Cationic Polymers Remarkably Boost Haloalkane Dehalogenase Activity in Organic Solvent Solutions and the Molecular Implications
Abstract
:1. Introduction
2. Results and Discussion
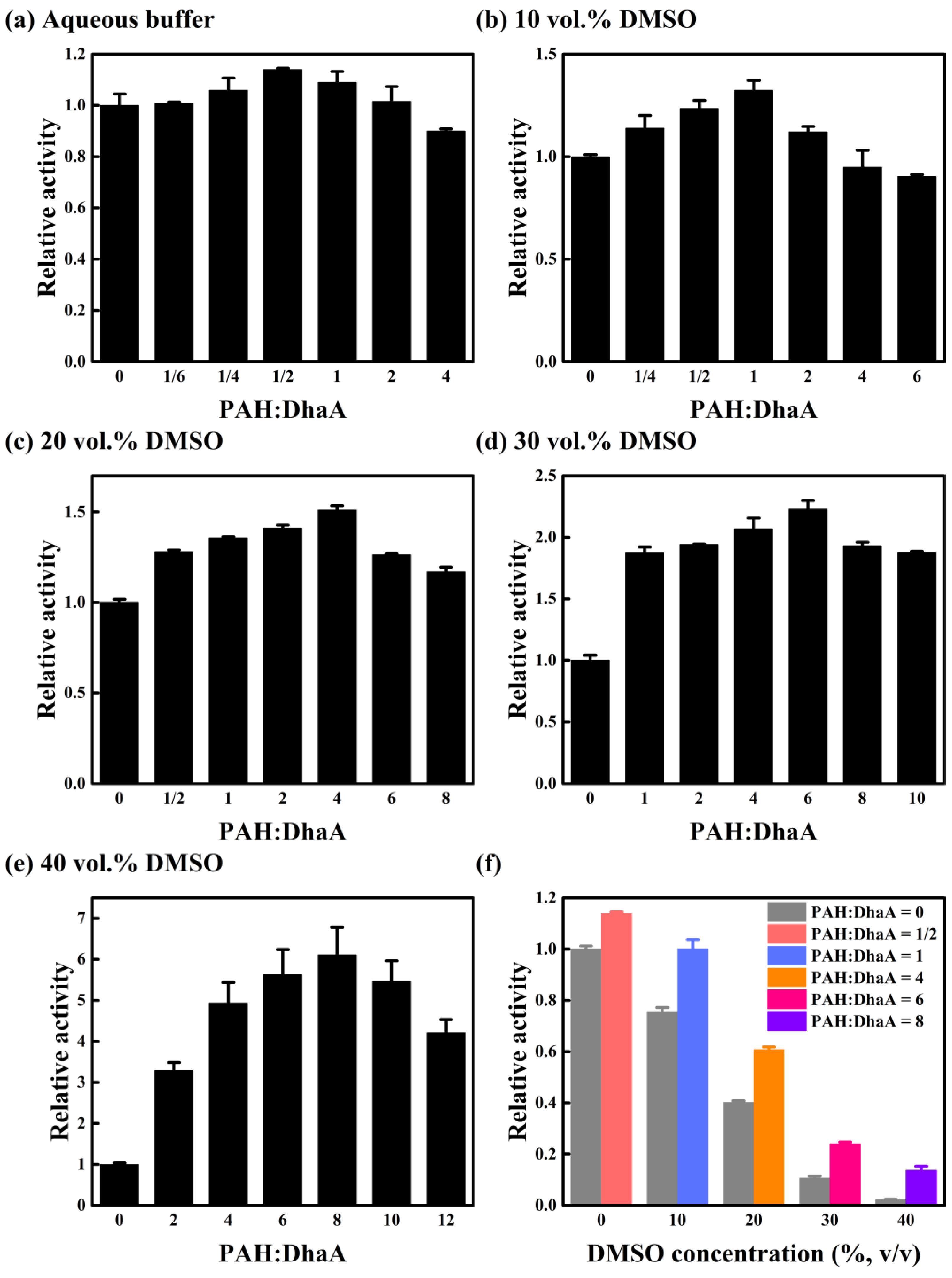
2.1. Effect of PAH Concentration on Catalytic Performance
2.2. Enzymatic Kinetics and Stability in OS Solution
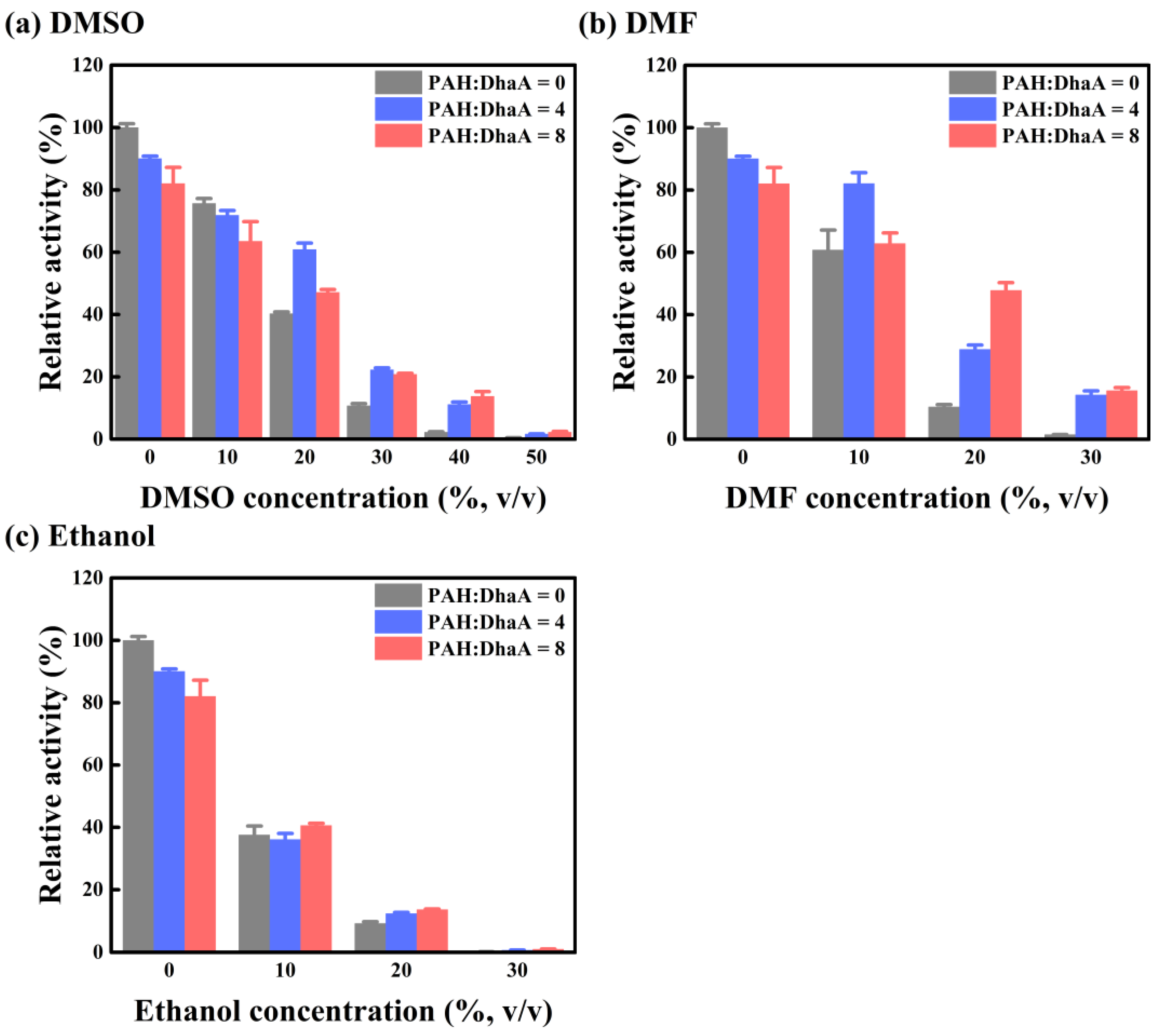
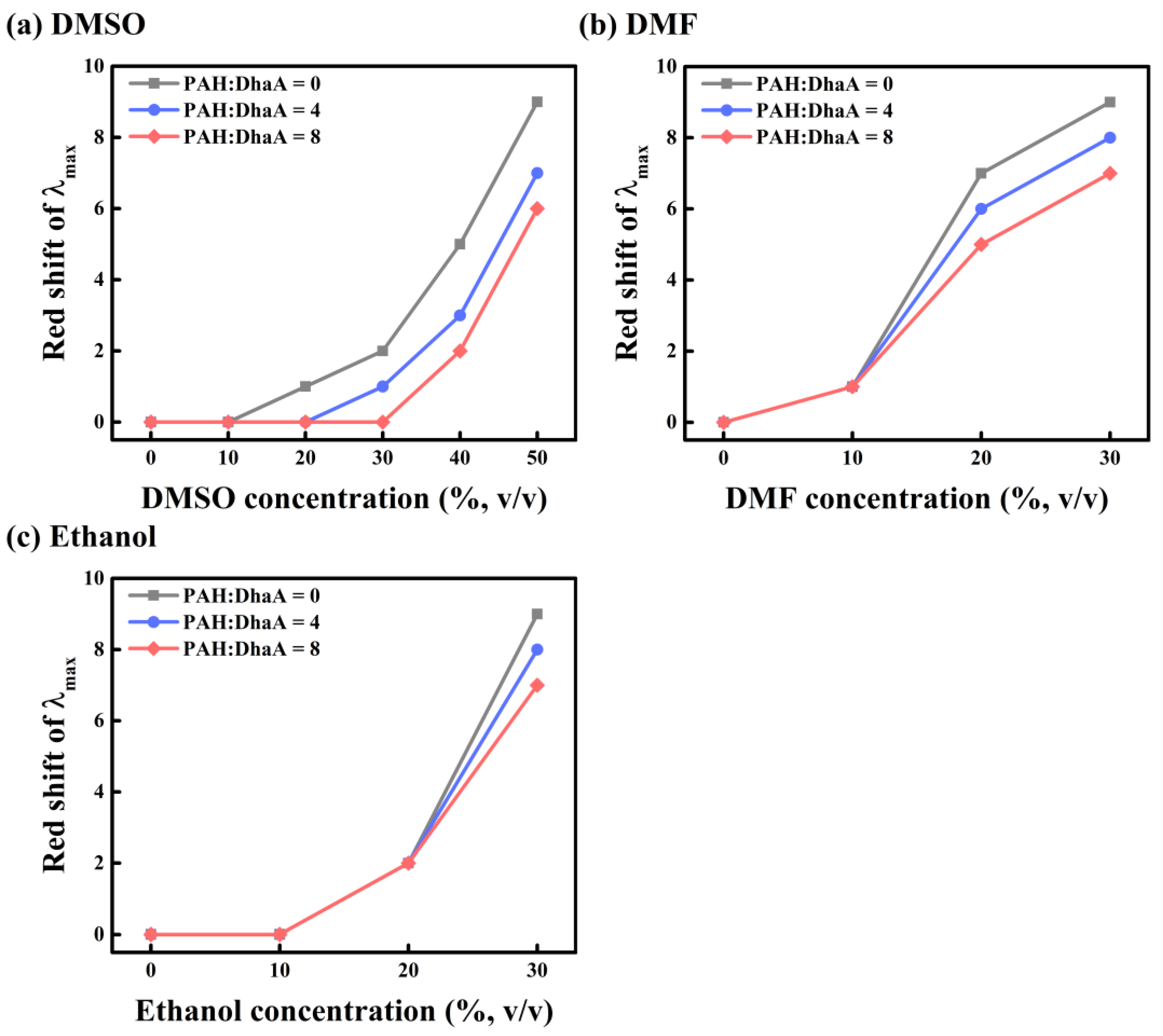
2.3. Enzymatic Performance and Conformational Change in Various OS Solutions
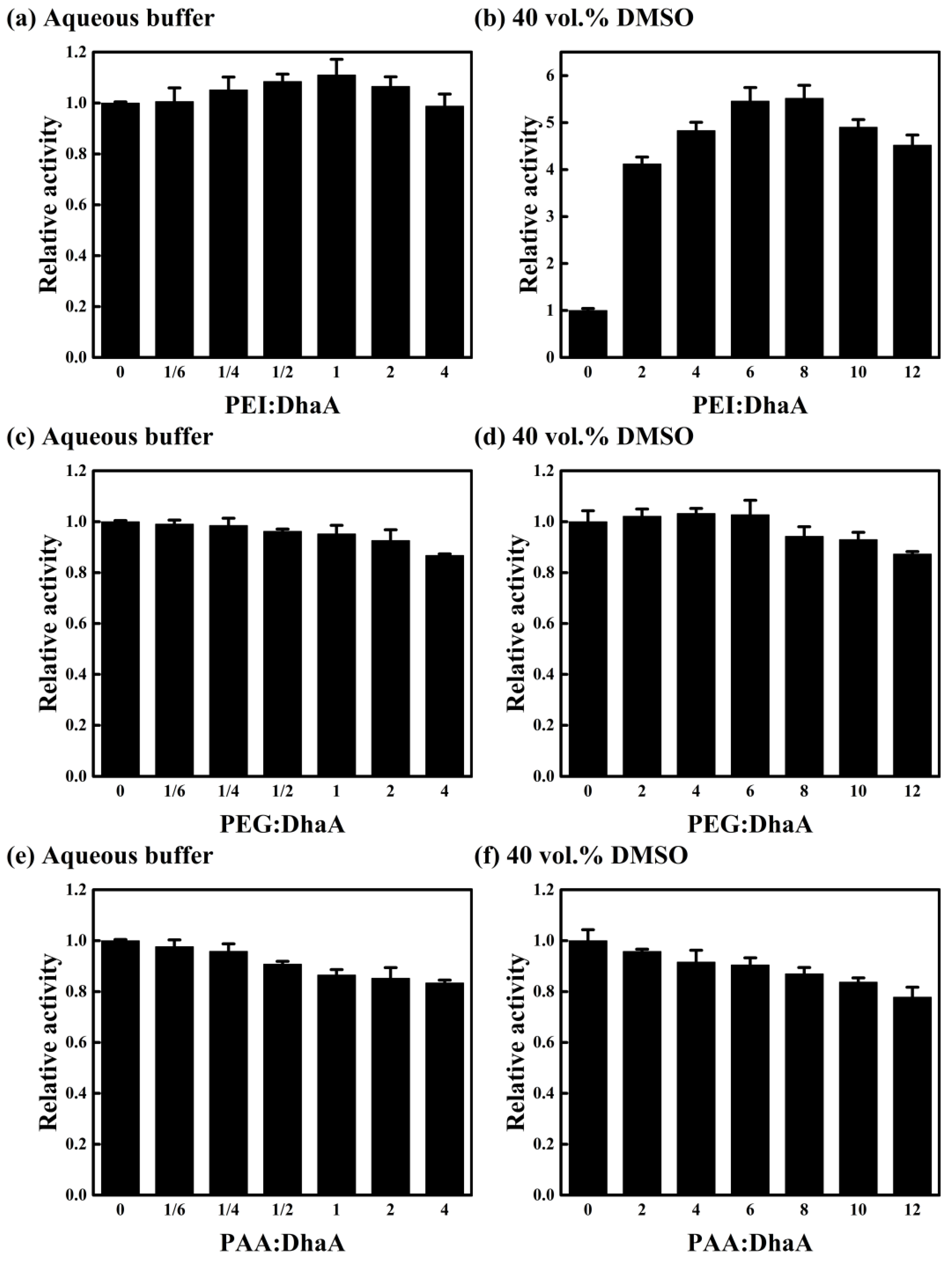
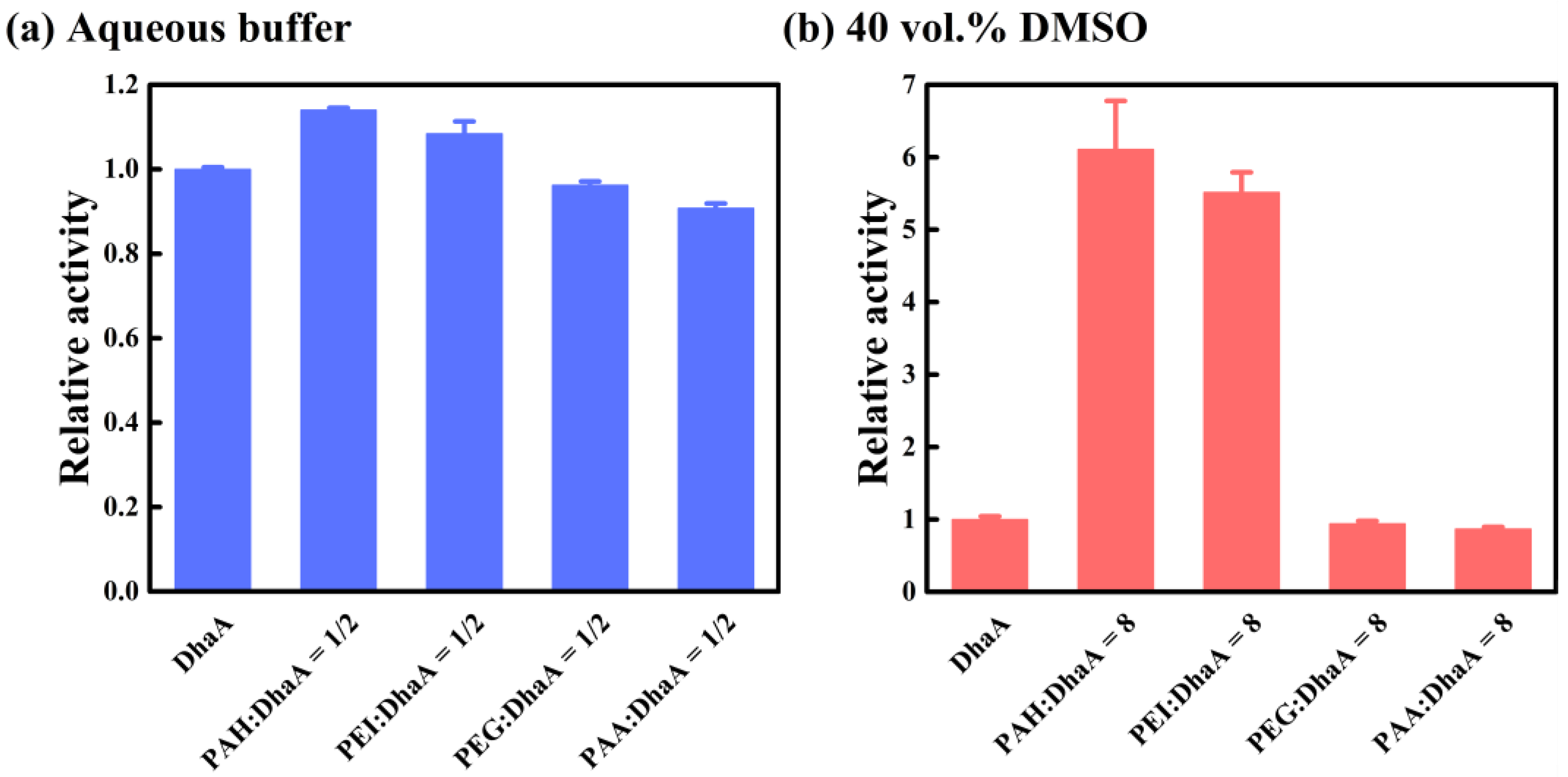
2.4. Effects of Polymer Additives with Different Charges
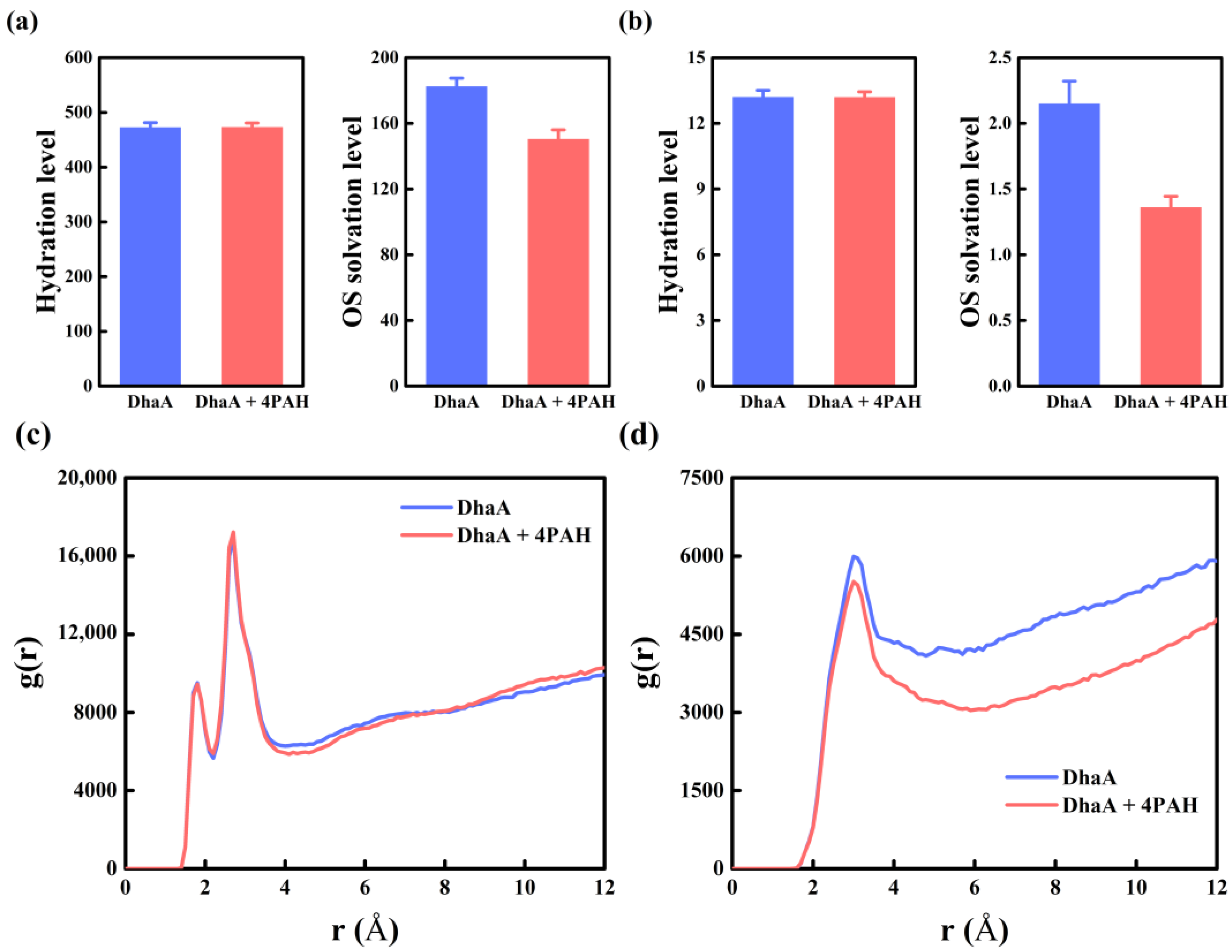
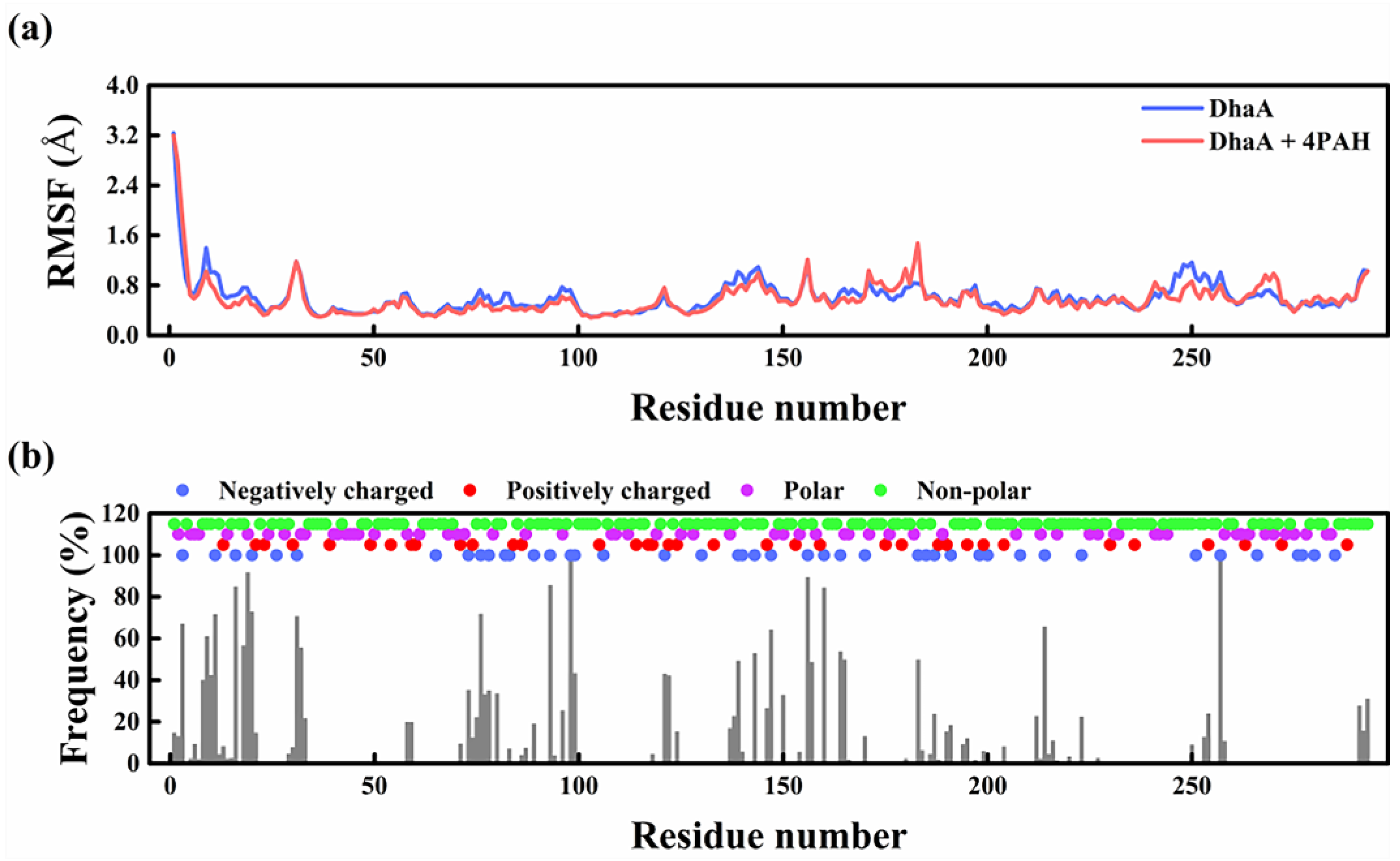
2.5. Molecular Mechanism Underlying the Improved Enzymatic Performance
3. Materials and Methods
3.1. Materials
3.2. Enzyme Preparation
3.3. Enzymatic Assays
3.4. Viscosity Assays
3.5. Fluorescence and CD Spectroscopy
3.6. Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Burow, K.R.; Floyd, W.D.; Landon, M.K. Factors affecting 1,2,3-trichloropropane contamination in groundwater in California. Sci. Total Environ. 2019, 672, 324–334. [Google Scholar] [CrossRef]
- Yang, L.; Mei, G.; Yang, Y.; Cui, J.; Peng, S.; Peng, Z.; Cheng, Y. Hexachlorocyclohexane impairs human sperm motility by affecting lysine glutarylation and mitochondrial functions. Food Chem. Toxicol. 2023, 179, 113991. [Google Scholar] [CrossRef]
- Koudelakova, T.; Bidmanova, S.; Dvorak, P.; Pavelka, A.; Chaloupkova, R.; Prokop, Z.; Damborsky, J. Haloalkane dehalogenases: Biotechnological applications. Biotechnol. J. 2013, 8, 32–45. [Google Scholar] [CrossRef]
- Koudelakova, T.; Chovancova, E.; Brezovsky, J.; Monincova, M.; Fortova, A.; Jarkovsky, J.; Damborsky, J. Substrate specificity of haloalkane dehalogenases. Biochem. J. 2011, 435, 345–354. [Google Scholar] [CrossRef]
- Janssen, D.B. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 2004, 8, 150–159. [Google Scholar] [CrossRef]
- Newman, J.; Peat, T.S.; Richard, R.; Kan, L.; Swanson, P.E.; Affholter, J.A.; Holmes, I.H.; Schindler, J.F.; Unkefer, C.J.; Terwilliger, T.C. Haloalkane dehalogenases: structure of a Rhodococcus enzyme. Biochemistry 1999, 38, 16105–16114. [Google Scholar] [CrossRef]
- Xue, F.; Li, C.; Xu, Q. Biocatalytic approaches for the synthesis of optically pure vic-halohydrins. Appl. Microbiol. Biotechnol. 2021, 105, 3411–3421. [Google Scholar] [CrossRef]
- Gul, I.; Le, W.; Jie, Z.; Ruiqin, F.; Bilal, M.; Tang, L. Recent advances on engineered enzyme-conjugated biosensing modalities and devices for halogenated compounds. TrAC Trends Anal. Chem. 2021, 134, 116145. [Google Scholar] [CrossRef]
- Mazzucchelli, S.; Colombo, M.; Verderio, P.; Rozek, E.; Andreata, F.; Galbiati, E.; Tortora, P.; Corsi, F.; Prosperi, D. Orientation-controlled conjugation of haloalkane dehalogenase fused homing peptides to multifunctional nanoparticles for the specific recognition of cancer cells. Angew. Chem. Int. Ed. 2013, 52, 3121–3125. [Google Scholar] [CrossRef]
- Prokop, Z.; Opluštil, F.; DeFrank, J.; Damborský, J. Enzymes fight chemical weapons. Biotechnol. J. 2006, 1, 1370–1380. [Google Scholar] [CrossRef]
- Stepankova, V.; Damborsky, J.; Chaloupkova, R. Organic co-solvents affect activity, stability and enantioselectivity of haloalkane dehalogenases. Biotechnol. J. 2013, 8, 719–729. [Google Scholar] [CrossRef]
- Stepankova, V.; Khabiri, M.; Brezovsky, J.; Pavelka, A.; Sykora, J.; Amaro, M.; Minofar, B.; Prokop, Z.; Hof, M.; Ettrich, R.; et al. Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents. ChemBioChem 2013, 14, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Z.; Yu, W.-L.; Zheng, H.; Guo, X.; Guo, N.; Hu, T.; Zhong, J.-Y. PEGylation with the thiosuccinimido butylamine linker significantly increases the stability of haloalkane dehalogenase DhaA. J. Biotechnol. 2017, 254, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ming, X.; Guo, Z.; Shi, Y.; Li, M.; Guo, Z.; Xin, Y.; Gu, Z.; Zhang, L.; Guo, X. Improvement in the environmental stability of haloalkane dehalogenase with self-assembly directed nano-hybrid with iron phosphate. Catalysts 2022, 12, 825. [Google Scholar] [CrossRef]
- Koudelakova, T.; Chaloupkova, R.; Brezovsky, J.; Prokop, Z.; Sebestova, E.; Hesseler, M.; Khabiri, M.; Plevaka, M.; Kulik, D.; Kuta Smatanova, I.; et al. Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel. Angew. Chem. Int. Ed. 2013, 52, 1959–1963. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, Y. Rational surface charge engineering of haloalkane dehalogenase for boosting the enzymatic performance in organic solvent solutions. Chin. J. Chem. Eng. 2023; in press. [Google Scholar]
- Behera, S.; Balasubramanian, S. Molecular simulations explain the exceptional thermal stability, solvent tolerance and solubility of protein–polymer surfactant bioconjugates in ionic liquids. Phys. Chem. Chem. Phys. 2022, 24, 21904–21915. [Google Scholar] [CrossRef]
- Suthiwangcharoen, N.; Nagarajan, R. Enhancing enzyme stability by construction of polymer–enzyme conjugate micelles for decontamination of organophosphate agents. Biomacromolecules 2014, 15, 1142–1152. [Google Scholar] [CrossRef]
- Tomita, S.; Nagasaki, Y.; Shiraki, K. Different mechanisms of action of poly(ethylene glycol) and arginine on thermal inactivation of lysozyme and ribonuclease A. Biotechnol. Bioeng. 2012, 109, 2543–2552. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Sun, Y.; Dong, X. Complicated effects of a zwitterionic polymer containing dimethyl chains on the structures, activities and stabilities of different enzymes. Biochem. Eng. J. 2021, 165, 107813. [Google Scholar] [CrossRef]
- Thiele, M.J.; Davari, M.D.; König, M.; Hofmann, I.; Junker, N.O.; Mirzaei Garakani, T.; Vojcic, L.; Fitter, J.; Schwaneberg, U. Enzyme–polyelectrolyte complexes boost the catalytic performance of enzymes. ACS Catal. 2018, 8, 10876–10887. [Google Scholar] [CrossRef]
- Yasuda, M.; Nikaido, H.; Glomm, W.R.; Ogino, H.; Ishimi, K.; Ishikawa, H. Enzyme immobilization on amphiphilic polymer particles having grafted polyionic polymer chains. Biochem. Eng. J. 2009, 48, 6–12. [Google Scholar] [CrossRef]
- Wilson, L.; Illanes, A.; Abián, O.; Pessela, B.C.C.; Fernández-Lafuente, R.; Guisán, J.M. Co-aggregation of penicillin G acylase and polyionic polymers: an easy methodology to prepare enzyme biocatalysts stable in organic media. Biomacromolecules 2004, 5, 852–857. [Google Scholar] [CrossRef]
- Betancor, L.; Fuentes, M.; Dellamora-Ortiz, G.; López-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Mateo, C.; Guisán, J.M.; Fernández-Lafuente, R. Dextran aldehyde coating of glucose oxidase immobilized on magnetic nanoparticles prevents its inactivation by gas bubbles. J. Mol. Catal. B Enzym. 2005, 32, 97–101. [Google Scholar] [CrossRef]
- Derham, B.K.; Harding, J.J. The effect of the presence of globular proteins and elongated polymers on enzyme activity. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2006, 1764, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Prud’homme, R.K. Reaction-diffusion of enzyme molecules in biopolymer matrices. Biocatal. Polym. Sci. 2003, 840, 265–284. [Google Scholar] [CrossRef]
- Magsumov, T.; Fatkhutdinova, A.; Mukhametzyanov, T.; Sedov, I. The effect of dimethyl sulfoxide on the lysozyme unfolding kinetics, thermodynamics, and mechanism. Biomolecules 2019, 9, 547. [Google Scholar] [CrossRef]
- Salehi, F.; Emamzadeh, R.; Nazari, M.; Rasa, S.M.M. Probing the emitter site of Renilla luciferase using small organic molecules; an attempt to understand the molecular architecture of the emitter site. Int. J. Biol. Macromol. 2016, 93, 1253–1260. [Google Scholar] [CrossRef]
- Prasad, A.R.S.; Luduena, R.F.; Horowitz, P.M. Detection of energy transfer between tryptophan residues in the tubulin molecule and bound bis(8-anilinonaphthalene-1-sulfonate), an inhibitor of microtubule assembly, that binds to a flexible region on tubulin. Biochemistry 1986, 25, 3536–3540. [Google Scholar] [CrossRef]
- Wang, M.; Cui, H.; Gu, C.; Li, A.; Qiao, J.; Schwaneberg, U.; Zhang, L.; Wei, J.; Li, X.; Huang, H. Engineering all-round cellulase for bioethanol production. ACS Synth. Biol. 2023, 12, 2187–2197. [Google Scholar] [CrossRef]
- Cui, H.; Vedder, M.; Schwaneberg, U.; Davari, M.D. Using molecular simulation to guide protein engineering for biocatalysis in organic solvents. In Enzyme Engineering: Methods in Molecular Biology; Magnani, F., Marabelli, C., Paradisi, F., Eds.; Springer: New York, NY, USA, 2022; pp. 179–202. [Google Scholar] [CrossRef]
- Cui, H.; Vedder, M.; Zhang, L.; Jaeger, K.-E.; Schwaneberg, U.; Davari, M.D. Polar substitutions on the surface of a lipase substantially improve tolerance in organic solvents. ChemSusChem 2022, 15, e202102551. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, Y. Co-encapsulating cofactor and enzymes in hydrogen-bonded organic frameworks for multienzyme cascade reactions with cofactor recycling. Angew. Chem. Int. Ed. 2023, e202308562. [Google Scholar] [CrossRef]
- Chen, K.; Dong, X.; Sun, Y. Sequentially co-immobilized PET and MHET hydrolases via Spy chemistry in calcium phosphate nanocrystals present high-performance PET degradation. J. Hazard. Mater. 2022, 438, 129517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, X.; Guo, Z.; Sun, Y. Remarkably enhanced activity and substrate affinity of lipase covalently bonded on zwitterionic polymer-grafted silica nanoparticles. J. Colloid Interface Sci. 2018, 519, 145–153. [Google Scholar] [CrossRef]
- Dockalova, V.; Sanchez-Carnerero, E.M.; Dunajova, Z.; Palao, E.; Slanska, M.; Buryska, T.; Damborsky, J.; Klan, P.; Prokop, Z. Fluorescent substrates for haloalkane dehalogenases: Novel probes for mechanistic studies and protein labeling. Comput. Struct. Biotechnol. J. 2020, 18, 922–932. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Y.; Dong, X.; Sun, Y. Molecular insights into the enhanced performance of EKylated PETase toward PET degradation. ACS Catal. 2021, 11, 7358–7370. [Google Scholar] [CrossRef]
- Chakraborty, A.; Ghosh, R.; Biswas, A. Interaction of constituents of MDT regimen for leprosy with Mycobacterium leprae HSP18: Impact on its structure and function. FEBS J. 2022, 289, 832–853. [Google Scholar] [CrossRef]
- Chakraborty, A.; Biswas, A. Structure, stability and chaperone function of Mycobacterium leprae Heat Shock Protein 18 are differentially affected upon interaction with gold and silver nanoparticles. Int. J. Biol. Macromol. 2020, 152, 250–260. [Google Scholar] [CrossRef]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. [Google Scholar] [CrossRef]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. Best bang for your buck: GPU nodes for GROMACS biomolecular simulations. J. Comput. Chem. 2015, 36, 1990–2008. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Lahoda, M.; Mesters, J.R.; Stsiapanava, A.; Chaloupkova, R.; Kuty, M.; Damborsky, J.; Kuta Smatanova, I. Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31. Acta Crystallogr. Sect. D 2014, 70, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
| PAH:DhaA | Km (μM) | kcat × 103 (s−1) | kcat/Km × 103 (s−1μM−1) |
|---|---|---|---|
| 0:1 | 75.7 ± 5.2 | 16 ± 1 | 0.21 ± 0.01 |
| 4:1 | 83.9 ± 8.3 | 84 ± 5 | 1.00 ± 0.05 |
| 8:1 | 91.0 ± 7.3 | 107 ± 5 | 1.17 ± 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Sun, Y. Cationic Polymers Remarkably Boost Haloalkane Dehalogenase Activity in Organic Solvent Solutions and the Molecular Implications. Molecules 2023, 28, 6795. https://doi.org/10.3390/molecules28196795
Wu Y, Sun Y. Cationic Polymers Remarkably Boost Haloalkane Dehalogenase Activity in Organic Solvent Solutions and the Molecular Implications. Molecules. 2023; 28(19):6795. https://doi.org/10.3390/molecules28196795
Chicago/Turabian StyleWu, Yin, and Yan Sun. 2023. "Cationic Polymers Remarkably Boost Haloalkane Dehalogenase Activity in Organic Solvent Solutions and the Molecular Implications" Molecules 28, no. 19: 6795. https://doi.org/10.3390/molecules28196795






