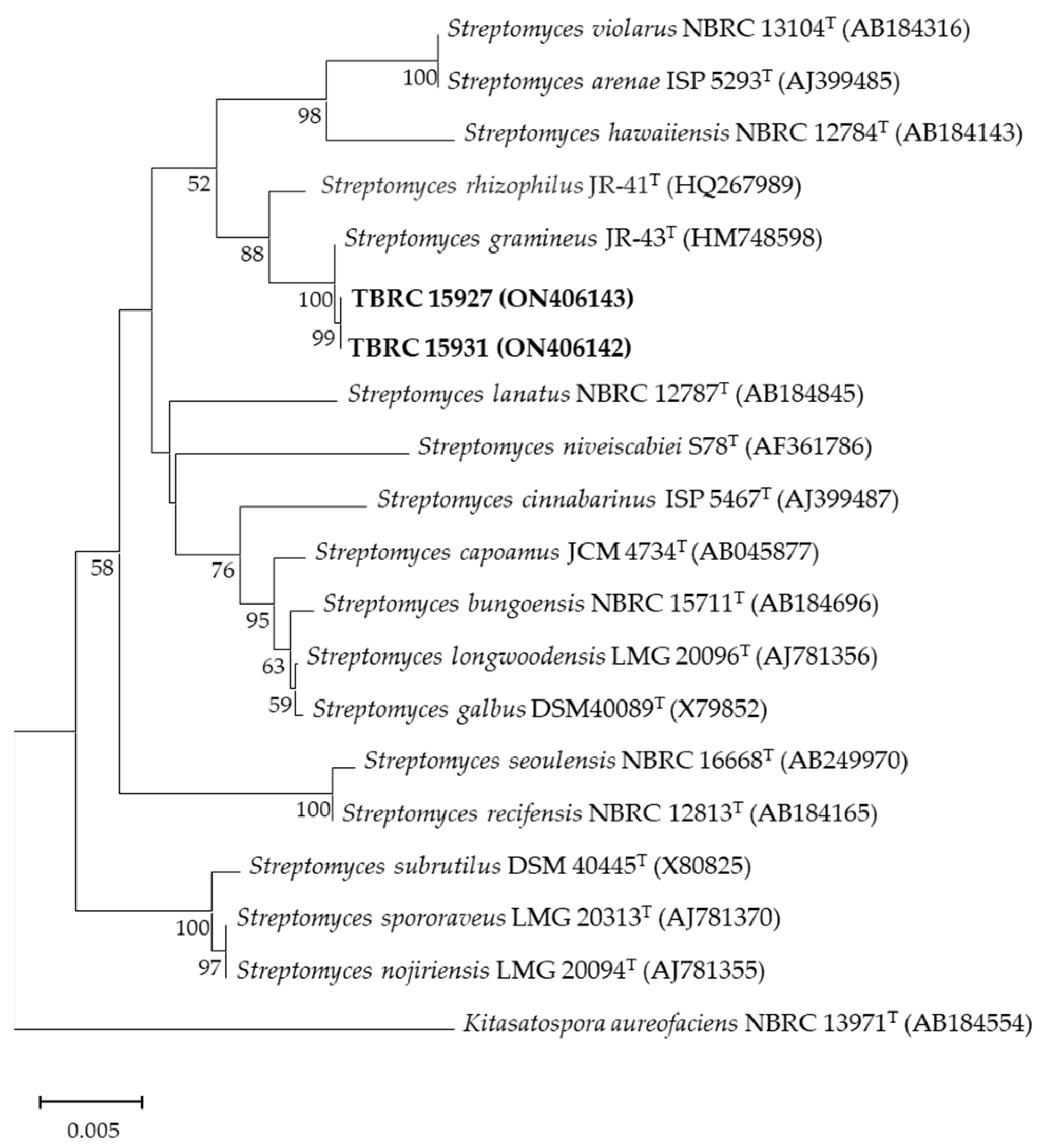
2.3. Identification of Actinobacteria and Phylogenetic Analysis
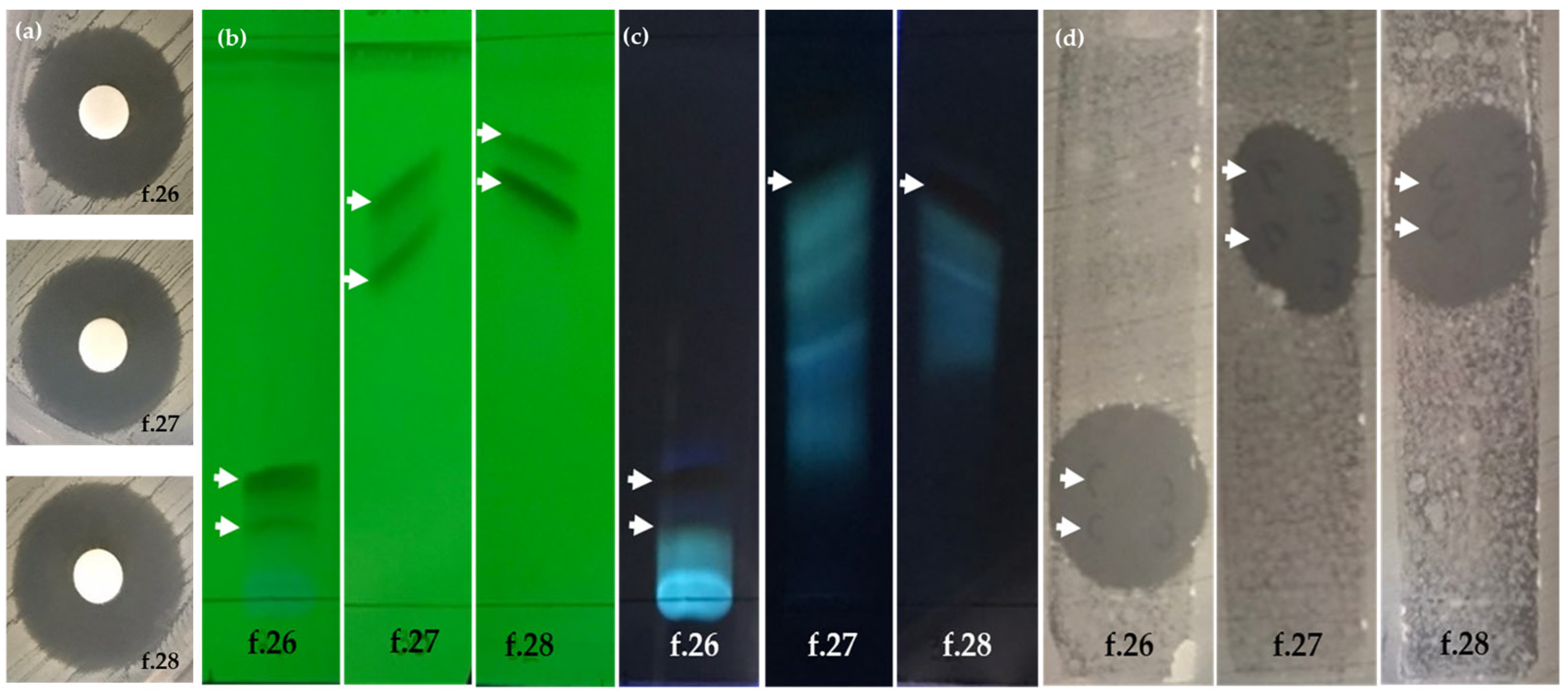
2.4. Separation and Bioassay-Guided Fractionation
2.4.1. Paper-Disc-Diffusion Assay
2.4.2. TLC-Bioautographic Assay
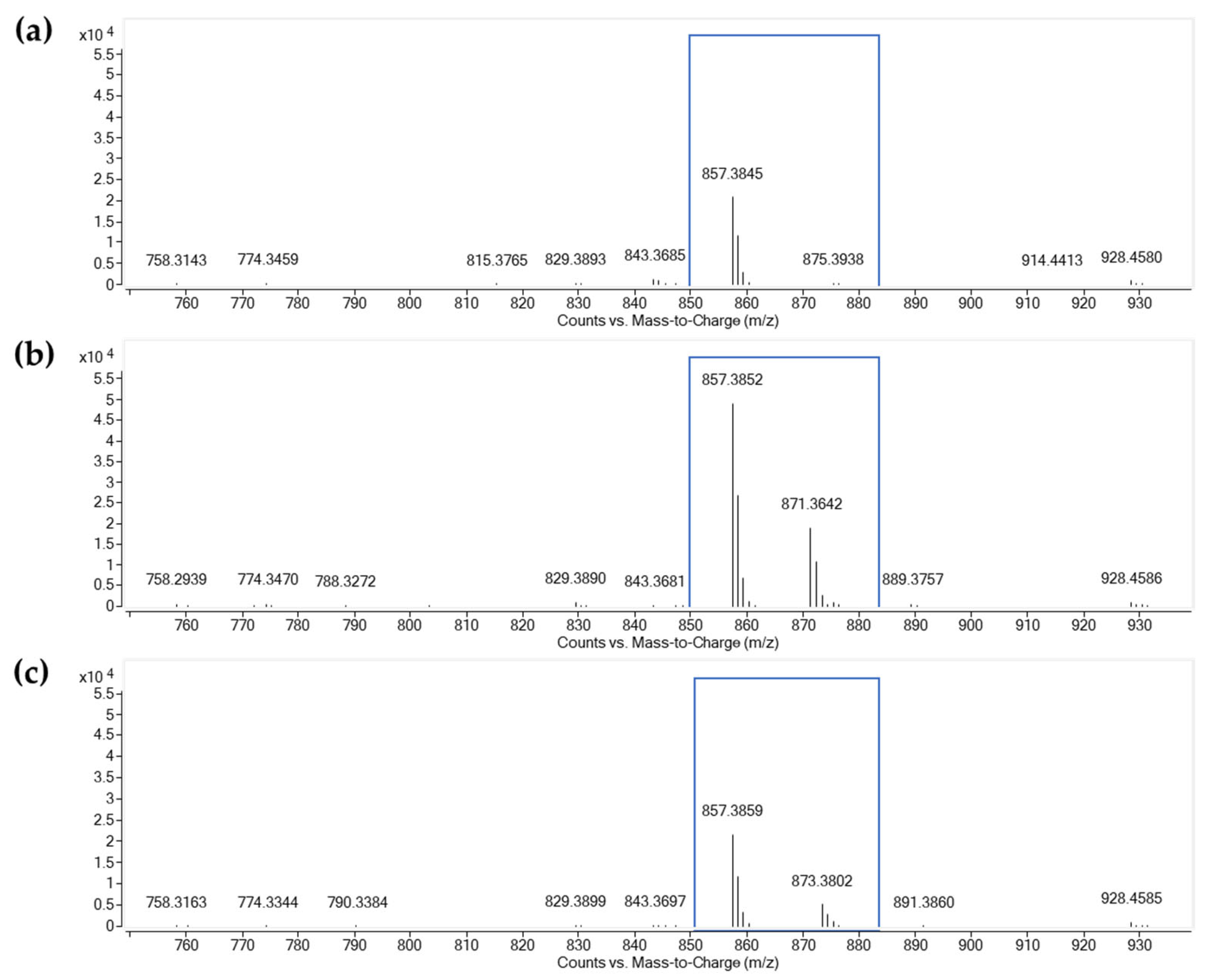
2.5. Detection of Actinomycins from Streptomyces gramineus TBRC 15927
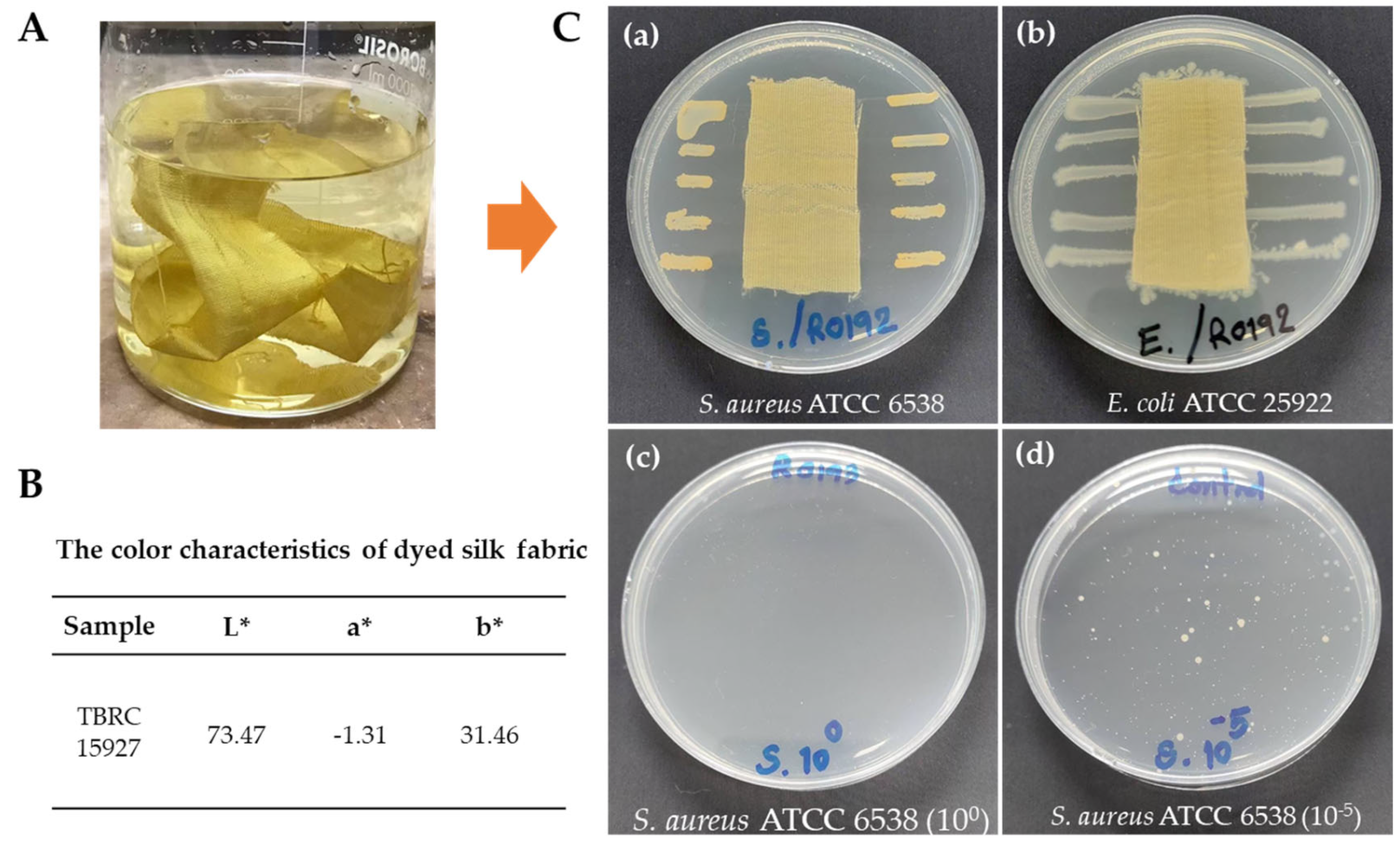
2.6. Color Characteristics
2.7. Antibacterial Activity of Dyed Silk Fabric
2.8. Cytotoxicity Assay
3. Discussion
4. Materials and Methods
4.1. Soil Sampling and Isolation of Actinobacteria
4.2. Screening for Pigment Production and Antibacterial Activity of Pigments
4.2.1. Solid-State Fermentation
4.2.2. Production of Ethyl Acetate Crude Extract
4.2.3. Preparation of Media
4.2.4. Test Microorganisms
4.2.5. Bioassay for Antibacterial Activity
4.3. Identification of Actinobacteria and Phylogenetic Analysis
4.4. Pigment Production and Extraction
4.5. Separation
4.6. Bioassay-Guided Fractionation
4.6.1. Paper-Disc-Diffusion Method
4.6.2. TLC-Bioautographic Assay
4.7. LC-ESI-Q-TOF-MS/MS
4.8. Color Characteristics and Determination of Antibacterial Activity on Dyed Silk Fabric
4.8.1. Color Characteristics
4.8.2. Determination of Antibacterial Activity on Dyed Silk Fabric
4.9. Cytotoxicity Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Saxena, S.; Raja, A.S.M. Natural dyes: Sources, chemistry, application and sustainability issues. In Roadmap to Sustainable Textiles and Clothing: Eco-Friendly Raw Materials, Technologies, and Processing Methods; Springer: Singapore, 2014; pp. 37–80. [Google Scholar] [CrossRef]
- Mohammad Azmin, S.N.H.; Sulaiman, N.S.; Mat Nor, M.S.; Abdullah, P.S.; Abdul Kari, Z.; Pati, S. A review on recent advances on natural plant pigments in foods: Functions, extraction, importance, and challenges. Appl. Biochem. Biotechnol. 2022, 194, 4655–4672. [Google Scholar] [CrossRef]
- Sarmiento-Tovar, A.A.; Silva, L.; Sánchez-Suárez, J.; Diaz, L. Streptomyces derived bioactive pigments: Ecofriendly source of bioactive compounds. Coatings 2022, 12, 1858. [Google Scholar] [CrossRef]
- Law, J.W.F.; Letchumanan, V.; Tan, L.T.H.; Ser, H.L.; Goh, B.H.; Lee, L.H. The rising of “modern actinobacteria” era. Prog. Microbes Mol. Biol. 2020, 3, 1–6. [Google Scholar] [CrossRef][Green Version]
- Bibb, M.J. Understanding and manipulating antibiotic production in actinomycetes. Biochem. Soc. Trans. 2013, 41, 1355–1364. [Google Scholar] [CrossRef]
- Dave, A.; Ingle, S. Potential of Streptomyces and its secondary metabolites for biocontrol of fungal plant pathogens. In Antifungal Metabolites of Rhizobacteria for Sustainable Agriculture; Fungal Biology; Sayyed, R., Singh, A., Ilyas, N., Eds.; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Wynn-Williams, D.D.; Edwards, H.G.M.; Newton, E.M.; Holder, J.M. Pigmentation as a survival strategy for ancient and modern photosynthetic microbes under high ultraviolet stress on planetary surfaces. Int. J. Astrobiol. 2002, 1, 39–49. [Google Scholar] [CrossRef]
- Suresh, M.; Renugadevi, B.; Brammavidhya, S.; Iyapparaj, P.; Anantharaman, P. Antibacterial activity of red pigment produced by Halolactibacillus alkaliphilus MSRD1-an isolate from seaweed. Appl. Biochem. Biotechnol. 2015, 176, 185–195. [Google Scholar] [CrossRef]
- Djemouai, N.; Meklat, A.; Gaceb-Terrak, R.; Youcef, K.O.H.; Nacer, A.; Saadi, S.A.; Saad, S.; Verheecke-Vaessen, C.; Bouras, N. Streptomyces species from the rhizosphere of the medicinal plant Artemisia herba-alba Asso: Screening for biological activities biological activities. Biologia 2022, 77, 2281–2299. [Google Scholar] [CrossRef]
- Peng, F.; Zhang, M.Y.; Hou, S.Y.; Chen, J.; Wu, Y.Y.; Zhang, Y.X. Insights into Streptomyces spp. isolated from the rhizospheric soil of Panax notoginseng: Isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptides. BMC Microbiol. 2020, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.; Zou, L.; Zhou, D.; Chen, Y.; Gao, Z.; Feng, R.; Zhang, M.; Li, K.; Xie, J.; Wang, W. Taxonomy and Broad-Spectrum Antifungal Activity of Streptomyces sp. SCA3-4 Isolated from Rhizosphere Soil of Opuntia stricta. Front. Microbiol. 2019, 10, 1390. [Google Scholar] [CrossRef]
- Janković, V.; Marković, D.; Nikodinovic-Runic, J.; Radetić, M.; Ilic-Tomic, T. Eco-friendly dyeing of polyamide and polyamide-elastane knits with living bacterial cultures of two Streptomyces sp. strains. World J. Microbiol. Biotechnol. 2023, 39, 32. [Google Scholar] [CrossRef]
- Bystrykh, L.V.; Fernández-Moreno, M.A.; Herrema, J.K.; Malpartida, F.; Hopwood, D.A.; Dijkhuizen, L. Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3 (2). J. Bacteriol. 1996, 178, 2238–2244. [Google Scholar] [CrossRef] [PubMed]
- Kramar, A.; Ilic-Tomic, T.; Petkovic, M.; Radulovic, N.; Kostic, M.; Jocic, D.; Nikodinovic-Runic, J. Crude bacterial extracts of two new Streptomyces sp. isolates as bio-colorants for textile dyeing. World J. Microbiol. Biotechnol. 2014, 30, 2231–2240. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.A.; El-Ewasy, S.M. Bioproduction, characterization, anticancer and antioxidant activities of extracellular melanin pigment produced by newly isolated microbial cell factories Streptomyces glaucescens NEAE-H. Sci. Rep. 2017, 7, 421919. [Google Scholar] [CrossRef]
- Jumpathong, J.; Nuengchamnong, N.; Masin, K.; Nakaew, N.; Suphrom, N. Thin layer chromatography-TLC-bioautographic assay for antibacterial compounds from Streptomyces sp. TBRC 8912, a newly isolated actinomycin D producer. Chiang Mai J. Sci. 2019, 46, 839–849. [Google Scholar]
- Charoenwiwattanakij, P.; Pratuangdejkul, J.; Chongruchiroj, S.; Suwanborirux, K.; Thawai, C.; Chingunpitak, J.; Satitpatipan, V. Effect of actinomycin D isolated from the cultured broth of marine Streptomyces spp. on cell division protein FtsZ. Chiang Mai J. Sci. 2020, 47, 362–377. [Google Scholar]
- Kanchanasin, P.; Saeng-in, P.; Nakashima, T.; Matsuo, H.; Takahashi, Y.; Tanasupawat, S. Actinomycins produced by Streptomyces lichenis LCR6-01T and its antibacterial activity. Chiang Mai J. Sci. 2020, 47, 864–871. [Google Scholar]
- Qureshi, K.A.; Bholay, A.D.; Rai, P.K.; Mohammed, H.A.; Khan, R.A.; Azam, F.; Jaremko, M.; Emwas, A.-H.; Stefanowicz, P.; Waliczek, M.; et al. Isolation, characterization, anti-MRSA evaluation, and in-silico multi-target anti-microbial validations of actinomycin X2 and actinomycin D produced by novel Streptomyces smyrnaeus UKAQ_23. Sci. Rep. 2021, 11, 14539. [Google Scholar] [CrossRef]
- Chen, W.; Ye, K.; Zhu, X.; Zhang, H.; Si, R.; Chen, J.; Chen, Z.; Song, K.; Yu, Z.; Han, B. Actinomycin X2, an antimicrobial depsipeptide from marine-derived Streptomyces cyaneofuscatus applied as a good natural dye for silk fabric. Mar. Drugs 2021, 20, 16. [Google Scholar] [CrossRef]
- Shah, A.M.; Hussain, A.; Mushtaq, S.; Rather, M.A.; Shah, A.; Ahmad, Z.; Khan, I.A.; Bhat, K.A.; Hassan, Q.P. Antimicrobial investigation of selected soil actinomycetes isolated from unexplored regions of Kashmir Himalayas, India. Microb. Pathog. 2017, 110, 93–99. [Google Scholar] [CrossRef]
- Dong, M.; Cao, P.; Ma, Y.T.; Luo, J.; Yan, Y.; Li, R.T.; Huang, S.X. A new actinomycin Z analogue with an additional oxygen bridge between chromophore and β-depsipentapeptide from Streptomyces sp. KIB-H714. Nat. Prod. Res. 2019, 33, 219–225. [Google Scholar] [CrossRef]
- Sharma, M.; Manhas, R.K. Purification and characterization of actinomycins from Streptomyces strain M7 active against methicillin resistant Staphylococcus aureus and vancomycin resistant Enterococcus. BMC Microbiol. 2019, 19, 44. [Google Scholar] [CrossRef]
- Adlin Jenifer, J.S.C.; Michaelbabu, M.; Eswaramoorthy Thirumalaikumar, C.L.; Jeraldin Nisha, S.R.; Uma, G.; Citarasu, T. Antimicrobial potential of haloalkaliphilic Nocardiopsis sp. AJ1 isolated from solar salterns in India. J. Basic Microbiol. 2019, 59, 288–301. [Google Scholar] [CrossRef]
- Waksman, S.A.; Woodruff, H.B. Bacteriostatic and bactericidal substances produced by a soil Actinomyces. Proc. Soc. Exp. Biol. Med. 1940, 45, 609–614. [Google Scholar] [CrossRef]
- Singh, S.B.; Genilloud, O.; Peláez, F. Terrestrial microorganisms–Filamentous bacteria. Compr. Nat. Prod. II 2010, 2, 109–140. [Google Scholar] [CrossRef]
- Farber, S.; D’Angio, G.; Evans, A.; Mitus, A. Clinical studies on actinomycin D with special reference to Wilms’ tumor in children. Ann. N. Y. Acad. Sci. 1960, 89, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Bensaude, O. Inhibiting eukaryotic transcription. Which compound to choose? How to evaluate its activity? Which compound to choose? How to evaluate its activity? Transcription 2011, 2, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Somphong, A.; Poengsungnoen, V.; Buaruang, K.; Sripreechasak, P.; Khantasup, K.; Intaraudom, C.; Pittayakhajonwut, P.; Tanasupawat, S.; Phongsopitanun, W. The lichen-derived Streptomyces isolated from Pyxine cocoes produces the antibiotic with potent antimicrobial and antitumor activities. ScienceAsia 2023, 49, 328. [Google Scholar] [CrossRef]
- Chen, C.; Song, F.; Wang, Q.; Abdel-Mageed, W.M.; Guo, H.; Fu, C.; Hou, W.; Dai, H.; Liu, X.; Yang, N.; et al. A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-tuberculosis activity. Appl. Microbiol. Biotechnol. 2012, 95, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Machushynets, N.V.; Elsayed, S.S.; Du, C.; Siegler, M.A.; de la Cruz, M.; Genilloud, O.; Hankemeier, T.; van Wezel, G.P. Discovery of actinomycin L, a new member of the actinomycin family of antibiotics. Sci. Rep. 2022, 12, 2813. [Google Scholar] [CrossRef]
- Zhou, W.; Xie, Z.; Si, R.; Chen, Z.; Javeed, A.; Li, J.; Wu, Y.; Han, B. Actinomycin-X2-immobilized silk fibroin film with enhanced antimicrobial and wound healing activities. Int. J. Mol. Sci. 2023, 24, 6269. [Google Scholar] [CrossRef]
- Ahn, S.-Y.; Jang, S.; Sudheer, P.D.V.N.; Choi, K.-Y. Microbial Production of Melanin Pigments from Caffeic Acid and L-Tyrosine Using Streptomyces glaucescens and FCS-ECH-Expressing Escherichia coli. Int. J. Mol. Sci. 2021, 22, 2413. [Google Scholar] [CrossRef] [PubMed]
- Zothanpuia, P.; Passari, A.; Singh, B.P. Molecular characterization of actinomycetes isolated from Tuichang river and their biosynthetic potential. Sci. Vis. 2015, 15, 136–144. [Google Scholar]
- Geetanjali; Jain, P. Antibiotic production by rhizospheric soil microflora-a review. Int. J. Pharm. Sci. Res. 2016, 7, 4304–4314. [Google Scholar] [CrossRef]
- Wei, Y.; Zhao, Y.; Zhou, D.; Qi, D.; Li, K.; Tang, W.; Chen, Y.; Jing, T.; Zang, X.; Xie, J.; et al. A newly isolated Streptomyces sp. YYS-7 with a broad-spectrum antifungal activity improves the banana plant resistance to Fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 2020, 11, 1712. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Isolation and characterization of a new endophytic actinobacterium Streptomyces californicus strain ADR1 as a promising source of anti-bacterial, anti-biofilm and antioxidant metabolites. Microorganisms 2020, 8, 929. [Google Scholar] [CrossRef]
- Mohseni, M.; Norouzi, H.; Hamedi, J.; Roohi, A. Screening of antibacterial producing actinomycetes from sediments of the Caspian Sea. Int. J. Mol. Cell. Med. 2013, 2, 64. [Google Scholar]
- Robinson, T.; Singh, D.; Nigam, P. Fermentación en estado sólido: Una tecnología microbiana promisoria para la producción de metabolitos secundarios. Vitae 2002, 9, 27–36. [Google Scholar] [CrossRef]
- Lee, H.J.; Han, S.I.; Whang, K.S. Streptomyces gramineus sp. nov., an antibiotic-producing actinobacterium isolated from bamboo (Sasa borealis) rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 4, 856–859. [Google Scholar] [CrossRef]
- Brockmann, H.; Pini, H. Actinorhodin, ein roter Farbstoff aus Actinomyceten. Naturwissenschaften 1947, 34, 190. [Google Scholar] [CrossRef]
- Hemeda, N.A.; Hegazy, G.E.; Abdelgalil, S.A.; Soliman, N.A.; Abdel-Meguid, D.I.; El-Assar, S.A. Maximization of red pigment production from Streptomyces sp. LS1 structure elucidation and application as antimicrobial/antifouling against human pathogens and marine microbes. J. Genet. Eng. Biotechnol. 2022, 20, 168. [Google Scholar] [CrossRef]
- Mitchell, A.; Spencer, M.; Edmiston, C., Jr. Role of healthcare apparel and other healthcare textiles in the transmission of pathogens: A review of the literature. J. Hosp. Infect. 2015, 90, 285–292. [Google Scholar] [CrossRef]
- Colclasure, V.J.; Soderquist, T.J.; Lynch, T.; Schubert, N.; McCormick, D.S.; Urrutia, E.; Knickerbocker, C.; McCord, D.; Kavouras, J.H. Coliform bacteria, fabrics, and the environment. Am. J. Infect. Control. 2015, 43, 154–158. [Google Scholar] [CrossRef]
- Li, N.; Wang, Q.; Zhou, J.; Li, S.; Liu, J.; Chen, H. Insight into the progress on natural dyes: Sources, structural features, health effects, challenges, and potential. Molecules 2022, 27, 3291. [Google Scholar] [CrossRef]
- Kramar, A.; Kostic, M.M. Bacterial secondary metabolites as biopigments for textile dyeing. Textiles 2022, 2, 252–264. [Google Scholar] [CrossRef]
- Ellaiah, P.; Srinivasulu, B.; Adinarayana, K. Optimisation studies on neomycin production by a mutant strain of Streptomyces marinensis in solid state fermentation. Process Biochem. 2004, 39, 529–534. [Google Scholar] [CrossRef]
- Liu, H.; Du, X.; Yuan, Q.; Zhu, L. Optimisation of enzyme assisted extraction of silybin from the seeds of Silybum marianum by Box–Behnken experimental design. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 2009, 20, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the actinomycin D biosynthetic pathway from marine-derived Streptomyces costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. [Google Scholar] [CrossRef]
- Wang, D.; Wang, C.; Gui, P.; Liu, H.; Khalaf, S.M.; Elsayed, E.A.; Wadaan, M.A.M.; Hozzein, W.N.; Zhu, W. Identification, bioactivity, and productivity of actinomycins from the marine-derived Streptomyces heliomycini. Front. Microbiol. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Wang, R.; Wang, Z.; Yang, B.; Kuang, H. An evolving technology that integrates classical methods with continuous technological developments: Thin-layer chromatography bioautography. Molecules 2021, 26, 4647. [Google Scholar] [CrossRef] [PubMed]
- Chandrakar, S.; Gupta, A.K. Actinomycin-producing endophytic Streptomyces parvulus associated with root of Aloe vera and optimization of conditions for antibiotic production. Probiotics Antimicrob. Proteins 2019, 11, 1055–1069. [Google Scholar] [CrossRef]
- Kirk, J.M. The mode of action of actinomycin D. Biochim. Biophys. Acta 1960, 42, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.G.; Lee, K.; Kim, C.J.; Park, D.J.; Ju, Y.; Lee, J.C.; Wood, T.K.; Lee, J. Streptomyces-derived actinomycin D inhibits biofilm formation by Staphylococcus aureus and its hemolytic activity. Biofouling 2016, 32, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.Q.; Xie, T.T.; Zeng, H.; Chen, W.; Wan, C.X.; Zhang, L.L. Streptomyces-derived actinomycin D inhibits biofilm formation via downregulating ica locus and decreasing production of PIA in Staphylococcus epidermidis. J. Appl. Microbiol. 2020, 128, 1201–1207. [Google Scholar] [CrossRef]
- Ramirez-Rodriguez, L.; Stepanian-Martinez, B.; Morales-Gonzalez, M.; Diaz, L. Optimization of the cytotoxic activity of three Streptomyces strains isolated from Guaviare river sediments (Colombia, South America). BioMed Res. Int. 2018, 2018, 2839356. [Google Scholar] [CrossRef]
- Benbelkhir, F.Z.; Medjekal, S. Microalgal carotenoids: A promising alternative to synthetic dyes. Algal Res. 2022, 66, 102823. [Google Scholar] [CrossRef]
- Venil, C.K.; Velmurugan, P.; Dufossé, L.; Devi, P.R.; Ravi, A.V. Fungal pigments: Potential coloring compounds for wide ranging applications in textile dyeing. J. Fungi 2020, 6, 68. [Google Scholar] [CrossRef]
- Hamaki, T.; Suzuki, M.; Fudou, R.; Jojima, Y.; Kajiura, T.; Tabuchi, A.; Sen, K.; Shibai, H. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J. Biosci. Bioeng. 2005, 5, 485–492. [Google Scholar] [CrossRef]
- Lee, L.H.; Zainai, N.; Azman, A.S.; End, S.K.; Goh, B.H.; Yin, W.F.; Mutalib, N.S.; Chan, K.G. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014, 2014, 698178. [Google Scholar] [CrossRef] [PubMed]
- Trusheva, B.; Trunkova, D.; Bankova, V. Different extraction methods of biologically active components from propolis: A preliminary study. Chem. Cent. J. 2007, 1, 13. [Google Scholar] [CrossRef]
- Lorian, V. Laboratory methods used to assess the activity of antimicrobial combinations. In Antibiotics in Laboratory Medicine, 3rd ed.; Lorian, V., Ed.; Williams & Wilkins Co.: Baltimore, MD, USA, 1991; pp. 434–444. [Google Scholar]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 55–63. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Jeyaseelan, E.C.; Jenothiny, S.; Pathmanathan, M.K.; Jeyadevan, J.P. Antibacterial activity of sequentially extracted organic solvent extracts of fruits, flowers and leaves of Lawsonia inermis L. from Jaffna. Asian Pac. J. Trop. Biomed. 2012, 2, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Suphrom, N.; Daowtak, K.; Potup, P.; Thongsri, Y.; Usuwanthim, K. Anticancer Effect of Citrus hystrix DC. Leaf Extract and Its Bioactive Constituents Citronellol and, Citronellal on the Triple Negative Breast Cancer MDA-MB-231 Cell Line. Pharmaceuticals 2020, 13, 476. [Google Scholar] [CrossRef]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. TLC-bioautographic assay and its scope in the field of natural product chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef]
- Yamaç, M.; Bilgili, F. Antimicrobial activities of fruit bodies and/or mycelial cultures of some mushroom isolates. Pharm. Biol. 2006, 44, 660–667. [Google Scholar] [CrossRef]
- Liu, C.W.; Lu, Y.Y.; Yang, Z.Z.; Xing, Y.Y.; Xi, T. Rapid screening and characterization of metabolites from a marine-derived actinomycete by high-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 3413–3418. [Google Scholar] [CrossRef]
- AATCC TM 147-2011; Parallel Streak: Assess. AATCC (American Association of Textile Chemists and Colorists): Research Triangle Park, NC, USA, 2011.
- AATCC TM 100-2019; Test Method for Antibacterial Finishes on Textile Materials: Assess. AATCC (American Association of Textile Chemists and Colorists): Research Triangle Park, NC, USA, 2019.
- Sriwiriyajan, S.; Ninpesh, T.; Sukpondma, Y.; Nasomyon, T.; Graidist, P. Cytotoxicity screening of plants of genus Piper in breast cancer cell lines. Trop. J. Pharm. Res. 2014, 13, 921–928. [Google Scholar] [CrossRef]

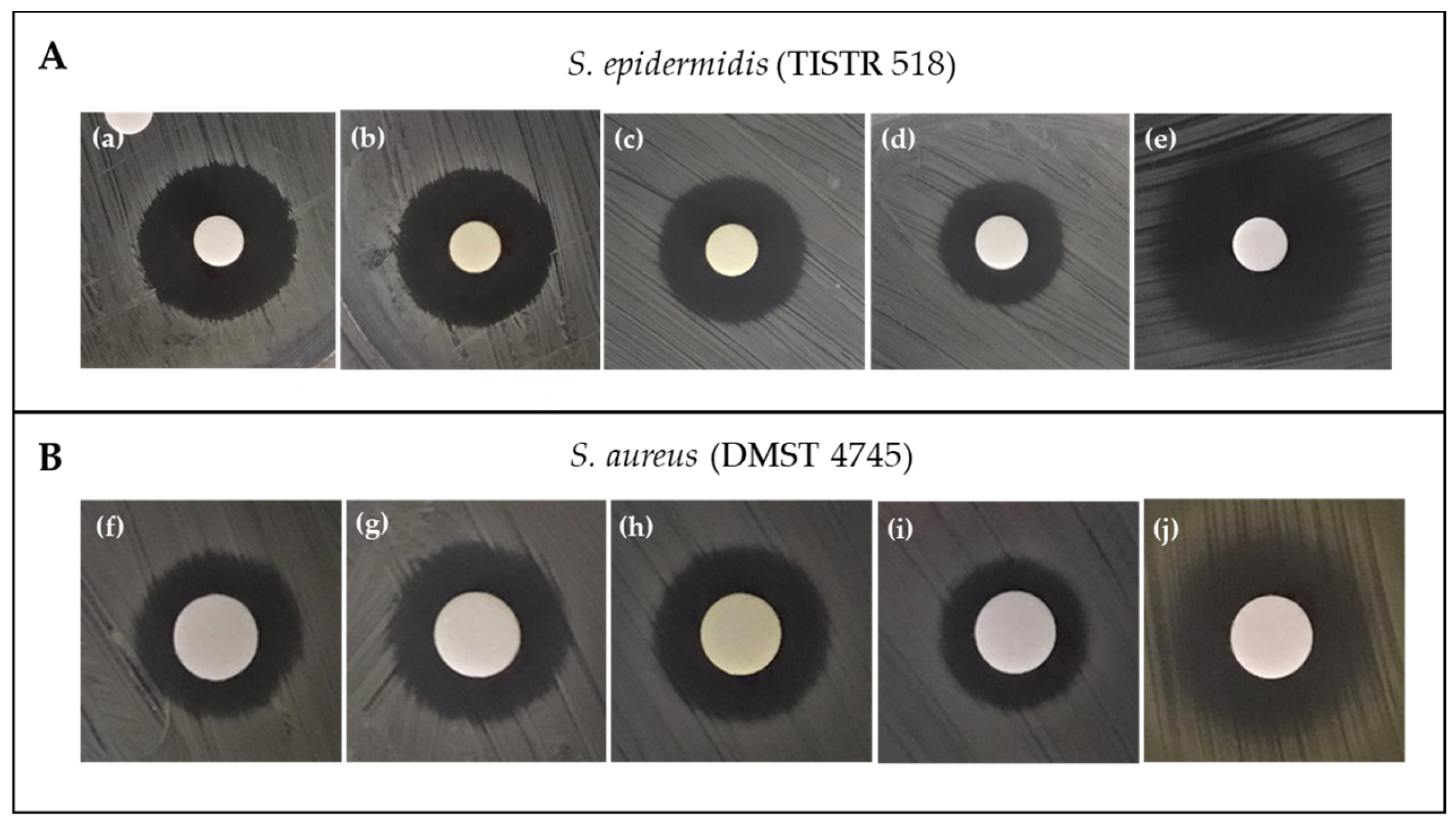
| Diameter of Inhibition Zone (mm) § | ||||||||
|---|---|---|---|---|---|---|---|---|
| Streptomyces Crude Pigment | S. epidermidis (TISTR 518) | S. aureus (DMST 4745) | ||||||
| Ethyl Acetate | Methanol | 95% Ethanol | 70% Ethanol | Ethyl Acetate | Methanol | 95% Ethanol | 70% Ethanol | |
| TBRC 15924 | - | - | 5.90 ± 0.44 d | - | - | - | - | - |
| TBRC 15925 | 16.50 ± 1.45 bc | - | - | - | 11.00 ± 1.32 aA | - | - | - |
| TBRC 15926 | 9.87 ± 0.32 deA | 7.30 ± 0.26 dC | 8.27 ± 0.25 cB | 6.83 ± 0.59 dC | 6.60 ± 0.36 cA | - | - | - |
| TBRC 15927 | 17.63 ± 1.48 bAB | 18.07 ± 1.01 bA | 16.20 ± 0.35 bB | 12.37 ± 0.35 cC | 11.50 ± 0.87 aAB | 11.75 ± 0.68 aAB | 12.17 ± 0.76 aA | 10.35 ± 1.02 bB |
| TBRC 15928 | 9.00 ± 0.30 ef | - | - | - | 6.30 ± 0.20 cA | - | - | - |
| TBRC 15929 | 15.77 ± 0.21 cA | 15.27 ± 0.75 cA | - | 15.50 ± 0.48 bA | 11.73 ± 0.59 aA | 11.97 ± 0.95 aA | 5.70 ± 0.17 bB | 11.47 ± 0.15 aA |
| TBRC 15930 | 8.27 ± 0.75 fA | 6.03 ± 0.21 dB | 8.03 ± 0.45 cA | 6.08 ± 0.41 dB | 6.50 ± 0.46 cA | - | 6.20 ± 0.53 bA | - |
| TBRC 15931 | 10.77 ± 0.12 d | - | - | - | 8.57 ± 0.85 bA | - | - | - |
| Control | 21.27 ± 0.58 a* | 21.27 ± 0.58 a | 21.27 ± 0.58 a | 21.27 ± 0.58 a | 12.27 ± 0.81 a | 12.27 ± 0.81 a | 12.27 ± 0.81 a | 12.27 ± 0.81 a |
| Isolate Code | NCBI Accession Numbers | Amplified 16S rRNA Gene (bps) | Closely Related Taxa and NCBI Accession Number | Similarity (%) | Chosen Nomenclature |
|---|---|---|---|---|---|
| TBRC 15924 | ON406138 | 1434 | Streptomyces cinnamoneus NBRC 12852T | 99.72 | Streptomyces sp. |
| TBRC 15925 | ON406139 | 1459 | Streptomyces shenzhenensis 172115T | 99.72 | S. shenzhenensis |
| TBRC 15926 | ON406140 | 1447 | Streptomyces aquilus GGCR-6T | 99.64 | Streptomyces sp. |
| TBRC 15927 | ON406141 | 1433 | Streptomyces gramineus JR-43T | 100.0 | S. gramineus |
| TBRC 15928 | ON406142 | 1366 | Streptomyces netropsis NBRC 3723T | 99.19 | Streptomyces sp. |
| TBRC 15929 | ON406143 | 1451 | Streptomyces adustus WH-9T | 99.93 | S. adustus |
| TBRC 15930 | ON406144 | 1436 | Streptomyces aquilus GGCR-6T | 99.64 | Streptomyces sp. |
| TBRC 15931 | ON406145 | 1471 | Streptomyces gramineus JR-43T | 100.0 | S. gramineus |
| Sample | Diameter of Inhibition Zone (mm) ¥ | |
|---|---|---|
| 24 h | 48 h | |
| Fraction 22 | 15.50 ± 0.78 e | 16.07 ± 0.12 f |
| Fraction 23 | 17.37 ± 0.35 c | 18.07 ± 0.42 c |
| Fraction 24 | 16.83 ± 0.12 cd | 17.07 ± 0.12 d |
| Fraction 25 | 16.70 ± 0.10 d | 16.77 ± 0.59 de |
| Fraction 26 | 18.23 ± 0.15 b | 18.33 ± 0.15 bc |
| Fraction 27 | 18.67 ± 0.21 b | 18.80 ± 0.17 b |
| Fraction 28 | 20.30 ± 0.30 a | 20.70 ± 0.40 a |
| Fraction 29 | 16.43 ± 0.29 d | 16.40 ± 0.10 ef |
| Fraction 30 | 15.50 ± 0.35 e | 15.37 ± 0.12 g |
| Fraction 31 | 10.83 ± 0.29 g | 9.23 ± 0.06 i |
| Chloramphenicol | 10.43 ± 0.57 g | - |
| Streptomycin | 11.90 ± 0.30 f* | 10.50 ± 0.10 h |
| Compound | tR (min) | Measurement (m/z) | Calculated (m/z) | Diff (ppm) |
|---|---|---|---|---|
| Compound I: Actinomycin D (X1) | ||||
| Fraction 26 | 28.430 | 1255.6372 | 1255.6358 | −1.15 |
| Fraction 27 | 28.294 | 1255.6385 | 1255.6358 | −2.19 |
| Fraction 28 | 28.249 | 1255.6379 | 1255.6358 | −1.71 |
| Standard Actinomycin D (X1) | 28.411 | 1255.6368 | 1255.6358 | −0.84 |
| Compound II: Actinomycin X2 | ||||
| Fraction 26 | 28.590 | 1269.6193 | 1269.6150 | −3.37 |
| Fraction 27 | 28.306 | 1269.6218 | 1269.6150 | −5.34 |
| Fraction 28 | 28.357 | 1269.6177 | 1269.6150 | −2.11 |
| Standard Actinomycin X2 | 28.383 | 1269.6173 | 1269.6150 | −1.80 |
| Compound III: Actinomycin X0β | ||||
| Fraction 27 | 28.129 | 1271.6326 | 1271.6307 | −1.52 |
| Fraction 28 | 28.167 | 1271.6332 | 1271.6307 | −1.99 |
| Sample | IC50 Values from MTT (µg/mL) |
|---|---|
| Crude extract TBRC15927 | 0.029 ± 0.008 * |
| Doxorubicin | 0.402 ± 0.040 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuanjohn, T.; Suphrom, N.; Nakaew, N.; Pathom-Aree, W.; Pensupa, N.; Siangsuepchart, A.; Dell, B.; Jumpathong, J. Actinomycins from Soil-Inhabiting Streptomyces as Sources of Antibacterial Pigments for Silk Dyeing. Molecules 2023, 28, 5949. https://doi.org/10.3390/molecules28165949
Nuanjohn T, Suphrom N, Nakaew N, Pathom-Aree W, Pensupa N, Siangsuepchart A, Dell B, Jumpathong J. Actinomycins from Soil-Inhabiting Streptomyces as Sources of Antibacterial Pigments for Silk Dyeing. Molecules. 2023; 28(16):5949. https://doi.org/10.3390/molecules28165949
Chicago/Turabian StyleNuanjohn, Tananya, Nungruthai Suphrom, Nareeluk Nakaew, Wasu Pathom-Aree, Nattha Pensupa, Apiradee Siangsuepchart, Bernard Dell, and Juangjun Jumpathong. 2023. "Actinomycins from Soil-Inhabiting Streptomyces as Sources of Antibacterial Pigments for Silk Dyeing" Molecules 28, no. 16: 5949. https://doi.org/10.3390/molecules28165949
APA StyleNuanjohn, T., Suphrom, N., Nakaew, N., Pathom-Aree, W., Pensupa, N., Siangsuepchart, A., Dell, B., & Jumpathong, J. (2023). Actinomycins from Soil-Inhabiting Streptomyces as Sources of Antibacterial Pigments for Silk Dyeing. Molecules, 28(16), 5949. https://doi.org/10.3390/molecules28165949






