Comprehensive Metabolite Identification of Genipin in Rats Using Ultra-High-Performance Liquid Chromatography Coupled with High Resolution Mass Spectrometry
Abstract
:1. Introduction
2. Results
2.1. Establishment of the Analytical Strategy
2.2. Establishment of the MMDF Data-Mining Method
2.3. DPI Based on the Mass Fragmentation Behaviors of Genipin and Genipinine
2.4. Structural Confirmation of Genipin Metabolites
2.5. Identification of Genipin and Its Metabolites
2.6. Identification of Genipinine and Its Metabolites
2.7. Identification of M16 and M30
2.8. Identification of Genipin In Vitro Fecal Fermentation and Its Metabolites
3. Discussion
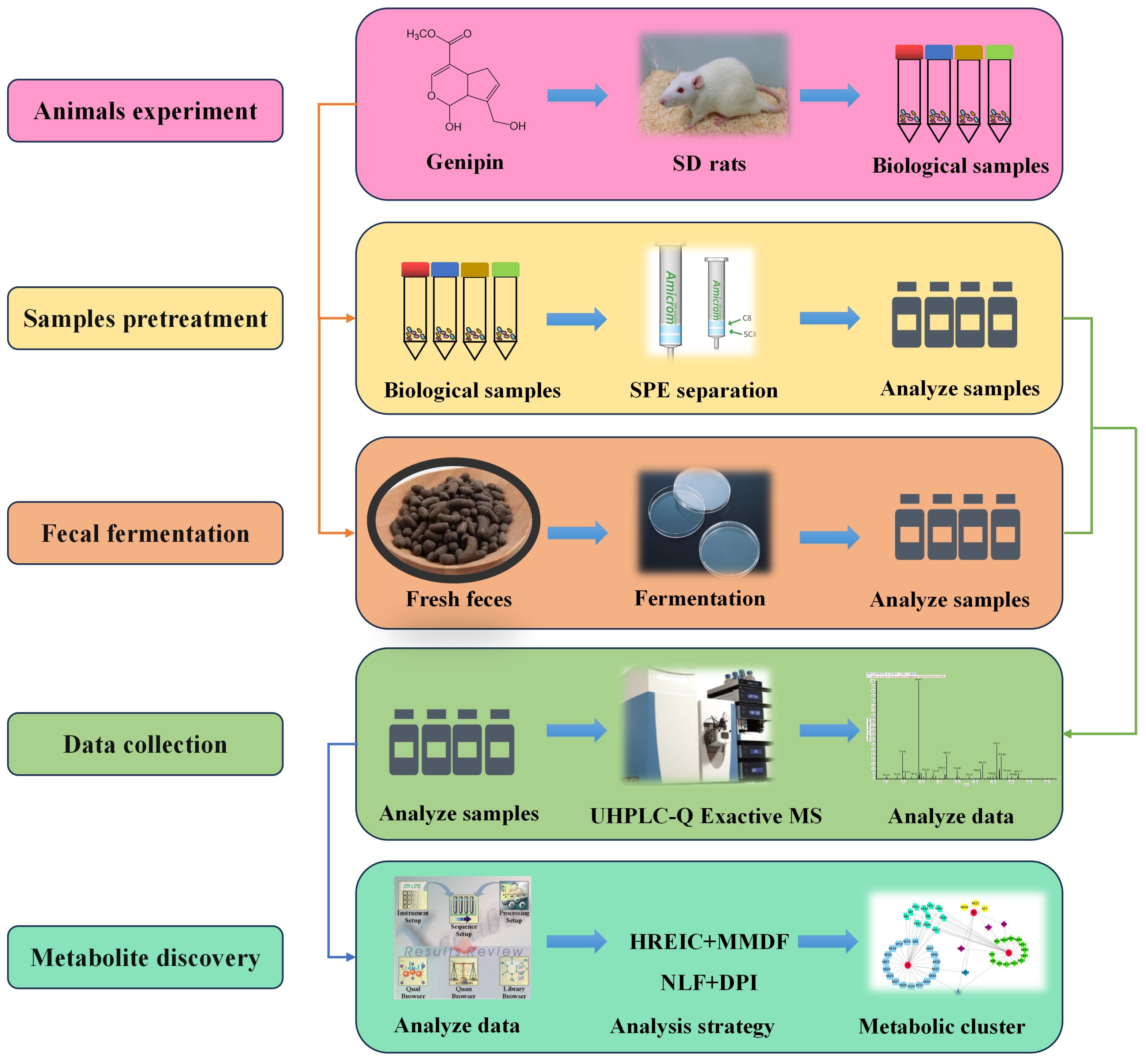
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Identification of Genipin Metabolites In Vivo
4.2.1. Animals
4.2.2. Drug Administration and Biological Sample Preparation
4.3. Biotransformation In Vitro of Genipin by SD Rat Feces
4.4. Instrument and Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zhang, X.; Liu, S.; Pi, Z.; Liu, Z.; Song, F. Simultaneous quantification method for comparative pharmacokinetics studies of two major metabolites from geniposide and genipin by online mircrodialysis-UPLC-MS/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1041, 11–18. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Kim, W. Genipin inhibits rotavirus-induced diarrhea by suppressing viral replication and regulating inflammatory responses. Sci. Rep. 2020, 10, 15836. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Wu, Y.; Liu, H.; Xie, C. Genipin Induces Autophagy and Suppresses Cell Growth of Oral Squamous Cell Carcinoma via PI3K/AKT/MTOR Pathway. Drug Des. Dev. Ther. 2020, 14, 395–405. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Li, X.; Jiang, L.; Chang, Z.; Li, Q. Biologically responsive, long-term release nanocoating on an electrospun scaffold for vascular endothelialization and anticoagulation. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 107, 110212. [Google Scholar] [CrossRef]
- Yu, D.; Shi, M.; Bao, J.; Yu, X.; Li, Y.; Liu, W. Genipin ameliorates hypertension-induced renal damage via the angiotensin II-TLR/MyD88/MAPK pathway. Fitoterapia 2016, 112, 244–253. [Google Scholar] [CrossRef]
- Xiao, W.; Li, S.; Wang, S.; Ho, C.T. Chemistry and bioactivity of Gardenia jasminoides. J. Food Drug Anal. 2017, 25, 43–61. [Google Scholar] [CrossRef]
- Wang, Y.; Mei, X.; Liu, Z.; Li, J.; Zhang, X.; Lang, S.; Dai, L.; Zhang, J. Drug Metabolite Cluster-Based Data-Mining Method for Comprehensive Metabolism Study of 5-hydroxy-6,7,3′,4′-tetramethoxyflavone in Rats. Molecules 2019, 18, 3278. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, X.; Fan, L.; Dong, H.; Ren, Y.; Chen, X. Stepwise Diagnostic Product Ions Filtering Strategy for Rapid Discovery of Diterpenoids in Scutellaria barbata Based on UHPLC-Q-Exactive-Orbitrap-MS. Molecules 2022, 23, 8185. [Google Scholar] [CrossRef]
- Li, Y.; Cai, W.; Cai, Q.; Che, Y.; Zhao, B.; Zhang, J. Comprehensive characterization of the in vitro and in vivo metabolites of geniposide in rats using ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer. Xenobiotica Fate Foreign Compd. Biol. Syst. 2016, 4, 357–368. [Google Scholar] [CrossRef]
- Ma, Q.; Li, Y.; Li, P.; Wang, M.; Wang, J.; Tang, Z.; Wang, T.; Luo, L.; Wang, C.; Wang, T.; et al. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 117, 109138. [Google Scholar] [CrossRef]
- Zhou, B.; Yuan, Y.; Zhang, S.; Guo, C.; Li, X.; Li, G.; Xiong, W.; Zeng, Z. Intestinal Flora and Disease Mutually Shape the Regional Immune System in the Intestinal Tract. Front. Immunol. 2020, 11, 575. [Google Scholar] [CrossRef]
- Sebastián Domingo, J.J.; Sánchez Sánchez, C. From the intestinal flora to the microbiome. Rev. Esp. De Enfermedades Dig. 2018, 110, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Che, Q.; Luo, T.; Shi, J.; He, Y.; Xu, D.L. Mechanisms by Which Traditional Chinese Medicines Influence the Intestinal Flora and Intestinal Barrier. Front. Cell. Infect. Microbiol. 2022, 12, 863779. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.W.; Bao, B.; Han, Z.Q.; Han, Q.Y.; Yang, X.L. Metabolic profile of Fructus Gardeniae in human plasma and urine using ultra high-performance liquid chromatography coupled with high-resolution LTQ-orbitrap mass spectrometry. Xenobiotica Fate Foreign Compd. Biol. Syst. 2016, 46, 901–912. [Google Scholar] [CrossRef]
- Dong, P.; Shi, L.; Wang, S.; Jiang, S.; Li, H.; Dong, F.; Xu, J.; Dai, L.; Zhang, J. Rapid Profiling and Identification of Vitexin Metabolites in Rat Urine, Plasma and Faeces after Oral Administration Using a UHPLC-Q-Exactive Orbitrap Mass Spectrometer Coupled with Multiple Data-mining Methods. Curr. Drug Metab. 2021, 22, 185–197. [Google Scholar] [PubMed]
- Wang, F.; Shang, Z.; Xu, L.; Wang, Z.; Zhao, W.; Mei, X.; Lu, J.; Zhang, J.Y. Profiling and identification of chlorogenic acid metabolites in rats by ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer. Xenobiotica Fate Foreign Compd. Biol. Syst. 2018, 48, 605–617. [Google Scholar] [CrossRef]
- Yang, B.; Li, H.; Ruan, Q.; Tong, Y.; Liu, Z.; Xuan, S.; Jin, J.; Zhao, Z. Rapid profiling and pharmacokinetic studies of multiple potential bioactive triterpenoids in rat plasma using UPLC/Q-TOF-MS/MS after oral administration of Ilicis Rotundae Cortex extract. Fitoterapia 2018, 129, 210–219. [Google Scholar] [CrossRef]
- Han, H.; Yang, L.; Xu, Y.; Ding, Y.; Bligh, S.W.; Zhang, T.; Wang, Z. Identification of metabolites of geniposide in rat urine using ultra-performance liquid chromatography combined with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Rapid Commun. Mass Spectrom. RCM 2011, 21, 3339–3350. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Che, Y.; Wang, F.; Shang, Z.; Lu, J.; Dai, S.; Zhang, J.; Cai, W. Identification of Metabolites of 6′-Hydroxy-3,4,5,2′,4′-pentamethoxychalcone in Rats by a Combination of Ultra-High-Performance Liquid Chromatography with Linear Ion Trap-Orbitrap Mass Spectrometry Based on Multiple Data Processing Techniques. Molecules 2016, 21, 1266. [Google Scholar] [CrossRef]
- Wu, Y.; Pan, L.; Chen, Z.; Zheng, Y.; Diao, X.; Zhong, D. Metabolite Identification in the Preclinical and Clinical Phase of Drug Development. Curr. Drug Metab. 2021, 22, 838–857. [Google Scholar] [CrossRef]
- Cai, W.; Zhang, J.Y.; Dong, L.Y.; Yin, P.H.; Wang, C.G.; Lu, J.Q.; Zhang, H.G. Identification of the metabolites of Ixerin Z from Ixeris sonchifolia Hance in rats by HPLC-LTQ-Orbitrap mass spectrometry. J. Pharm. Biomed. Anal. 2015, 107, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.C.; Tsai, S.Y.; Lai, P.Y.; Chen, Y.S.; Chao, P.D. Metabolism and pharmacokinetics of genipin and geniposide in rats. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2008, 46, 2764–2769. [Google Scholar] [CrossRef]
- Chen, C.; Han, F.; Zhang, Y.; Lu, J.; Shi, Y. Simultaneous determination of geniposide and its metabolites genipin and genipinine in culture of Aspergillus niger by HPLC. Biomed. Chromatogr. BMC 2008, 22, 753–757. [Google Scholar] [CrossRef]
- Shang, Z.; Wang, F.; Dai, S.; Lu, J.; Wu, X.; Zhang, J. Profiling and identification of (-)-epicatechin metabolites in rats using ultra-high performance liquid chromatography coupled with linear trap-Orbitrap mass spectrometer. Drug Test. Anal. 2017, 8, 1224–1235. [Google Scholar] [CrossRef]
- Pähler, A.; Brink, A. Software aided approaches to structure-based metabolite identification in drug discovery and development. Drug Discov. Today. Technol. 2013, 10, e207–e217. [Google Scholar] [CrossRef]
- Ding, Y.; Hou, J.W.; Zhang, Y.; Zhang, L.Y.; Zhang, T.; Chen, Y.; Cai, Z.Z.; Yang, L. Metabolism of Genipin in Rat and Identification of Metabolites by Using Ultraperformance Liquid Chromatography/Quadrupole Time-of-Flight Tandem Mass Spectrometry. Evid. -Based Complement. Altern. Med. Ecam 2013, 2013, 957030. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Wang, Y.; Li, J.; Liu, Z.; Lang, S.; Ouyang, W.; Zhang, J. Comprehensive metabolism study of polydatin in rat plasma and urine using ultra-high performance liquid chromatography coupled with high-resolution mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1117, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Pan, H.; Li, Y.; Huang, L.; Wu, Q.; Lu, Y. A sensitive LC-MS/MS method for simultaneous quantification of geniposide and its active metabolite genipin in rat plasma and its application to a pharmacokinetic study. Biomed. Chromatogr. BMC 2018, 32, e4126. [Google Scholar] [CrossRef]
- Li, Z.; Ma, T.; Zhang, W.; Shang, Y.; Zhang, Y.; Ma, Y. Genipin attenuates dextran sulfate sodium-induced colitis via suppressing inflammatory and oxidative responses. Inflammopharmacology 2020, 28, 333–339. [Google Scholar] [CrossRef]
- Xing, S.; Hu, Y.; Yin, Z.; Liu, M.; Tang, X.; Fang, M.; Huan, T. Retrieving and Utilizing Hypothetical Neutral Losses from Tandem Mass Spectra for Spectral Similarity Analysis and Unknown Metabolite Annotation. Anal. Chem. 2020, 21, 14476–14483. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Liang, X. Application of ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry in identification of three isoflavone glycosides and their corresponding metabolites. Rapid Commun. Mass Spectrom. RCM 2018, 32, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Z.; Tang, Y.; Cui, X.; Luo, R.; Guo, S.; Zheng, Y.; Huang, C. Isolation, identification and antiviral activities of metabolites of calycosin-7-O-β-D-glucopyranoside. J. Pharm. Biomed. Anal. 2011, 56, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Prasain, J.K.; Reppert, A.; Jones, K.; Moore, D.R.; Barnes, S., 2nd; Lila, M.A. Identification of isoflavone glycosides in Pueraria lobata cultures by tandem mass spectrometry. Phytochem. Anal. PCA 2007, 18, 50–59. [Google Scholar] [CrossRef]
- Luo, L.; Wu, S.; Zhu, X.; Su, W. The profiling and identification of the absorbed constituents and metabolites of Naoshuantong capsule in mice biofluids and brain by ultra- fast liquid chromatography coupled with quadrupole-time-of-flight tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1129, 121791. [Google Scholar] [CrossRef]
- Polson, C.; Sarkar, P.; Incledon, B.; Raguvaran, V.; Grant, R. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 785, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Ötles, S.; Kartal, C. Solid-Phase Extraction (SPE): Principles and Applications in Food Samples. Acta Sci. Pol. Technol. Aliment. 2016, 15, 5–15. [Google Scholar] [CrossRef]


| Peak | Product | tR /min | Formula | Theoretical Mass (m/z) | Experimental Mass (m/z) | Error (ppm) | MS/MS Fragment Ions | Identification | Vivo | Vitro | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | B | F | Different Time (h) | |||||||||
| M0 | Genipin | 6.38 | C11H13O5 | 225.07685 | 225.07623 | 2.133 | 101(100), 69(68), 123(37), 147(24), 207(10), 119(4.5), 111(2.4) |  | N | 0 h(N), 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| 6.38 | C11H15O5 | 227.09140 | 227.09050 | −3.963 | 116(100), 70(92), 209(23), 157(5), 131(4), 85(3) | 0 h(P), 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||||||
| M3 | Sulfation | 0.99 | C11H13O8S | 305.03365 | 305.03381 | 4.085 | 101(100), 123(64), 69(39), 305(27), 225(16), 147(15), 97(14), 81(13),207(12), 273(9.3), 111(8.7) |  | N | N | ||
| M4 | Hydroxylation | 1.03 | C11H13O6 | 241.07121 | 241.07164 | 4.046 | 69(100), 165(53), 209(33), 162(29), 101(12), 241(10), 80(6.3) |  | N | N | 0 h(N), 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | |
| M6 | Dehydroxylation and demethylation | 1.20 | C10H11O4 | 195.06629 | 195.06563 | 2.280 | 123(100), 133(51), 81(43), 135(32), 177(30), 59(21), 97(17), 119(15) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N) | ||
| M9 | Sulfation and hydroxylation | 5.37 | C11H13O9S | 321.02855 | 321.02856 | 3.367 | 101(100), 97(81), 127(74), 109(55), 321(45), 67(28),139(20), 79(18), 241(16), 81(15), 209(12), 111(12),121(10) |  | N | N | ||
| M11 | Genipin-1-O-glucuronide | 5.63 | C17H20O11 | 401.10895 | 401.10883 | 2.474 | 101(100), 123(67), 401(63), 69(57), 113(48), 85(28), 207(18), 147(16), 369(13), 105(11), 87(11), 325(10), 193(10), 225(2.3) |  | N | N | ||
| M12 | Sulfation | 6.10 | C11H13O8S | 305.03365 | 305.03357 | 3.296 | 101(100), 123(63), 69(39), 305(27), 96(15), 147(14), 225(13),207(13) |  | N | N | N | |
| M15 | Demethylation | 6.29 | C10H9O4 | 193.05069 | 193.04991 | 1.941 | 133(100), 178(53), 193(22), 149(19), 137(17), 74(7.7), 109(1.9) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N) | ||
| M16 | Amination | 6.43 | C11H16O4N | 226.10738 | 226.10699 | −1.745 | 194(100), 151(96), 226(48), 106(34), 80(25), 82(23), 120(22), 114(22), 148(20), 55(17),101(10),122(13), 180(14), 79(10) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P) | |
| M17 | Dehydrogenation and demethylation | 6.45 | C10H9O5 | 209.04549 | 209.04498 | 2.536 | 121(100), 62(15), 209(11), 120(4.5), 181(3.1), 91(1.7), 79(1.4), 69(1.0), 136(1.0) |  | N | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | |
| M21 | Decarbonylation | 6.82 | C10H13O4 | 197.08195 | 197.08128 | 2.256 | 153(100), 197(48), 135(38), 59(24), 111(16), 83(7.6), 69(7.2), 179(7.0), 123(5.9) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N) | ||
| M23 | Dehydroxylation | 6.89 | C11H15O3 | 195.10265 | 195.10201 | 2.521 | 151(100), 95(71), 135(53), 195(39), 149(5.8), 80(3.2), 177(2.9), 130(2.5),165(1.6) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M25 | Glucuronidation | 7.35 | C17H25O10 | 389.14480 | 389.14548 | 3.229 | 80(100), 389(50), 81(34), 113(34), 85(25), 59(15), 71(15), 180(14), 235(13),73(10), 213(7.9) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M28 | Decarbonylation | 7.39 | C11H13O4 | 197.08195 | 197.08119 | 1.799 | 153(100), 59(46), 74(45), 135(12), 75(12), 137(7), 197(5.1), 179(0.99) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M29 | Glucuronidation | 7.43 | C17H25O10 | 389.14480 | 389.14532 | 2.818 | 80(100), 389(51), 81(32), 113(16), 235(14), 85(13), 180(12), 59(9), 213(5.6) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M30 | Aldehyde | 7.45 | C11H13O5 | 225.07685 | 225.07642 | 2.977 | 225(100), 59 (44), 207(24), 137(23), 163(19), 69(15), 109(12), 181(10) |  | N | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | |
| M32 | Oxygen-containing heterocycle | 7.76 | C10H15O4 | 199.09765 | 199.09765 | 0.515 | 155(100), 199(60), 137(37), 83(24), 181(14), 59(11), 130(4.9) |  | N | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | |
| M34 | Dehydrogenation | 8.00 | C11H11O5 | 223.06115 | 223.06070 | 2.690 | 223(100), 179(99), 155(37), 59(30), 161(12), 143(11), 195(8), 144(6.5) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M35 | Dehydroxylation and hydrogenation | 8.04 | C11H15O4 | 211.09819 | 211.09695 | 2.201 | 167(100), 211(58), 81(49), 80(49), 132(29), 149(20), 59(15), 106(15), 99(1.0) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M37 | Dehydroxylation and hydrogenation | 8.18 | C11H15O4 | 211.09819 | 211.09697 | 0.485 | 167(100), 59(14), 80(14), 149(13), 81(13), 211(10), 132(4.6), 99(1.2) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M38 | Deoxygenation and demethylation | 8.21 | C11H15O2 | 179.10775 | 179.10689 | 0.234 | 179(100), 59(50), 135(21), 91(14), 161(11), 71(10) |  | N | N | 24 h(N), 36 h(N), 48 h(N), 60 h(N) | |
| M39 | Dehydroxylation and hydrogenation | 8.27 | C11H15O4 | 211.09819 | 211.09694 | 0.455 | 99(100), 167(75), 80(18), 211(13), 59(10), 149(9), 132(8.7) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M40 | 3-Methyl-genipin | 8.30 | C12H15O5 | 239.09255 | 239.09215 | 3.317 | 239(100), 59(24), 221(22), 83(18), 177(17), 151(12), 195(10), 123(6.7) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N) | ||
| M41 | Hydrogenation | 8.32 | C11H17O4 | 213.11270 | 213.11269 | 0.554 | 169(100), 213(53), 151(23), 195(4), 73(2.8), 83(1.7), 113(1.5) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M43 | Hydrogenation | 8.43 | C11H17O4 | 213.11270 | 213.11270 | 0.564 | 169(100), 213(27), 151(19), 73(2.0),195(1.7), 170(1.4) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M44 | Hydrogenation | 8.53 | C11H17O4 | 213.11270 | 213.11269 | 0.554 | 213(100), 169(92), 151(56), 195(20), 97(10), 83(8.8), 59(7.0), 113(3.3), 73(1.4) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M45 | Hydrogenation | 8.69 | C11H17O4 | 213.11270 | 213.11272 | 0.584 | 213(100), 169(92), 151(56), 195(20), 97(10), 83(8.8), 59(7.0), 123(6.5), 73(1.7) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| M46 | Dehydroxylation | 8.82 | C11H13O4 | 209.08087 | 209.08057 | −1.269 | 121(100), 93(63), 149(56), 91(47), 103(37), 79(26), 95(20), 85(19), 81(18), 105(16), 177(15), 131(13), 53(11), 209(7.7) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | |
| M48 | Glucuronic acid | 9.45 | C15H19O10 | 359.09839 | 359.09708 | −0.538 | 297(100), 82(86), 215(70), 171(70), 359(48), 81(27), 143(24),125(24), 163(15) |  | N | 48 h(N), 60 h(N) | ||
| M49 | 3-Methyl-genipin | 9.59 | C12H15O5 | 239.09255 | 239.09219 | 3.304 | 195(100), 151(69), 195(41), 239(33), 177(13), 59(7.1), 123(4.5) |  | N | 12 h(N), 24 h(N), 36 h(N), 48 h(N), 60 h(N) | ||
| Peak | Product | tR /min | Formula | Theoretical Mass (m/z) | Experimental Mass (m/z) | Error (ppm) | MS/MS Fragment Ions | Identification | Vivo | Vitro | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | B | F | Different Time (h) | |||||||||
| M1 | 3-Hydroxy-3-(methylamino)propanoic acid | 0.81 | C4H10O3N | 120.06567 | 120.06549 | −0.247 | 120(100), 103(53), 93(11), 91(5.9), 95(2), 53(1.6), 106(1.2) |  | P | 0 h(P), 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M2 | 5-(Hydroxymethyl) nicotinic acid | 0.92 | C7H8O3N | 154.05007 | 154.04953 | −2.205 | 154(100), 92(21), 136(13), 112(8.3), 108(1.1), 95(3.0), 110(1.5) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M5 | Hydroxylation | 1.06 | C11H12O4N | 222.07602 | 222.07555 | −0.534 | 222(100), 134(34), 190(29), 176(28), 162(26), 144(13), 177(12), 106(7.0), 204(5.4) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P) | ||
| M7 | Demethylation and dehydrogenation | 4.86 | C10H12O3N | 194.08112 | 194.08081 | −0.360 | 91(100), 76(23), 57(1.9), 146(1.8), 134(1.8), 130(1.8), 108(1.7), 67(1.6), 194(1.0) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | |
| M8 | Dehydrogenation | 5.33 | C11H12O3N | 206.08107 | 206.08069 | −2.328 | 146(100), 174(64), 206(58), 118(44), 91(25), 154(19), 178(10), 156(7.7) |  | p | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M10 | Demethylation and dehydrogenation | 5.48 | C10H12O3N | 194.08112 | 194.08086 | −0.310 | 91(100), 76(23), 134(7.4), 194(3.5), 150(3.4), 106(3.1), 57(2.2), 92(2.1), 130(2.0) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | |
| M13 | Dehydrogenation | 6.12 | C11H14NO3 | 208.09682 | 208.09636 | −2.210 | 120(100), 85(21), 166(6.4), 103(6.0), 162(2.4), 148(1.7), 208(1.5) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | |
| M14 | Hydrogenation | 6.20 | C9H12O4N | 198.07617 | 198.07578 | −1.537 | 138(100), 123(32), 96(19), 126(15), 110(12), 120(12), 130(10), 55(9.4), 108(2.6) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P) | ||
| M18 | Dehydrogenation | 6.49 | C11H12O3N | 206.08107 | 206.08080 | −1.794 | 118(100), 130(68), 146(37), 170(34), 188(27), 160(23), 132(22), 206(18), 142(11) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M19 | Glycine conjugation | 6.56 | C13H19O4N2 | 267.13397 | 267.13339 | −2.035 | 134(100), 162(17), 161(15), 84(13), 197(6.6), 239(5.5), 135(5.4), 80(3.1),130(2.4), 207(1.9) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M20 | Carbonylation | 6.66 | C11H14O4N | 224.09167 | 224.09128 | −2.208 | 146(100), 192(54), 118(53), 82(48), 178(42), 174(36), 114(32), 224(31), 206(26), 91(19), 105(12), 60(11) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M22 | Demethylation and hydroxylation | 6.88 | C10H14O4N | 212.09167 | 212.09132 | −1.954 | 109(100), 76(15), 137(1.7), 81(1.7), 95(1.5), 79(1.3), 166(1.0),134(1.0) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M24 | Dehydrogenation | 7.00 | C11H12O3N | 206.08107 | 206.08084 | −1.600 | 118(100), 130(68), 146(37), 170(34), 95(1.4), 146(1.2), 116(1.1), 206(1.1) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M26 | Dehydrogenation | 7.35 | C11H10O3N | 204.06542 | 204.06512 | −0.400 | 204(100), 144(71), 116(50), 92(29), 134(15), 65(14), 120(12), 176(11), 175(4.7) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | |
| M27 | Sulfation | 7.36 | C11H16O7NS | 306.06417 | 306.06418 | −0.062 | 147(100), 81(60), 289(43), 119(42), 105(40), 175(38), 157(30), 67(28), 165(27), 123(23), 210(15) |  | P | 48 h(P) | ||
| M31 | Genipinine | 7.72 | C11H16O3N | 210.11247 | 210.11214 | −1.457 | 178(100), 210(36), 109(32), 81(21), 79(17), 84(12), 132(12), 160(9), 133(8.8) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P) | ||
| M33 | Hydroxylation | 7.83 | C11H12O4N | 222.07602 | 222.07562 | −0.464 | 119(100), 204(70), 222(37), 162(34), 144(31), 116(27), 118(26), 172(23), 105(19) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P) | ||
| M36 | Hydrogenation | 8.06 | C11H20O3N | 214.14322 | 214.14326 | −2.382 | 70(100), 116(68), 81(38), 71(10), 214(4.1), 99(4.04), 79(3.5), 168(2.4) |  | P | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | |
| M42 | Dehydroxylation | 8.32 | C11H12O2N | 190.08632 | 190.08589 | −0.365 | 130(100), 55(21), 172(8.1), 190(6.0), 149(3.4), 144(1.5), 72(1.4), 162(1.2) |  | P | 12 h(P), 24 h(P), 36 h(P), 48 h(P), 60 h(P) | ||
| M47 | Dehydroxylation and glucuronidation | 9.40 | C16H22O8N | 356.13402 | 356.13348 | −0.508 | 135(100), 93(40), 107(38), 356(35), 67(15), 109(15), 60(12), 180(5.6) |  | P | 48 h(P) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Z.; Li, Z.; Dong, W.; Qiu, L.; Zhang, J.; Wang, S. Comprehensive Metabolite Identification of Genipin in Rats Using Ultra-High-Performance Liquid Chromatography Coupled with High Resolution Mass Spectrometry. Molecules 2023, 28, 6307. https://doi.org/10.3390/molecules28176307
Cui Z, Li Z, Dong W, Qiu L, Zhang J, Wang S. Comprehensive Metabolite Identification of Genipin in Rats Using Ultra-High-Performance Liquid Chromatography Coupled with High Resolution Mass Spectrometry. Molecules. 2023; 28(17):6307. https://doi.org/10.3390/molecules28176307
Chicago/Turabian StyleCui, Zhifeng, Zhe Li, Weichao Dong, Lili Qiu, Jiayu Zhang, and Shaoping Wang. 2023. "Comprehensive Metabolite Identification of Genipin in Rats Using Ultra-High-Performance Liquid Chromatography Coupled with High Resolution Mass Spectrometry" Molecules 28, no. 17: 6307. https://doi.org/10.3390/molecules28176307
APA StyleCui, Z., Li, Z., Dong, W., Qiu, L., Zhang, J., & Wang, S. (2023). Comprehensive Metabolite Identification of Genipin in Rats Using Ultra-High-Performance Liquid Chromatography Coupled with High Resolution Mass Spectrometry. Molecules, 28(17), 6307. https://doi.org/10.3390/molecules28176307







