The Genomic-Driven Discovery of Glutarimide-Containing Derivatives from Burkholderia gladioli
Abstract
:1. Introduction
2. Results
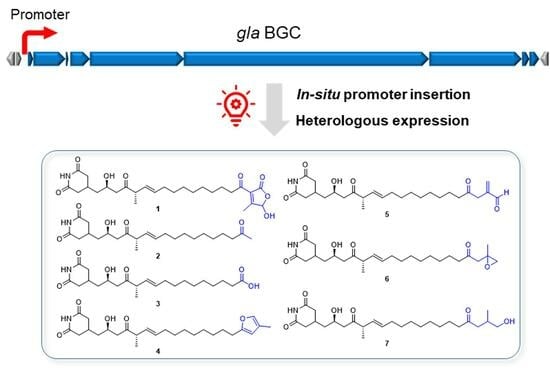
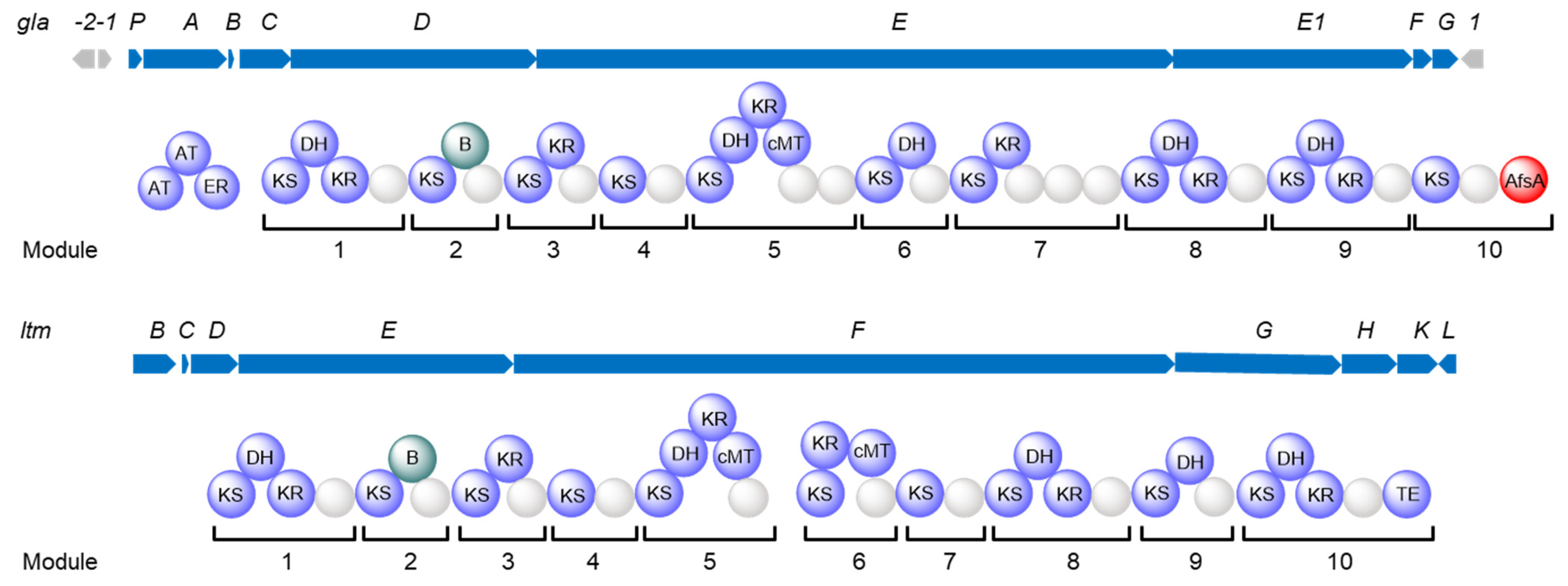
2.1. Characterization of the Silent trans-AT Biosynthetic Gene Cluster
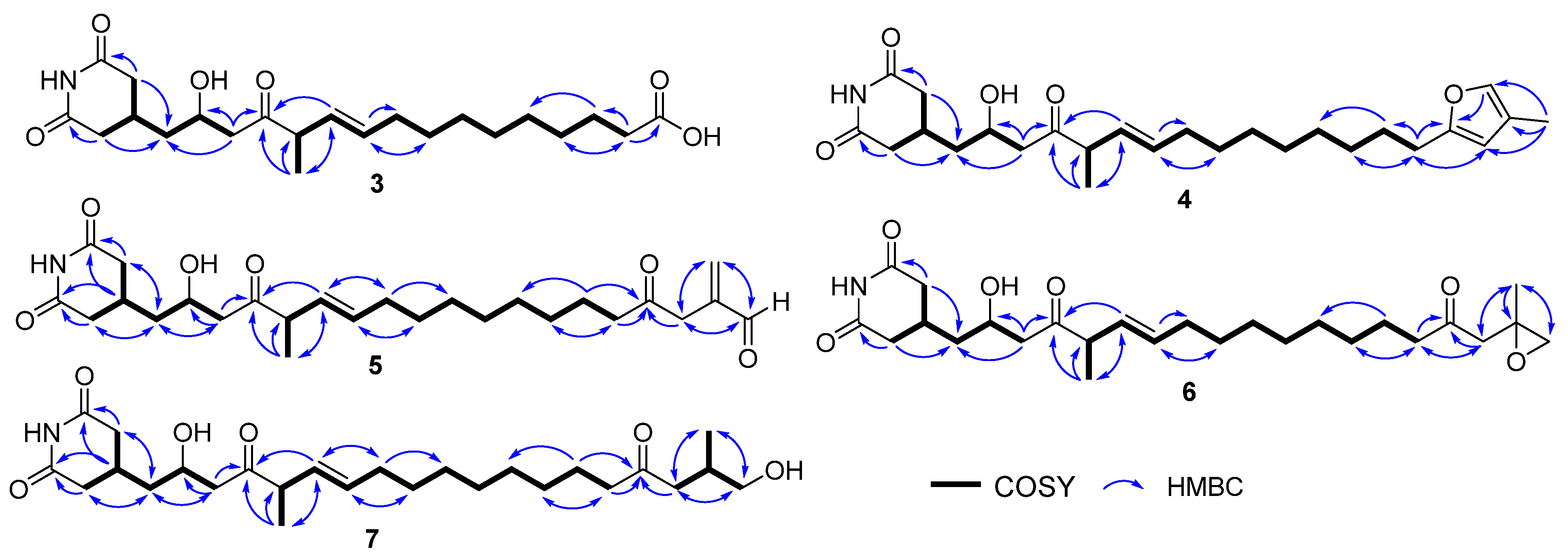
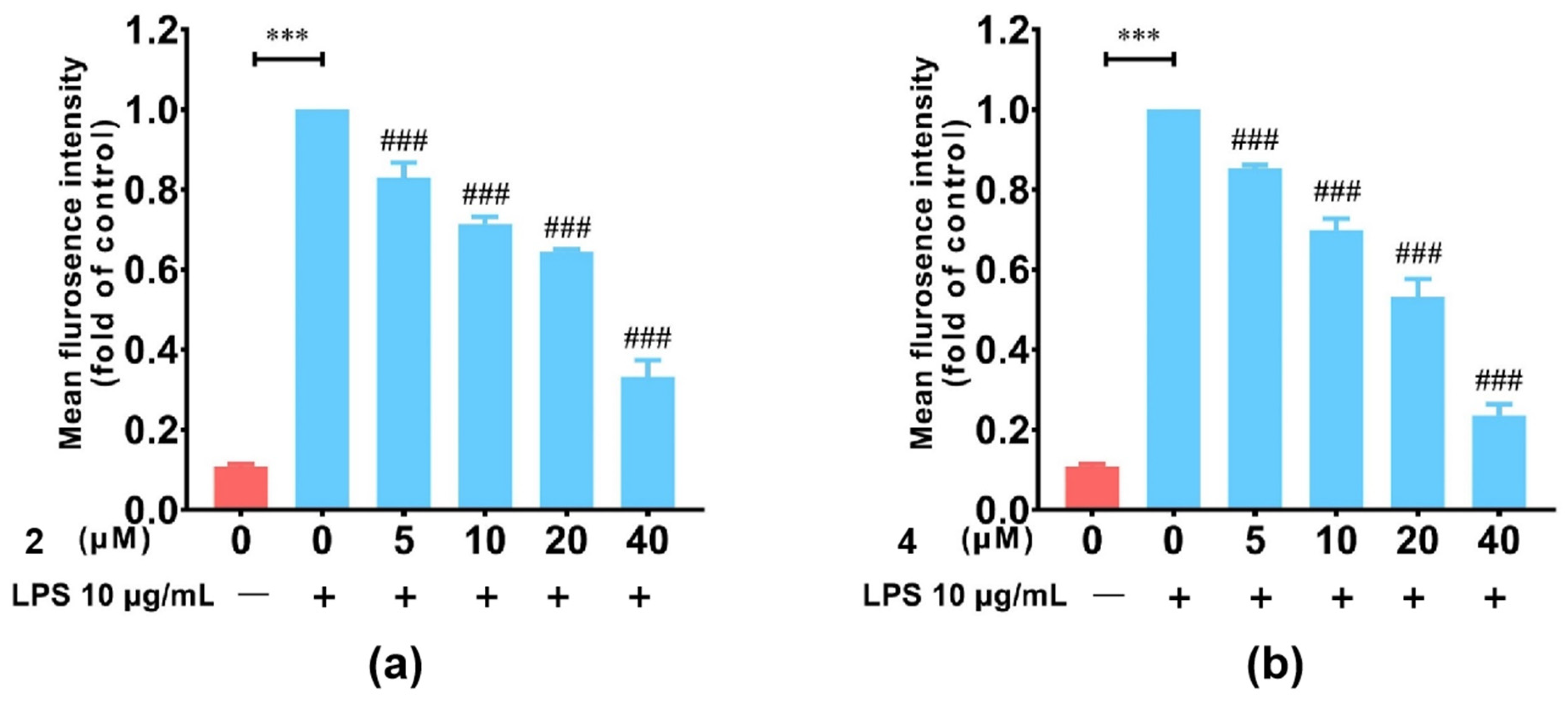
2.2. Structural Elucidation and Bioactivities Assay of Compounds 1–7
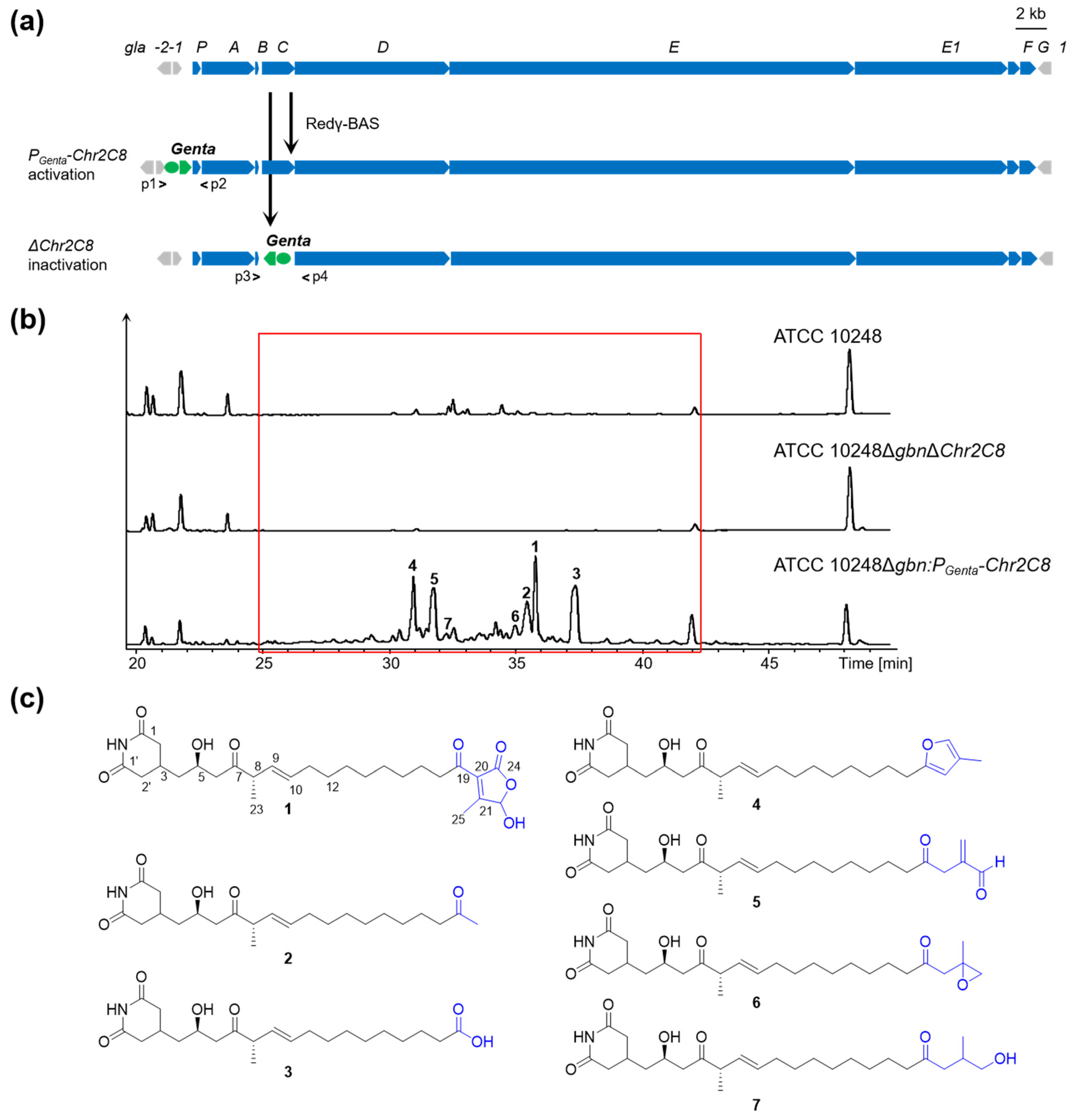
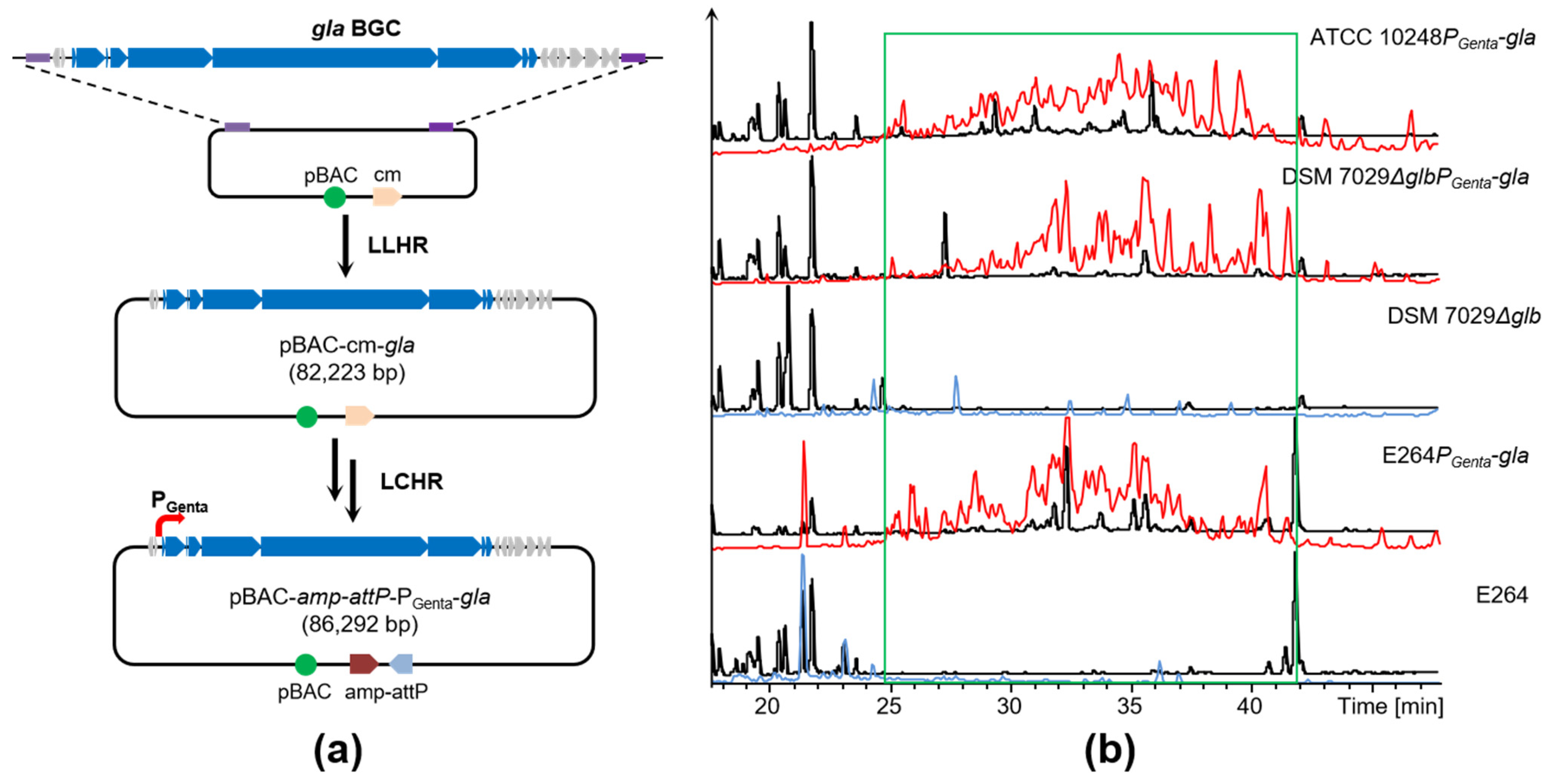
2.3. Direct Cloning and Heterologous Expression of the gla BGC
3. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sugawara, K.; Nishiyama, Y.; Toda, S.; Komiyama, N.; Hatori, M.; Moriyama, T.; Sawada, Y.; Kamei, H.; Konishi, M.; Oki, T. Lactimidomycin, a new glutarimide group antibiotic. Production, isolation, structure and biological activity. J. Antibiot. 1992, 45, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.H.; Seo, J.W.; Her, Y.; Lim, S.K.; Shen, B. New lactimidomycin congeners shed insight into lactimidomycin biosynthesis in Streptomyces amphibiosporus. Org. Lett. 2007, 9, 5183–5186. [Google Scholar] [CrossRef] [PubMed]
- Nakae, K.; Yoshimoto, Y.; Sawa, T.; Homma, Y.; Hamada, M.; Takeuchi, T.; Imoto, M. Migrastatin, a new inhibitor of tumor cell migration from Streptomyces sp. MK929-43F1. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2000, 53, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Woo, E.J.; Starks, C.M.; Carney, J.R.; Arslanian, R.; Cadapan, L.; Zavala, S.; Licari, P. Migrastatin and a new compound, isomigrastatin, from Streptomyces Platensis. J. Antibiot. 2002, 55, 141–146. [Google Scholar] [CrossRef]
- Yin, M.; Yan, Y.J.; Lohman, J.R.; Huang, S.X.; Ma, M.; Zhao, G.R.; Xu, L.H.; Xiang, W.S.; Shen, B. Cycloheximide and actiphenol production in Streptomyces sp. YIM56141 governed by single biosynthetic machinery featuring an acyltransferase-less type I polyketide synthase. Org. Lett. 2014, 16, 3072–3075. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Moon, S.S.; Hwang, B.K. Isolation, antifungal activity, and structure elucidation of the glutarimide antibiotic, streptimidone, produced by Micromonospora coerulea. J. Agric. Food. Chem. 1999, 47, 3372–3380. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.L.; Wang, H.; Xue, Z.L.; Li, J.S.; Qi, H.; Zhang, H.; Zhao, T.; Wang, J.D.; Xiang, W.S. Two new glutarimide antibiotics from Streptomyces sp. HS-NF-780. J. Antibiot. 2019, 72, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Son, S.; Lee, J.K.; Jang, M.; Heo, K.T.; Ko, S.K.; Park, D.J.; Park, C.S.; Kim, C.J.; Ahn, J.S.; et al. Isolation of new streptimidone derivatives, glutarimide antibiotics from Streptomyces sp. W3002 using LC-MS-guided screening. J. Antibiot. 2020, 73, 184–188. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Ueoka, R.; Chevrette, M.G.; Hemmerling, F.; Lu, X.W.; Leopold-Messer, S.; Minas, H.A.; Burch, A.Y.; Lindow, S.E.; Piel, J.; et al. Evolution of combinatorial diversity in trans-acyltransferase polyketide synthase assembly lines across bacteria. Nat. Commun. 2021, 12, 1422. [Google Scholar] [CrossRef]
- Niehs, S.P.; Kumpfmüller, J.; Dose, B.; Little, R.F.; Ishida, K.; Flórez, L.V.; Kaltenpoth, M.; Hertweck, C. Insect-associated bacteria assemble the antifungal butenolide gladiofungin by non-canonical polyketide chain termination. Angew. Chem. Int. Ed. Engl. 2020, 59, 23122–23126. [Google Scholar] [CrossRef]
- Nakou, I.T.; Jenner, M.; Dashti, Y.; Romero-Canelón, I.; Masschelein, J.; Mahenthiralingam, E.; Challis, G.L. Genomics-driven discovery of a novel glutarimide antibiotic from Burkholderia gladioli reveals an unusual polyketide synthase chain release mechanism. Angew. Chem. Int. Ed. Engl. 2020, 59, 23145–23153. [Google Scholar] [CrossRef]
- Kačar, D.; Cañedo, L.M.; Rodríguez, P.; González, E.G.; Galán, B.; Schleissner, C.; Leopold-Messer, S.; Piel, J.; Cuevas, C.; Calle, F.; et al. Identification of trans-AT polyketide clusters in two marine bacteria reveals cryptic similarities between distinct symbiosis factors. Environ. Microbiol. 2021, 23, 2509–2521. [Google Scholar] [CrossRef]
- Awal, R.P.; Haack, P.A.; Bader, C.D.; Riese, C.N.; Schüler, D.; Müller, R. Sesbanimide, R, a novel cytotoxic polyketide produced by Magnetotactic Bacteria. mBio 2021, 12, e00591-21. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, E.J.N.; Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016, 33, 231–316. [Google Scholar] [CrossRef]
- Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2010, 27, 996–1047. [Google Scholar] [CrossRef] [PubMed]
- Kosol, S.; Jenner, M.; Lewandowski, J.R.; Challis, G.L. Protein-protein interactions in trans-AT polyketide synthases. Nat. Prod. Rep. 2018, 35, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Haines, A.S.; Dong, X.; Song, Z.S.; Farmer, R.; Williams, C.; Hothersall, J.; Płoskoń, E.; Wattana-Amorn, P.; Stephens, E.R.; Yamada, E.; et al. A conserved motif flags acyl carrier proteins for β-branching in polyketide synthesis. Nat. Chem. Biol. 2013, 9, 685–692. [Google Scholar] [CrossRef]
- Seo, J.W.; Ma, M.; Kwong, T.; Ju, J.H.; Lim, S.K.; Jiang, H.; Lohman, J.R.; Yang, C.Y.; Cleveland, J.; Zazopoulos, E.; et al. Comparative characterization of the lactimidomycin and iso-migrastatin biosynthetic machineries revealing unusual features for acyltransferase-less type I polyketide synthases and providing an opportunity to engineer new analogues. Biochemistry 2014, 53, 7854–7865. [Google Scholar] [CrossRef]
- Heine, D.; Bretschneider, T.; Sundaram, S.; Hertweck, C. Enzymatic polyketide chain branching to give substituted lactone, lactam, and glutarimide heterocycles. Angew Chem. Int. Ed. Engl. 2014, 53, 11645–11649. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef]
- Li, L. Next-generation synthetic biology approaches for the accelerated discovery of microbial natural products. Eng. Microbiol. 2023, 3, 100060. [Google Scholar] [CrossRef]
- Covington, B.C.; Xu, F.; Seyedsayamdost, M.R. A Natural product chemist’s guide to unlocking silent biosynthetic gene clusters. Annu. Rev. Biochem. 2021, 90, 763–788. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, H.B.; Chen, H.N.; Jing, X.S.; Zheng, W.T.; Li, R.J.; Sun, T.; Liu, J.Q.; Fu, J.; Huo, L.J.; et al. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.N.; Sun, T.; Bai, X.P.; Yang, J.; Yan, F.; Yu, L.; Tu, Q.; Li, A.Y.; Zhang, Y.M.; Bian, X.Y.; et al. Genomics-driven activation of silent biosynthetic gene clusters in Burkholderia gladioli by screening recombineering system. Molecules 2021, 26, 700. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Zhou, H.B.; Ren, X.M.; Chen, H.N.; Zhong, L.; Bai, X.P.; Bian, X.Y. Recombineering enables genome mining of novel siderophores in a non-model Burkholderiales strain. Eng. Microbiol. 2023, 3, 100106. [Google Scholar] [CrossRef]
- Huo, L.J.; Hug, J.J.; Fu, C.Z.; Bian, X.Y.; Zhang, Y.M.; Müller, R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436. [Google Scholar] [CrossRef]
- Zhang, J.J.; Tang, X.Y.; Moore, B.S. Genetic platforms for heterologous expression of microbial natural products. Nat. Prod. Rep. 2019, 36, 1313–1332. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Göker, M. Validation List no. 212. Valid publication of new names and new combinations effectively published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 2023, 73, 005709. [Google Scholar] [CrossRef]
- Liu, J.Q.; Zhou, H.B.; Yang, Z.Y.; Wang, X.; Chen, H.N.; Zhong, L.; Zheng, W.T.; Niu, W.J.; Wang, S.; Ren, X.M.; et al. Rational construction of genome-reduced Burkholderiales chassis facilitates efficient heterologous production of natural products from proteobacteria. Nat. Commun. 2021, 12, 4347. [Google Scholar] [CrossRef]
- Liu, X.Y.; Cheng, Y.Q. Genome-guided discovery of diverse natural products from Burkholderia sp. J. Ind. Microbiol. Biotechnol. 2014, 41, 275–284. [Google Scholar] [CrossRef]
- Mao, D.N.; Bushin, L.B.; Moon, K.; Wu, Y.H.; Seyedsayamdost, M.R. Discovery of scmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc. Natl. Acad. Sci. USA 2017, 114, E2920–E2928. [Google Scholar] [CrossRef]
- Kim, H.S.; Schell, M.A.; Yu, Y.; Ulrich, R.L.; Sarria, S.H.; Nierman, W.C.; DeShazer, D. Bacterial genome adaptation to niches: Divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genom. 2005, 6, 174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Liu, X.T.; Zhou, H.B.; Liu, Y.; Zhong, L.; Wang, X.; Tu, Q.; Huo, L.J.; Yan, F.; Gu, L.C.; et al. Engineering of Burkholderia thailandensis strain E264 serves as a chassis for expression of complex specialized metabolites. Front. Microbiol. 2022, 13, 1073243. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Rankin, D.J.; Rocha, E.P.C.; Brown, S.P. What traits are carried on mobile genetic elements, and why? Heredity 2011, 106, 1–10. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.L.; Chen, J.W.; Zhang, H.W.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Li, R.J.; Shi, H.B.; Zhao, X.Y.; Liu, X.Q.; Duan, Q.; Song, C.Y.; Chen, H.N.; Zheng, W.T.; Shen, Q.Y.; Wang, M.Q.; et al. Development and application of an efficient recombineering system for Burkholderia glumae and Burkholderia plantarii. Microb. Biotechnol. 2021, 14, 1809–1826. [Google Scholar] [CrossRef]
- Wang, H.L.; Li, Z.; Jia, R.N.; Yin, J.; Li, A.Y.; Xia, L.Q.; Yin, Y.L.; Müller, R.; Fu, J.; Stewart, A.F.; et al. ExoCET: Exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 2018, 46, e28. [Google Scholar]
- Fu, J.; Teucher, M.; Anastassiadis, K.; Skarnes, W.; Stewart, A.F. A recombineering pipeline to make conditional targeting constructs. Methods Enzymol. 2010, 477, 125–144. [Google Scholar] [PubMed]
- Bonis, B.M.; Gralnick, J.A. Marinobacter subterrani, a genetically tractable neutrophilic Fe (II)-oxidizing strain isolated from the Soudan Iron Mine. Front. Microbiol. 2015, 6, 719. [Google Scholar] [CrossRef] [PubMed]
- Stulberg, E.R.; Lozano, G.L.; Morin, J.B.; Park, H.; Baraban, E.G.; Mlot, C.; Heffelfinger, C.; Phillips, G.M.; Rush, J.S.; Phillips, A.J.; et al. Genomic and secondary metabolite analyses of Streptomyces sp. 2AW provide insight into the evolution of the cycloheximide pathway. Front. Microbiol. 2016, 7, 573. [Google Scholar] [CrossRef]
- Rajski, S.R.; Shen, B. Multifaceted modes of action for the glutarimide-containing polyketides revealed. Chembiochem 2010, 11, 1951–1954. [Google Scholar] [CrossRef]
- Takayasu, Y.; Tsuchiya, K.; Aoyama, T.; Sukenaga, Y. NK30424A and B, novel inhibitors of lipopolysaccharide-induced tumour necrosis factor alpha production, produced by Streptomyces sp. NA30424. J. Antibiot. 2001, 54, 1111–1115. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Tachibana, M.; Matsui, C.; Obata, R.; Umezawa, K.; Nishiyama, S. Synthesis and biological evaluation on novel analogs of 9-methylstreptimidone, an inhibitor of NF-κB. Bioorg. Med. Chem. Lett. 2009, 19, 1726–1728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yi, W.W.; Ge, H.J.; Zhang, Z.Z.; Wu, B. Bioactive streptoglutarimides A-J from the marine-derived Streptomyces sp. ZZ741. J. Nat. Prod. 2019, 82, 2800–2808. [Google Scholar] [CrossRef]
- Ju, J.H.; Lim, S.K.; Jiang, H.; Shen, B. Migrastatin and dorrigocins are shunt metabolites of iso-migrastatin. J. Am. Chem. Soc. 2005, 127, 1622–1623. [Google Scholar] [CrossRef]
- Kato, J.-Y.; Funa, N.; Watanabe, H.; Ohnishi, Y.; Horinouchi, S. Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl. Acad Sci. USA 2007, 104, 2378–2383. [Google Scholar] [CrossRef]
- Braun, D.; Pauli, N.; Séquin, U.; Zähner, H. New butenolides from the photoconductivity screening of Streptomyces antibioticus (Waksman and Woodruff) Waksman and Henrici 1948. FEMS Microbiol. Lett. 1995, 126, 37–42. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T.; Clardy, J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. USA 2008, 105, 4601–4608. [Google Scholar] [CrossRef] [PubMed]
- Ruzzini, A.C.; Clardy, J. Gene flow and molecular innovation in Bacteria. Curr. Biol. 2016, 26, R859–R864. [Google Scholar] [CrossRef]
- Stasiak, M.; Mackiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent genes: Antimicrobial resistance and antibiotic production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Ongley, S.E.; Bian, X.Y.; Zhang, Y.M.; Chau, R.; Gerwick, W.H.; Müller, R.; Neilan, B.A. High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chem. Biol. 2013, 8, 1888–1893. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Li, Z.; Jia, R.N.; Hou, Y.; Yin, J.; Bian, X.Y.; Li, A.Y.; Müller, R.; Stewart, A.F.; Fu, J.; et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc. 2016, 11, 1175–1190. [Google Scholar] [CrossRef] [PubMed]
| No | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|
| 2a | 2.77, m a | 2.76, m a | 2.76, m a | 2.77, m a | 2.76, m a |
| 2b | 2.33, m a | 2.33, m a | 2.32, m a | 2.33, m a | 2.33, m a |
| 3 | 2.49, m | 2.47, m | 2.48, m | 2.46, m a | 2.48, m |
| 4a | 1.60, m | 1.60, m a | 1.60, m a | 1.58, m a | 1.60, m |
| 4b | 1.34, m | 1.33, m a | 1.34, m a | 1.34, m | 1.34, m a |
| 5 | 4.10, m | 4.09, m | 4.09, m | 4.09, m | 4.09, m |
| 6a | 2.65, dd (8.8, 18.1) | 2.62, dd (8.8, 18.1) | 2.63, dd (8.5, 18.1) | 2.63, dd (8.7, 18.0) | 2.63, dd (8.7, 18.1) |
| 6b | 2.57, dd (2.8, 18.1) | 2.58, dd (2.8, 18.1) | 2.57, dd (3.1, 18.1) | 2.55, dd (2.7, 18.0) | 2.57, dd (2.9, 18.1) |
| 8 | 3.12, m | 3.11, m | 3.11, m | 3.12, m | 3.12, m |
| 9 | 5.32, dd (8.3, 15.3) | 5.32, dd (8.3, 15.3) | 5.32, dd (8.4, 15.3) | 5.32, dd (8.3, 15.2) | 5.32, dd (8.4, 15.3) |
| 10 | 5.59, dt (6.8, 15.3) | 5.59, dt (6.7, 15.3) | 5.58, dt (6.8, 15.3) | 5.58, dt (6.7, 15.2) | 5.59, dt (6.8, 15.3) |
| 11 | 2.01, m | 2.00, m | 2.00, m | 2.00, m | 2.01, m |
| 12 | 1.27, m a | 1.33, m a | 1.34, m a | 1.26, m a | 1.34, m a |
| 13 | 1.27, m a | 1.27, m a | 1.26, m a | 1.26, m a | 1.26, m a |
| 14 | 1.27, m a | 1.27, m a | 1.26, m a | 1.26, m a | 1.26, m a |
| 15 | 1.27, m a | 1.27, m a | 1.26, m a | 1.26, m a | 1.26, m a |
| 16 | 1.27, m a | 1.27, m a | 1.26, m a | 1.26, m a | 1.26, m a |
| 17 | 1.62, m | 1.60, m a | 1.60, m a | 1.58, m a | 1.55, m |
| 18 | 2.33, t (7.4) | 2.54, t (7.5) | 2.48, t (7.4) | 2.46, m a | 2.41, m |
| 20a | 1.15, d (6.9) | 5.83, s | 3.35, s | 2.79, d (16.6) | 2.53, m |
| 20b | 2.54, d (16.6) | 2.33, m a | |||
| 21 | 2.21, m | ||||
| 22a | 7.04, s | 9.52, s | 3.45, d (11.2) | 3.55, m | |
| 22b | 3.37, d (11.2) | 3.83, m | |||
| 23 | 1.14, d (6.8) | 1.14, d (6.9) | 1.14, d (6.8) | 1.15, d (6.8) | |
| 24a | 1.97, s | 6.37, s | 1.19, s | 0.92, d (7.0) | |
| 24b | 6.22, s | ||||
| 2′a | 2.77, m a | 2.76, m a | 2.76, m a | 2.77, m a | 2.76, m a |
| 2′b | 2.33, m a | 2.33, m a | 2.32, m a | 2.33, m a | 2.33, m a |
| 1-NH | 8.36, s | 8.21, s | 8.08, s | 8.29, s | 8.04, s |
| No | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|
| 1 | 172.7, C | 172.4, C | 172.3, C | 172.5, C | 172.3, C |
| 2 | 38.5, CH2 | 38.5, CH2 | 38.5, CH2 | 38.5, CH2 | 38.5, CH2 |
| 3 | 27.2, CH | 27.2, CH | 27.2, CH | 27.2, CH | 27.2, CH |
| 4 | 40.8, CH2 | 40.8, CH2 | 40.9, CH2 | 40.8, CH2 | 40.8, CH2 |
| 5 | 65.0, CH | 64.9, CH | 64.9, CH | 64.9, CH | 64.9, CH |
| 6 | 47.2, CH2 | 47.2, CH2 | 47.1, CH2 | 47.2, CH2 | 47.1, CH2 |
| 7 | 213.4, C | 213.4, C | 213.3, C | 213.3, C | 213.4, C |
| 8 | 51.0, CH | 51.0, CH | 51.0, CH | 51.0, CH | 51.1, CH |
| 9 | 128.4, CH | 128.3, CH | 128.3, CH | 128.4, CH | 128.4, CH |
| 10 | 134.6, CH | 134.7, CH | 134.6, CH | 134.6, CH | 134.7, CH |
| 11 | 32.6, CH2 | 32.7, CH2 | 32.6, CH2 | 32.6, CH2 | 32.6, CH2 |
| 12 | 29.2, CH2 | 29.3, CH2 | 29.2, CH2 | 29.4, CH2 | 29.1, CH2 |
| 13 | 29.0, CH2 | 29.4, CH2 | 29.2, CH2 | 29.1, CH2 | 29.1, CH2 |
| 14 | 29.2, CH2 | 29.4, CH2 | 29.4, CH2 | 29.3, CH2 | 29.3, CH2 |
| 15 | 29.2, CH2 | 29.3, CH2 | 29.3, CH2 | 29.2, CH2 | 29.3, CH2 |
| 16 | 29.0, CH2 | 29.2, CH2 | 29.1, CH2 | 29.2, CH2 | 29.3, CH2 |
| 17 | 24.8, CH2 | 28.2, CH2 | 23.8, CH2 | 23.5, CH2 | 23.9, CH2 |
| 18 | 33.9, CH2 | 28.2, CH2 | 43.1, CH2 | 44.9, CH2 | 43.6, CH2 |
| 19 | 178.4, C | 156.7, C | 206.9, C | 214.3, C | 212.1, C |
| 20 | 16.0, CH3 | 107.6, CH | 41.7, CH2 | 48.3, CH2 | 47.3, CH2 |
| 21 | 120.5, C | 143.5, C | 72.6, C | 32.1, CH | |
| 22 | 137.3, CH | 193.6, CH | 70.0, CH2 | 68.1, CH2 | |
| 23 | 16.1, CH3 | 16.0, CH3 | 16.0, CH3 | 16.0, CH3 | |
| 24 | 9.9, CH3 | 137.1, CH2 | 24.5, CH3 | 17.1, CH3 | |
| 1′ | 172.5, C | 172.3, C | 172.3, C | 172.4, C | 172.2, C |
| 2′ | 37.2, CH2 | 37.3, CH2 | 37.3, CH2 | 37.3, CH2 | 37.3, CH2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Bai, X.; Sun, T.; Wang, X.; Zhang, Y.; Bian, X.; Zhou, H. The Genomic-Driven Discovery of Glutarimide-Containing Derivatives from Burkholderia gladioli. Molecules 2023, 28, 6937. https://doi.org/10.3390/molecules28196937
Chen H, Bai X, Sun T, Wang X, Zhang Y, Bian X, Zhou H. The Genomic-Driven Discovery of Glutarimide-Containing Derivatives from Burkholderia gladioli. Molecules. 2023; 28(19):6937. https://doi.org/10.3390/molecules28196937
Chicago/Turabian StyleChen, Hanna, Xianping Bai, Tao Sun, Xingyan Wang, Youming Zhang, Xiaoying Bian, and Haibo Zhou. 2023. "The Genomic-Driven Discovery of Glutarimide-Containing Derivatives from Burkholderia gladioli" Molecules 28, no. 19: 6937. https://doi.org/10.3390/molecules28196937
APA StyleChen, H., Bai, X., Sun, T., Wang, X., Zhang, Y., Bian, X., & Zhou, H. (2023). The Genomic-Driven Discovery of Glutarimide-Containing Derivatives from Burkholderia gladioli. Molecules, 28(19), 6937. https://doi.org/10.3390/molecules28196937