Assessing Fermentation Broth Quality of Pineapple Vinegar Production with a Near-Infrared Fiber-Optic Probe Coupled with Stability Competitive Adaptive Reweighted Sampling
Abstract
:1. Introduction
2. Results and Discussion
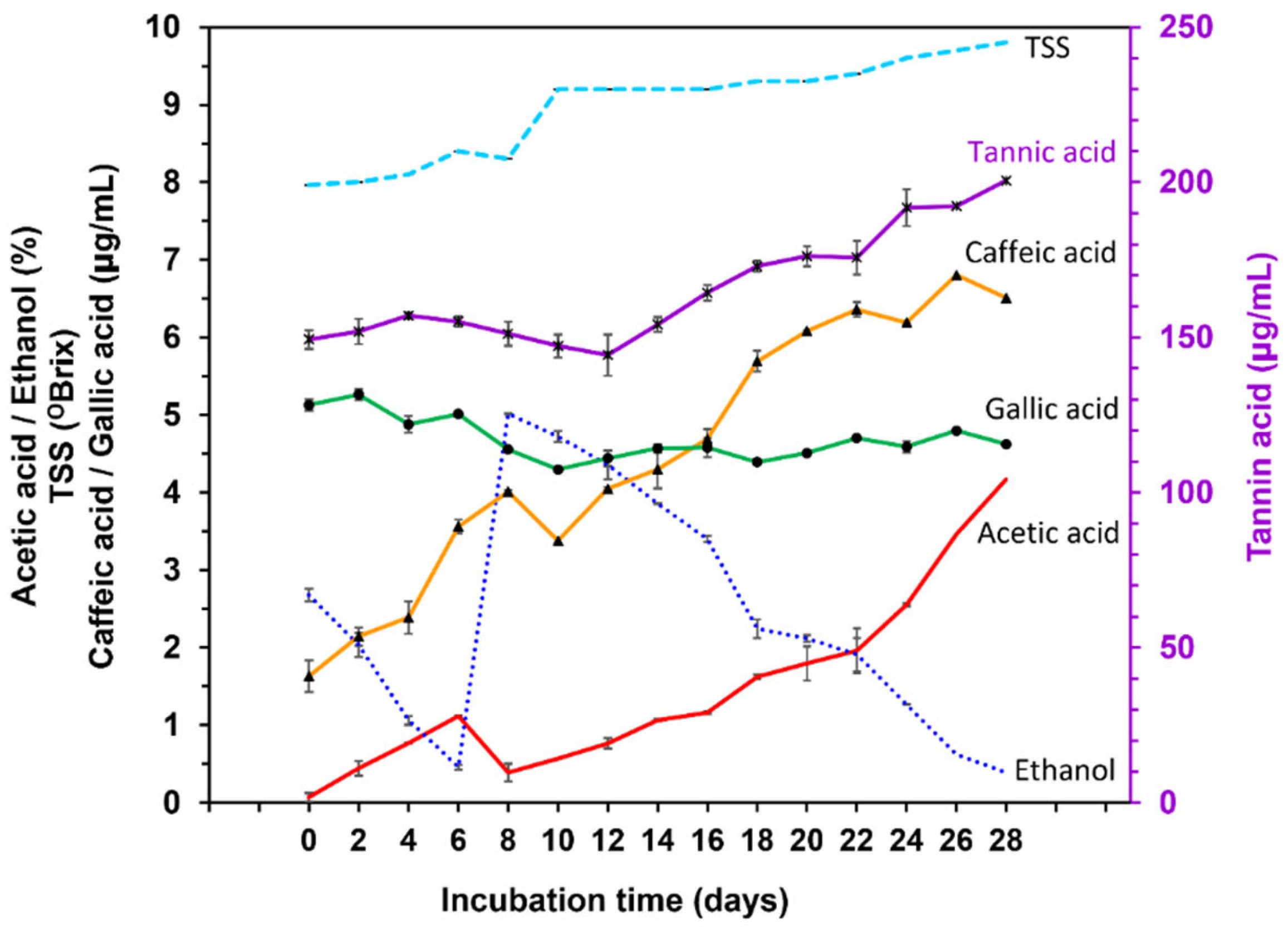
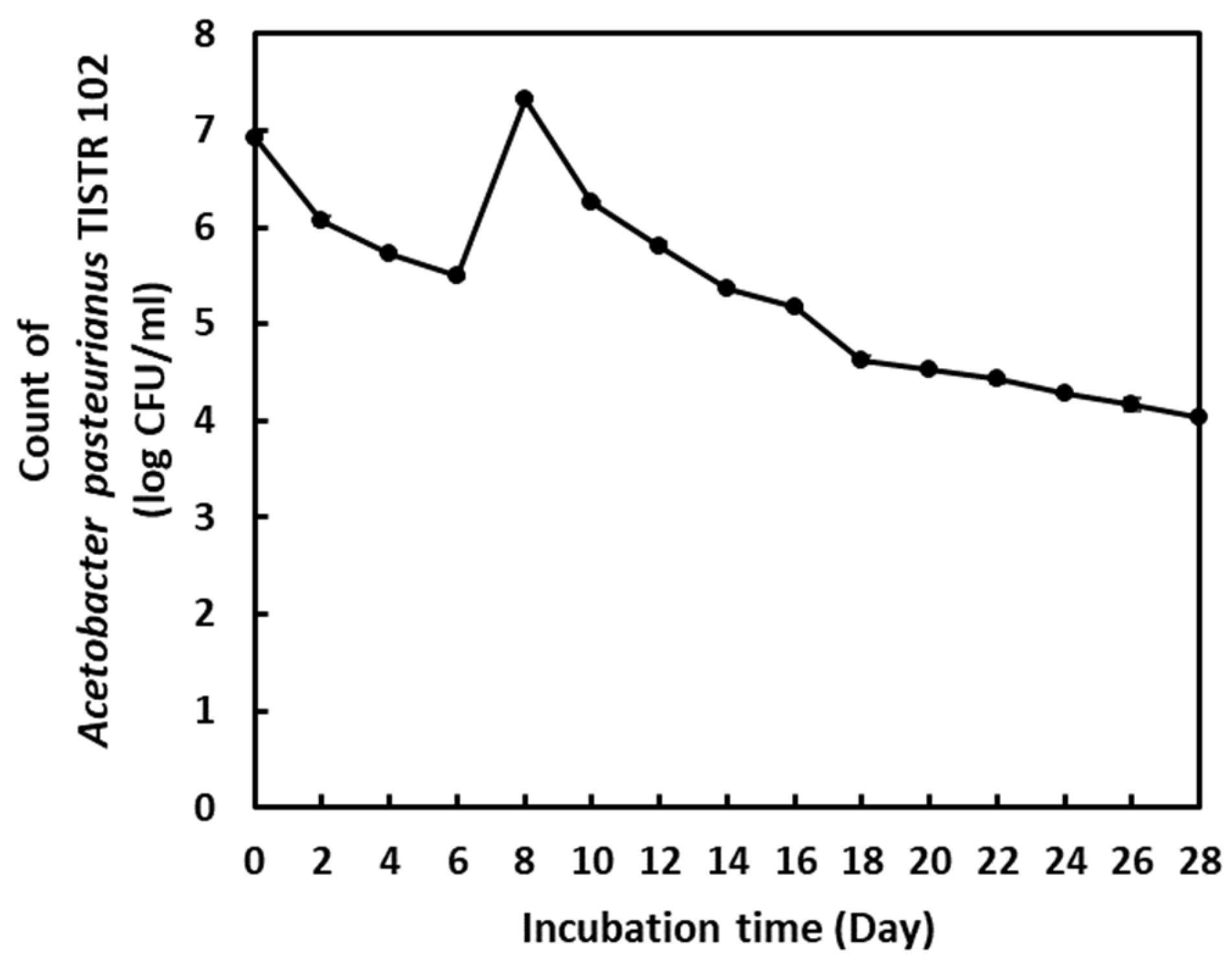
2.1. Chemical Change in Vinegar Fermentation
2.2. Statistical Parameters of Acetic Acid, Ethanol, TSS, Caffeic Acid, Gallic Acid and Tannic Acid in Calibration and Prediction Sets for NIR Analysis
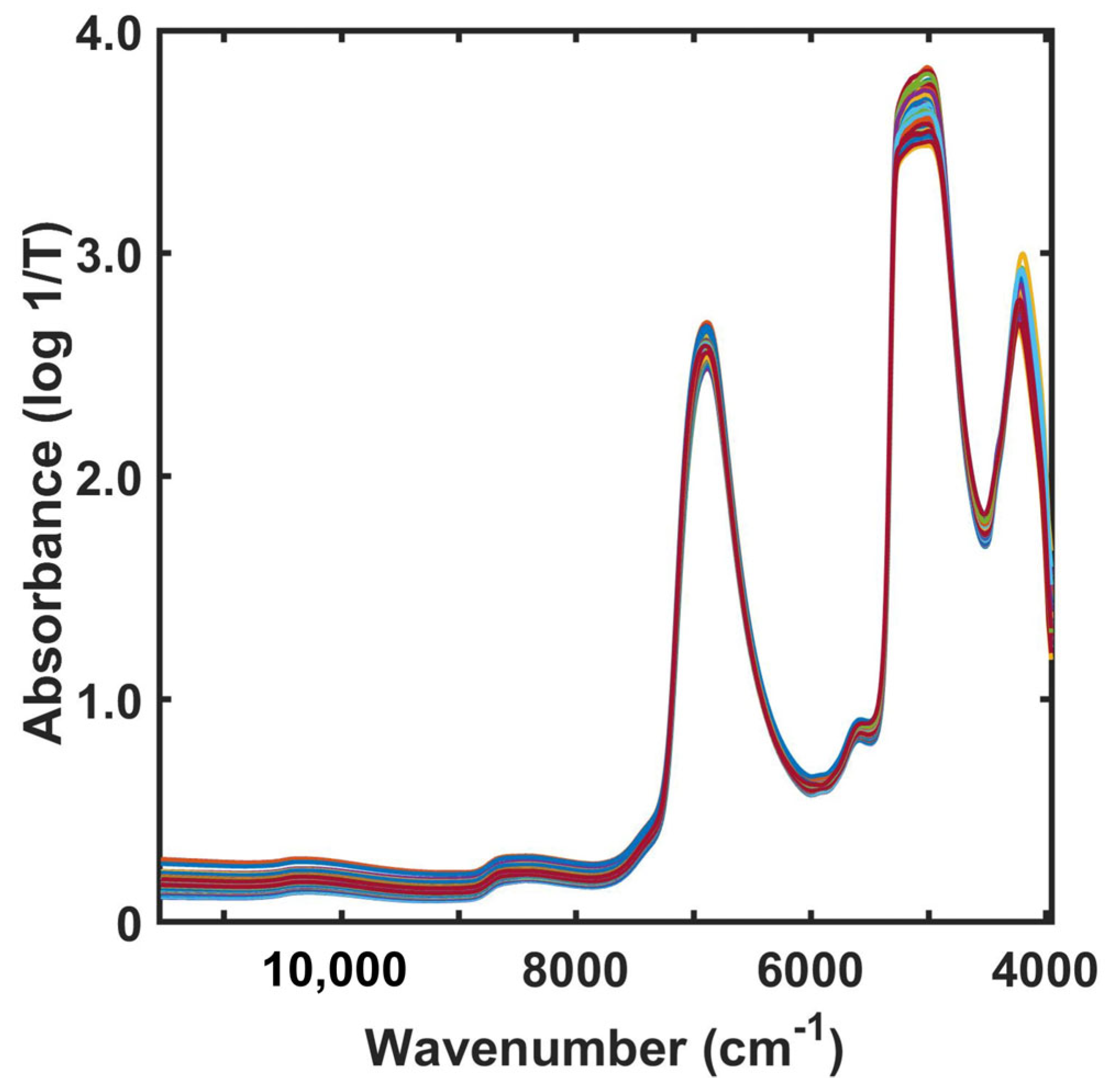
2.3. NIR Spectra of Fermentation Broth in Pineapple Vinegar Production
2.4. Comparison of PLS Models
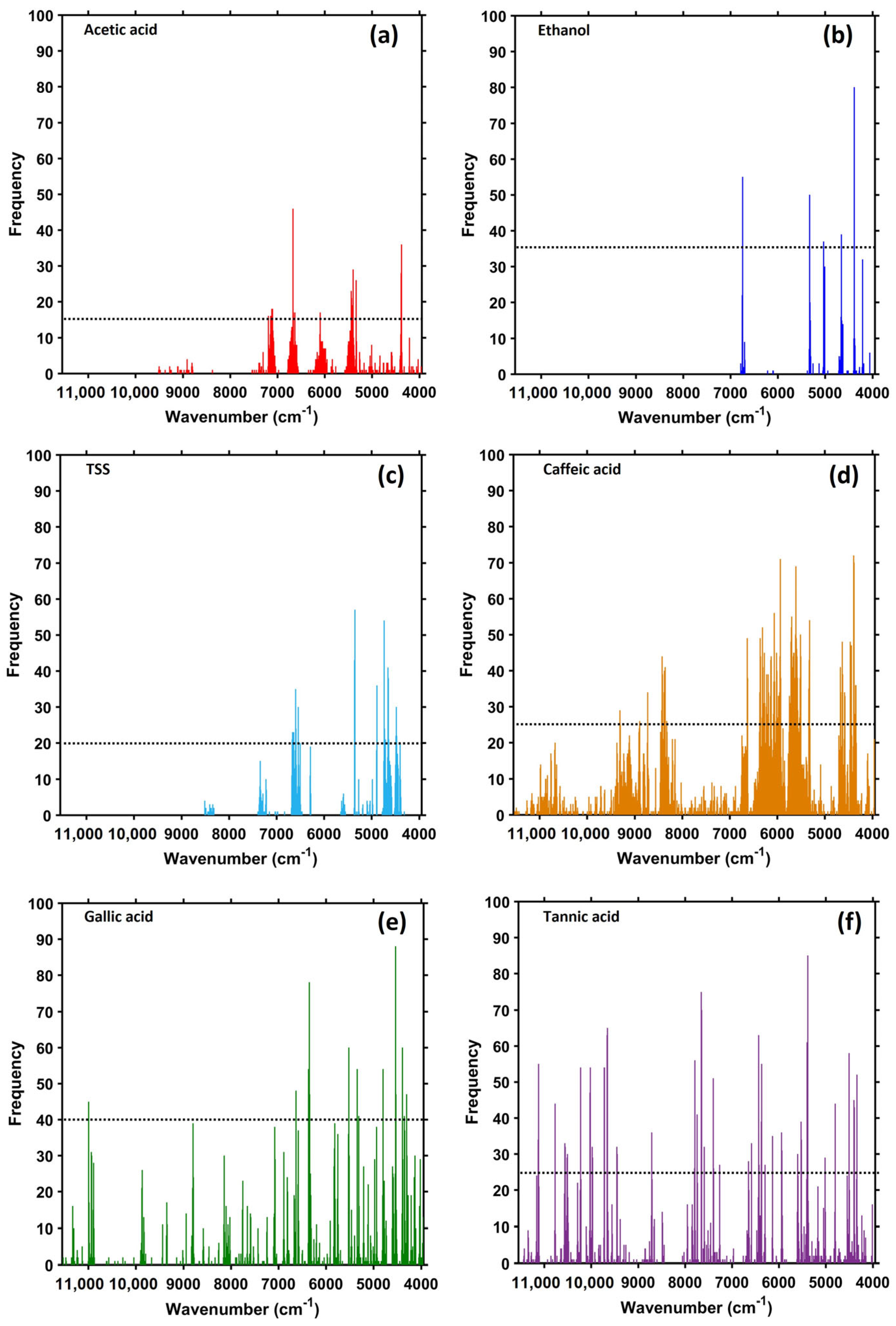
2.5. Spectral Variables Selected by SCARS
2.6. Comparison of PLS and SCARS–PLS Models
3. Materials and Methods
3.1. Sample Preparation
3.1.1. Preparation of Pineapple Juice
3.1.2. Pineapple Wine Fermentation
3.1.3. Pineapple Vinegar Fermentation
3.2. Reference Methods for Quantitative Analysis of the Target Constituents in Fermented Broth of Pineapple Vinegar
- (1)
- Analysis of acetic acid content
- (2)
- Analysis of ethanol content
- (3)
- Analysis of the TSS
- (4)
- Analysis of phenolic compounds
- (5)
- Reference method validation
3.3. NIR Fiber-Optic Probe Measurement
3.4. Calibration and Prediction Samples
3.5. Model Development by PLS
3.6. Model Development by SCARS–PLS
3.7. Model Performance Evaluation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Adams, M.R. Vinegar. In Microbiology of Fermenter; Batt, C.A., Tortorello, M.L., Eds.; Blackie Acadimic and Professional: London, UK, 2014; pp. 717–721. [Google Scholar]
- Garcia-Parrilla, M.C.; Torija, M.J.; Mas, A.; Cerezo, A.B.; Troncoso, A.M. Chapter 25—Vinegars and other fermented condiments. In In Fermented Foods in Health and Disease Prevention; Frias, J., Martinez-Villaluenga, C., Peñas, E., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 577–591. [Google Scholar]
- Kondo, S.; Tayama, K.; Tsukamoto, Y.; Nagasava, T.; Ikeda, K.; Ymori, Y. Antihypertensive effects of acetic acid and vinegar on spontaneously hypertensive rats. Biosci. Biotechnol. Biochem. 2001, 65, 2690–2694. [Google Scholar] [CrossRef]
- Entani, E.; Asai, M.; Tsujihata, S.; Tsukamoto, Y.; Ohata, M. Antibacterial action of vinegar against food-borne pathogenic bacteria including Escherichia coli O157: H7. J. Food Prot. 1998, 61, 953–959. [Google Scholar] [CrossRef]
- Ndoye, B.; Weekers, F.; Diawara, B.; Guiro, T.A.; Thonart, P. Survival and preservation after freeze-drying process of thermoresistant acetic acid bacteria isolated from tropical products of sub-Saharan Africa. J. Food Eng. 2007, 79, 1374–1382. [Google Scholar] [CrossRef]
- Newett, S.; Rigden, P. The pineapple problem solver field guide. In Tropical Fruits: Pineapples; Department of Agriculture and Fisheries, Queensland Government: Brisbane City, Australia, 2015. [Google Scholar]
- Thai Ministry of Public Health, (No. 204) B.E. 2543 (2000) Re: Vinegar. Available online: https://www.fao.org/faolex/results/details/en/c/LEX-FAOC160070 (accessed on 7 July 2023).
- Vasiljevic, T. Chapter 10 Pineapple. In Valorization of Fruit Processing By-Products; Galanakis, C.M., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 203–225. [Google Scholar]
- Cozzolino, D.; Kwiatkowski, M.J.; Parker, M.; Cynkar, W.U.; Dambergs, R.G.; Gishen, M.; Herderich, M.J. Prediction of phenolic compounds in red wine fermentations by visible and near infrared spectroscopy. Anal. Chim. Acta 2004, 513, 73–80. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, E.; Long, J.; Pan, X.; Xu, X.; Jin, Z.; Jiao, A. Comparison between ATR-IR, Raman, concatenated ATR-IR and Raman spectroscopy for the determination of total antioxidant capacity and total phenolic content of Chinese rice wine. Food Chem. 2016, 194, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Aye-Ayire Sedjoah, R.C.; Ma, Y.; Yan, H. Fast monitoring total acids and total polyphenol contents in fermentation broth of mulberry vinegar using MEMS and optical fiber near-infrared spectrometers, Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 260, 119938. [Google Scholar] [CrossRef]
- Liu, F.; He, Y.; Wang, L.; Sun, G. Detection of organic acids and pH of fruit vinegars using near-infrared spectroscopy and multivariate calibration. Food Bioproc. Technol. 2011, 4, 1331–1340. [Google Scholar] [CrossRef]
- Liu, F.; He, Y.; Wang, L. Comparison of calibrations for the determination of soluble solids content and pH of rice vinegars using visible and short-wave near infrared spectroscopy. Anal. Chim. Acta 2008, 610, 196–204. [Google Scholar] [CrossRef]
- Chen, Q.; Ding, J.; Cai, J.; Sun, Z.; Zhao, J. Simultaneous measurement of total acid content and soluble salt-free solids content in Chinese vinegar using near-infrared spectroscopy. J. Food Sci. 2012, 77, 222–227. [Google Scholar] [CrossRef]
- Phanomsophon, T.; Sirisomboon, P.; Lapcharoensuk, R.; Shrestha, B.; Krusong, W. Evaluation of acetic acid and ethanol concentration in a rice vinegar internal venturi injector bioreactor using Fourier transform near infrared spectroscopy. J. Near Infrared Spectrosc. 2019, 27, 414–421. [Google Scholar] [CrossRef]
- Saiz-Abajo, M.J.; Gonza’lez-Sa’iz, J.M.; Pizarro, C. Prediction of organic acids and other quality parameters of wine vinegar by near-infrared spectroscopy. A feasibility study. Food Chem. 2006, 99, 615–621. [Google Scholar] [CrossRef]
- Sánchez, M.T.; Márquez, R.; Torres, I.; De la Haba, M.J.; Marín, D.P.; López, M.I. Characterization of wine vinegars belonging to the vinagre de montilla-moriles protected designation of origin, using near-infrared spectroscopy. Food Anal. 2020, 13, 802–810. [Google Scholar] [CrossRef]
- Guan, B.; Zhao, J.; Jin, H.; Lin, H. The qualitative and quantitative analysis of aromatic vinegar produced during different seasons by near infrared spectroscopy. Anal. Methods 2014, 6, 9634. [Google Scholar] [CrossRef]
- Chen, Q.; Ding, J.; Cai, J.; Zhao, J. Rapid measurement of total acid content (TAC) in vinegar using near infrared spectroscopy based on efficient variables selection algorithm and nonlinear regression tools. Food Chem. 2012, 135, 590–595. [Google Scholar] [CrossRef]
- Shi, J.Y.; Zou, X.B.; Huang, X.W.; Zhao, J.W.; Li, Y.; Hao, L.; Zhang, J. Rapid detecting total acid content and classifying different types of vinegar based on near infrared spectroscopy and least-squares support vector machine. Food Chem. 2013, 138, 192–199. [Google Scholar]
- Zheng, K.; Li, Q.; Wang, J.; Geng, J.; Cao, P.; Sui, T.; Wang, X.; Du, Y. Stability competitive adaptive reweighted sampling (SCARS) and its applications to multivariate calibration of NIR spectra. Chemom. Intell. Lab. Syst. 2012, 112, 48–54. [Google Scholar] [CrossRef]
- ISO 12099:2017(E); Statistics for Performance Measurement: Animal Feeding Stuffs, Cereals and Milled Cereal Products–Guidelines for Application of Near Infrared Spectrometer. International Standard: Geneva, Switzerland, 2017; pp. 5–12.
- Saichana, N. Acetic acid bacteria: Physiology and industrial applications. Huachiew Chalermprakiet Sci. Technol. J. 2015, 1, 75–89. [Google Scholar]
- Food and Drug Administration. FDA/ORA Compliance Policy Guides, Sec. 525.825 Vinegar, Definitions: Adulteration with Vinegar Eels (CPG 7109.22). 2007. Available online: https://www.fda.gov/ora/compliance_ref/cpg/cpgfod/cpg525-825.html (accessed on 7 July 2023).
- Mohamad, N.E.; Yeap, S.K.; Lim, K.L.; Yusof, H.M.; Beh, B.K.; Tan, S.W.; Ho, W.Y.; Sharifuddin, S.A.; Jamaluddin, A.; Long, K.; et al. Antioxidant effects of pineapple vinegar in reversing of paracetamol-induced liver damage in mice. Chin. Med. 2015, 10, 3. [Google Scholar] [CrossRef]
- Chiet, C.H.; Zulkifli, R.M.; Hidayat, T.; Yaakob, H. Bioactive compounds and antioxidant activity analysis of Malaysian pineapple cultivars. In AIP Conference Proceedings. Am. Inst. Phys. 2014, 1589, 398–399. [Google Scholar]
- Margreet Olthof, R.; Katan Martijn, B.; Hollman Peter, C.H. Chlorogenic acid and caffeic acid are absorbed in humans. J. Nutr. 2001, 131, 66–71. [Google Scholar] [CrossRef]
- Roda, A.; Lucini, L.; Torchio, F.; Dordoni, R.; De Faveri, D.M.; Lambri, M. Metabolite profiling and volatiles of pineapple wine and vinegar obtained from pineapple waste. Food Chem. 2017, 229, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Antoniszyn, J.; Manley, M. Near Infrared Technology: Getting the Best out of Light; African Sun Media, Sun Press: Stellenbosch, South Africa, 2019. [Google Scholar]
- Ríos-Reina, R.; Gonzalez, D.L.G.; Callejon, R.M.; Amigo, J.M. NIR spectroscopy and chemometrics for the typification of Spanish wine vinegars with a protected designation of origin. Food Control 2018, 89, 108–116. [Google Scholar] [CrossRef]
- Barbosa Mascareli, V.A.; Galvan, D.; Andrade, J.C.D.; Lelis, C.; Junior, C.A.C.; Lopes, G.M.G.; Junior, F.C.D.M.; Spinosa, W.A. Spectralprint techniques coupled with chemometric tools for vinegar classifications. Food Chem. 2023, 410, 135373. [Google Scholar] [CrossRef]
- Næs, T.; Isaksson, T.; Fearn, T.; Davies, T. A User-Friendly Guide to Multivariate Calibration and Classification; NIR Publications: Chichester, UK, 2002. [Google Scholar]
- Yano, T.; Aimi, T.; Nakano, Y.; Tamai, M. Prediction of the concentrations of ethanol and acetic acid in the culture broth of a rice vinegar fermentation using near-infrared spectroscopy. J. Ferment. Bioeng. 1997, 84, 461–465. [Google Scholar] [CrossRef]
- Kasemsumran, S.; Boondaeng, A.; Ngowsuwa, N.K.; Jungtheerapanich, S.; Apiwatanapiwat, W.; Janchai, P.; Vaithanomsat, P. Mid-infrared and near-infrared spectroscopies to classify improper fermentation of pineapple wine. Chem. Pap. 2023, 77, 335–349. [Google Scholar] [CrossRef]
- Dambergs, R.G.; Kambouris, A.; Francis, I.L.; Gishen, M. Rapid analysis of methanol in grape derived distillation products using near infrared transmission spectroscopy. J. Agric. Food Chem. 2002, 50, 3079–3084. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, D.; Parker, M.; Dambergs, R.G.; Herderich, M.; Gishen, M. Chemometrics and visible-near infrared spectroscopic monitoring of red wine fermentation in a pilot. Biotechnol. Bioeng. 2006, 95, 1101–1107. [Google Scholar] [CrossRef]
- Casale, M.; Abajo, M.J.S.; Saiz, J.M.G.; Pizarro, C.; Forina, M. Study of the aging and oxidation processes of vinegar samples from different origins during storage by near-infrared spectroscopy. Anal. Chim. Acta 2006, 557, 360–366. [Google Scholar] [CrossRef]
- Krepelka, P.; Pérez-Rodríguez, F.; Bartusek, K. Bacterial pattern identification in near-infrared spectrum. IAPGOS 2014, 4, 58–60. [Google Scholar] [CrossRef]
- Boondaeng, A.; Kasemsumran, S.; Ngowsuwan, K.; Vaithanomsat, P.; Apiwatanapiwat, W.; Trakunjae, C.; Janchai, P.; Jungtheerapanic, S.; Niyomvong, N. Fermentation condition and quality evaluation of pineapple fruit wine. Fermentation 2021, 8, 11. [Google Scholar] [CrossRef]
- Niyomvong, N.; Sritawan, R.; Keabpimai, J.; Trakunjae, C.; Boondaeng, A. Comparison of the chemical properties of vinegar obtained via one-step fermentation and sequential fermentation from dragon fruit and pineapple. Beverages 2022, 8, 74. [Google Scholar] [CrossRef]
- Seal, T. Quantitative HPLC analysis of phenolic acids, flavonoids and ascorbic acid in four different solvent extracts of two wild edible leaves, Sonchus arvensis and Oenanthe linearis of North-Eastern region in India. J. Appl. Pharm. Sci. 2016, 6, 157–166. [Google Scholar] [CrossRef]
- Deshpande, S.S.; Cheryan, M.; Salunkhe, D.K. Tannin analysis of food products. Crit. Rev. Food Sci. Nutr. 1986, 24, 401–449. [Google Scholar] [CrossRef] [PubMed]
- ICH Harmonised Tripartite Guideline: Validation of Analytical Procedures: Text and Methodology Q2(R1). In International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use; ICH: Geneva, Switzerland, 2005; pp. 1–13.
- Jiang, H.; Zhang, H.; Chen, Q.; Mei, C.; Liu, G. Identification of solid state fermentation degree with FT-NIR spectroscopy: Comparison of wavelength variable selection methods of CARS and SCARS. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 149, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, X.; Chen, Y.; Li, M.; Han, D.; Liu, W. Quantitative determination of acacia honey adulteration by terahertz-frequency dielectric properties as an alternative technique. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 274, 121106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Duan, C.; Wang, Y.; Gao, H.; Hu, L.; Wang, X. Research on a nondestructive model for the detection of the nitrogen content of tomato. Front. Plant Sci. 2023, 13, 1093671. [Google Scholar] [CrossRef]
| Sample | Instrument/ Spectral Region/Measurement Mode | Sample Cell | Chemometric Method | Quantitative Result |
|---|---|---|---|---|
| Aromatic vinegar (n = 120) [18] | FT–NIR spectrometer/ 10,000–4000 cm−1/Transmission mode | A standard glass colorimetric ware | PLS | RMSEP 0.3310 mg/mL lactic acid 0.0557 mg/mL malic acid 0.0062 mg/mL L-pyroglutamic acid |
| Chinese vinegar (n = 160) [19] | FT–NIR spectrometer/ 10,000–4000 cm−1/Transmission mode | Glass tube 5 mm | Synergy Interval (Si)–PLS | RMSEP 0.26 g/100 mL total acids 1.93 g/100 mL soluble salt-free solids |
| Fermentation broth of mulberry vinegar [11] | A digital Micro-Mirror-based NIR spectrometer/900–1700 nm/Transmission mode | Cuvette | PLS | RMSEP 0.22% v/v total acids 8.11 mg GAE/L total polyphenol |
| Fruit vinegars (n = 180) [12] (apple, lemon, and peach vinegars) | FT–IR–NIR spectrometer/ 7800–4000 cm−1/Transmission mode | Liquid cell 1 mm | Least Squares–Support Vector Machine (LS–SVM) | RMSEP 0.35 g/L acetic acid 0.19 g/L tartaric acid 0.17 g/L formic acid 0.0842 pH |
| Rice vinegar (n = 325) [13] | A handheld Vis/NIR spectrometer/ 550–1000 nm/Transmission mode | Cuvette 2 mm | Effective wavelengths–LS–SVM | RMSEP 0.189 °Brix soluble solids 0.008 pH |
| Rice vinegar (n = 150) [15] | FT–NIR spectrometer/ 12,500–4000 cm−1/Transflectance mode | 0.1 mm glass vial with an aluminum reflector | PLS | RMSECV 2.44 g/L acetic acid 2.73 g/L ethanol |
| Vinegar sold in China (n = 120) [14] (mature, aromatic, and rice vinegars) | FT–NIR spectrometer/ 10,000–4000 cm−1/Transmission mode | Quartz cuvette 5 mm | Si–extreme learning machine (ELM) | RMSEP: 0.25 g/100 mL total acids |
| Vinegar on the market made from different raw materials (n = 95) [20] | FT–NIR spectrometer/ 10,000–4000 cm−1/Transmission mode | A standard glass colorimetric ware | PLS | RMSEP 0.32 g/mL total acids |
| Wine vinegar (n = 64) [16] | NIR spectrometer/1100–2500 nm/Transflection mode | Quartz liquid cell 2 mm | PLS | Prediction errors ranged 0.008% to 1.15%. Total, non-volatile, and volatile acids; chloride; solids; ash; L-proline; L(+)-tartaric acid; L(−)-malic acid; lactic acid; acetic acid; citric acid; succinic acid; D-malic acid |
| Wine vinegar (n = 107) [17] | Vis/NIR spectrometer/ 400–2500 nm/Transflectance mode | Gold circular reflector cup 0.1 mm | PLS | SEP 3.23 g/L volumic mass 13.97 g/L reducing sugars 1.42 g/100 mL total acidity 0.22 pH |
| Analyte | Sample Set | Range | Mean | SD | n |
|---|---|---|---|---|---|
| Acetic acid (%w/v) | Calibration set | 4.69 × 10−2–4.24 | 1.27 | 1.12 | 162 |
| Prediction set | 5.00 × 10−2–4.19 | 1.67 | 1.29 | 30 | |
| Ethanol (%v/v) | Calibration set | 5.00 × 10−3–7.00 | 3.68 | 2.17 | 162 |
| Prediction set | 2.10 × 10−2–4.76 | 2.12 | 1.46 | 30 | |
| TSS (°Brix) | Calibration set | 7.90–10.80 | 9.65 | 0.68 | 161 a |
| Prediction set | 7.97–9.80 | 9.07 | 0.63 | 30 | |
| Caffeic acid (µg/mL) | Calibration set | 1.23–7.46 | 3.78 | 1.64 | 162 |
| Prediction set | 1.63–6.85 | 4.69 | 1.75 | 30 | |
| Gallic acid (µg/mL) | Calibration set | 3.46–5.98 | 4.90 | 0.66 | 162 |
| Prediction set | 3.99–5.35 | 4.67 | 0.33 | 30 | |
| Tannic acid (µg/mL) | Calibration set | 138.82–288.30 | 198.09 | 39.08 | 162 |
| Prediction set | 144.32–204.69 | 168.03 | 19.34 | 30 |
| Analyte | Spectral Preprocessing | LVs | Rc2 | RMSEP |
|---|---|---|---|---|
| Acetic acid (%w/v) | None | 5 | 0.870 | 0.419 |
| 2D | 5 | 0.888 | 0.509 | |
| SNV | 4 | 0.855 | 0.532 | |
| Ethanol (%v/v) | None | 6 | 0.876 | 0.500 |
| 2D | 6 | 0.974 | 0.602 | |
| SNV | 5 | 0.969 | 0.632 | |
| TSS (°Brix) | None | 9 | 0.960 | 1.057 |
| 2D | 8 | 0.956 | 1.107 | |
| SNV | 9 | 0.947 | 1.080 | |
| Caffeic acid (µg/mL) | None | 8 | 0.846 | 0.974 |
| 2D | 6 | 0.832 | 0.914 | |
| SNV | 7 | 0.825 | 0.877 | |
| Gallic acid (µg/mL) | None | 10 | 0.638 | 0.881 |
| 2D | 12 | 0.755 | 0.902 | |
| SNV | 8 | 0.567 | 1.064 | |
| Tannic acid (µg/mL) | None | 10 | 0.682 | 61.48 |
| 2D | 10 | 0.694 | 59.15 | |
| SNV | 9 | 0.641 | 66.78 |
| Analyte | Selected Informative Spectral Variable by SCARS (cm−1) | Optimal SCARS Parameter | ||
|---|---|---|---|---|
| N | M | Frequency Level | ||
| Acetic acid (%w/v) | 7192, 7144, 7120, 7104, 6672, 6664, 6632, 6096, 5440, 5432, 5408, 5400, 5336, 4384, 4376 | 200 | 20 | 15 |
| Ethanol (%v/v) | 6744, 5328, 5032, 4656, 4384 | 500 | 50 | 35 |
| TSS (°Brix) | 6672, 6664, 6648, 6640, 6600, 6592, 6544, 6504, 5360, 5352, 4888, 4736, 4728, 4712, 4656, 4648, 4640, 4488, 4480, 4472, 4400 | 500 | 200 | 20 |
| Caffeic acid (µg/mL) | 11,520, 9312, 9304, 8896, 8728, 8432, 8424, 8416, 8384, 8376, 8368, 8360, 8328, 6632, 6624, 6368, 6360, 6352, 6320, 6312, 6304, 6280, 6272, 6264, 6232, 6224, 6216, 6192, 6184, 6144, 6136, 6128, 6080, 6072, 6064, 6056, 6024, 6016, 6008, 6000, 5976, 5968, 5960, 5952, 5944, 5936, 5928, 5744, 5720, 5712, 5704, 5696, 5688, 5672, 5664, 5656, 5648, 5640, 5632, 5624, 5616, 5608, 5600, 5592, 5584, 5576, 5568, 5528, 5520, 5512, 5504, 5336, 5328, 5320, 4672, 4664, 4632, 4624, 4584, 4576, 4464, 4448, 4440, 4392, 4384, 4376, 4352, 4344 | 200 | 100 | 25 |
| Gallic acid (µg/mL) | 10,992, 6632, 6368, 6360, 6352, 6344, 5520, 5344, 5312, 4800, 4536, 4528, 4392, 4352, 4304 | 500 | 100 | 40 |
| Tannic acid (µg/mL) | 11,128, 11,120, 10,768, 10,560, 10,552, 10,512, 10,504, 10,224, 10,024, 10,016, 9976, 9968, 9720, 9664, 9656, 9456, 9448, 8712, 7800, 7792, 7744, 7656, 7648, 7592, 7400, 7392, 7264, 6648, 6584, 6432, 6424, 6376, 6368, 6296, 6136, 5944, 5936, 5600, 5528, 5520, 5400, 5384, 5016, 4800, 4504, 4400, 4392, 4336 | 200 | 200 | 25 |
| Analyte | Spectral Preprocessing | Method | Number of Variables | LVs | Rc2 | RMSEP |
|---|---|---|---|---|---|---|
| Acetic acid (%) | None | SCARS–PLS | 15 | 4 | 0.874 | 0.137 |
| PLS | 949 a | 5 | 0.870 | 0.419 | ||
| Ethanol (%) | None | SCARS–PLS | 5 | 5 | 0.973 | 0.178 |
| PLS | 949 a | 6 | 0.876 | 0.500 | ||
| TSS (°Brix) | None | SCARS–PLS | 21 | 3 | 0.903 | 0.875 |
| PLS | 949 a | 9 | 0.960 | 1.057 | ||
| Caffeic acid (µg/mL) | SNV | SCARS–PLS | 88 | 8 | 0.938 | 0.637 |
| PLS | 949 a | 7 | 0.825 | 0.877 | ||
| Gallic acid (µg/mL) | None | SCARS–PLS | 15 | 12 | 0.752 | 0.340 |
| PLS | 949 a | 10 | 0.638 | 0.881 | ||
| Tannic acid (µg/mL) | 2D | SCARS–PLS | 48 | 10 | 0.891 | 31.12 |
| PLS | 935 b | 10 | 0.694 | 59.15 |
| Best Model | Method | Statistic | Obtained Result | Criterion | Performance |
|---|---|---|---|---|---|
| Acetic acid (%) | SCARS–PLS | SEP | 0.136 | TUE = 0.480 | accepted |
| bias | 0.023 | Tb = ±0.051 | accepted | ||
| PLS | SEP | 0.424 | TUE = 0.487 | accepted | |
| bias | 0.043 | Tb = ±0.158 | accepted | ||
| Ethanol (%) | SCARS–PLS | SEP | 0.173 | TUE = 0.426 | accepted |
| bias | −0.053 | Tb = ±0.065 | accepted | ||
| PLS | SEP | 0.421 | TUE = 0.464 | accepted | |
| bias | −0.275 | Tb = ±0.157 | not accepted | ||
| TSS (°Brix) | SCARS–PLS | SEP | 0.662 | TUE = 0.256 | not accepted |
| bias | −0.586 | Tb = ±0.247 | not accepted | ||
| PLS | SEP | 0.754 | TUE = 0.165 | not accepted | |
| bias | −0.754 | Tb = ±0.282 | not accepted | ||
| Caffeic acid (µg/mL) | SCARS–PLS | SEP | 0.630 | TUE = 0.653 | accepted |
| bias | 0.148 | Tb = ±0.235 | accepted | ||
| PLS | SEP | 0.890 | TUE = 0.829 | not accepted | |
| bias | −0.068 | Tb = ±0.332 | accepted | ||
| Gallic acid (µg/mL) | SCARS–PLS | SEP | 0.342 | TUE = 0.396 | accepted |
| bias | −0.049 | Tb = ±0.128 | accepted | ||
| PLS | SEP | 0.615 | TUE = 0.478 | not accepted | |
| bias | −0.641 | Tb = ±0.230 | not accepted | ||
| Tannic acid (µg/mL) | SCARS–PLS | SEP | 27.051 | TUE = 15.584 | not accepted |
| bias | 16.163 | Tb = ±10.101 | not accepted | ||
| PLS | SEP | 53.433 | TUE = 26.106 | not accepted | |
| bias | −27.176 | Tb = ±19.952 | not accepted |
| Analytes | Analytical Method | Response | Linear Range | R2 | LODs | LOQs | %RSD |
|---|---|---|---|---|---|---|---|
| Acetic acid (μg/mL) | HPLC | Rt = 14.5 min. | 100–10,000 | 0.9993 | 0.13 | 0.38 | 0.48 |
| Ethanol (%) | GC | Rt = 1.853 min (Ethanol) Rt = 3.246 min (n-propanol; internal standard) | 0.25–10 | 0.9986 | 0.02 | 0.05 | 1.01 |
| Caffeic acid (μg/mL) | HPLC | Rt = 14.559 min | 3.125–50 | 1.0000 | 0.02 | 0.07 | 0.11 |
| Gallic acid (μg/mL) | HPLC | Rt = 6.523 min | 3.125–50 | 1.0000 | 0.03 | 0.09 | 0.17 |
| Tannic acid (μg/mL) | UV spectrometry | Abs, 280 nm | 2–18 | 0.9997 | 0.09 | 0.28 | 0.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasemsumran, S.; Boondaeng, A.; Jungtheerapanich, S.; Ngowsuwan, K.; Apiwatanapiwat, W.; Janchai, P.; Vaithanomsat, P. Assessing Fermentation Broth Quality of Pineapple Vinegar Production with a Near-Infrared Fiber-Optic Probe Coupled with Stability Competitive Adaptive Reweighted Sampling. Molecules 2023, 28, 6239. https://doi.org/10.3390/molecules28176239
Kasemsumran S, Boondaeng A, Jungtheerapanich S, Ngowsuwan K, Apiwatanapiwat W, Janchai P, Vaithanomsat P. Assessing Fermentation Broth Quality of Pineapple Vinegar Production with a Near-Infrared Fiber-Optic Probe Coupled with Stability Competitive Adaptive Reweighted Sampling. Molecules. 2023; 28(17):6239. https://doi.org/10.3390/molecules28176239
Chicago/Turabian StyleKasemsumran, Sumaporn, Antika Boondaeng, Sunee Jungtheerapanich, Kraireuk Ngowsuwan, Waraporn Apiwatanapiwat, Phornphimon Janchai, and Pilanee Vaithanomsat. 2023. "Assessing Fermentation Broth Quality of Pineapple Vinegar Production with a Near-Infrared Fiber-Optic Probe Coupled with Stability Competitive Adaptive Reweighted Sampling" Molecules 28, no. 17: 6239. https://doi.org/10.3390/molecules28176239
APA StyleKasemsumran, S., Boondaeng, A., Jungtheerapanich, S., Ngowsuwan, K., Apiwatanapiwat, W., Janchai, P., & Vaithanomsat, P. (2023). Assessing Fermentation Broth Quality of Pineapple Vinegar Production with a Near-Infrared Fiber-Optic Probe Coupled with Stability Competitive Adaptive Reweighted Sampling. Molecules, 28(17), 6239. https://doi.org/10.3390/molecules28176239







