Circular Dichroism Study of Orexin B under Oxidative Stress Conditions
Abstract
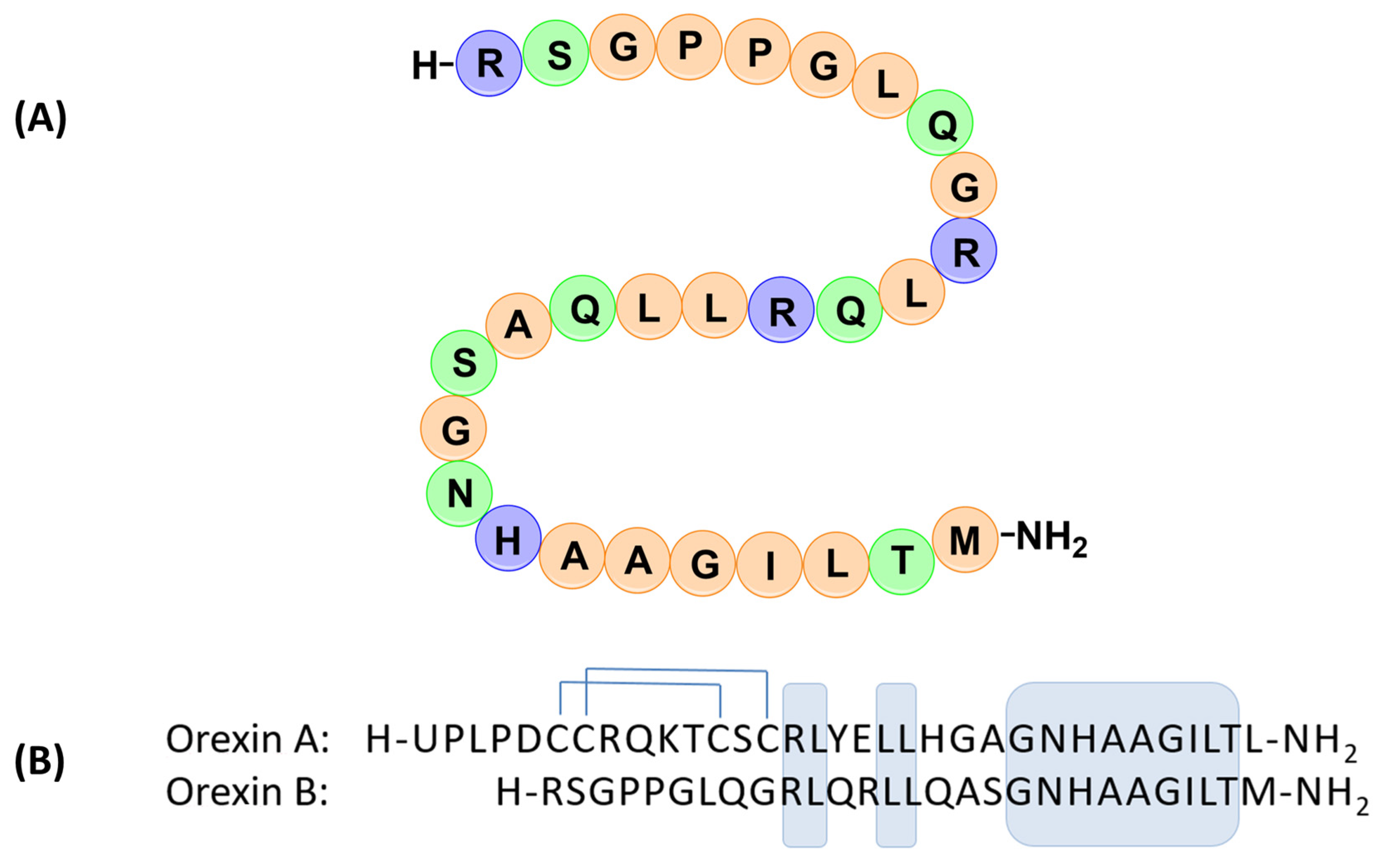
:1. Introduction
2. Results
2.1. Peptide Synthesis
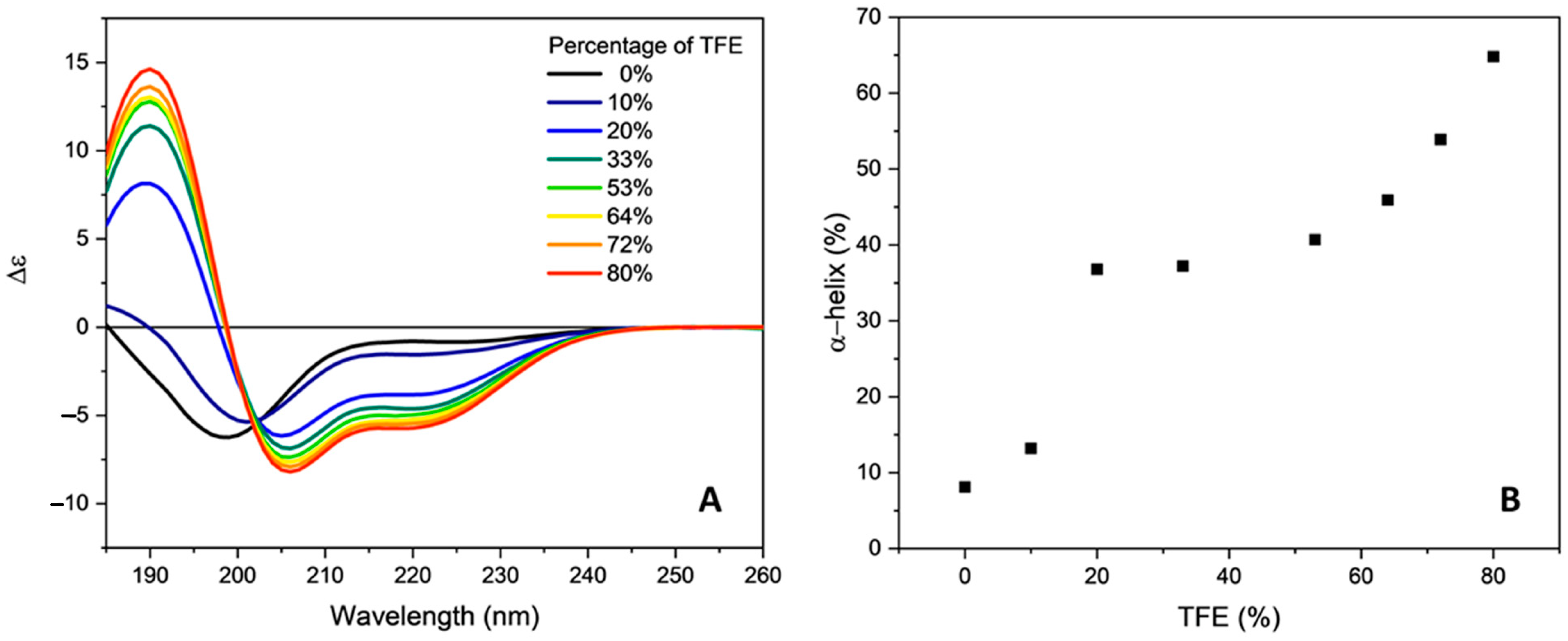
2.2. Conformational Studies
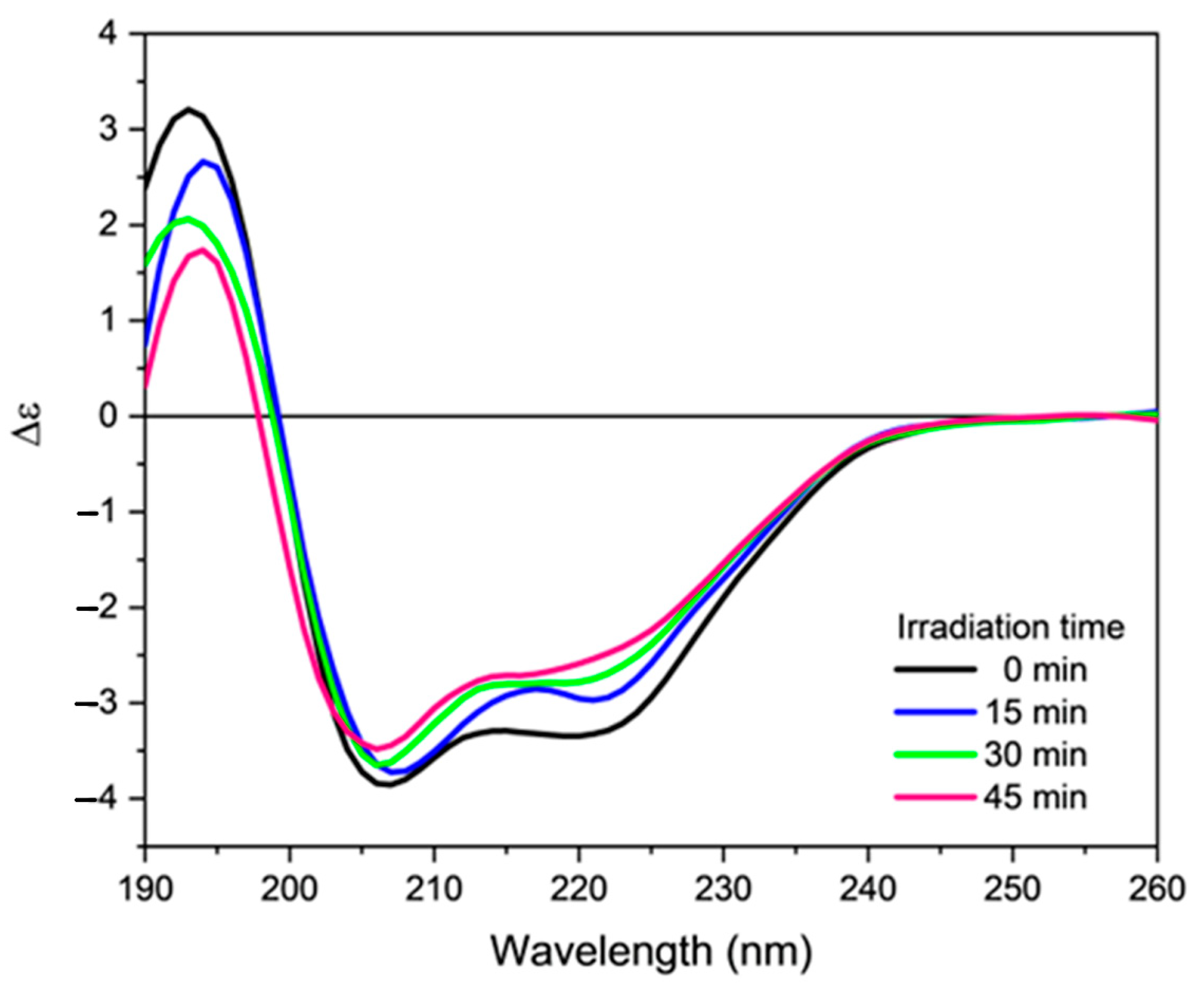
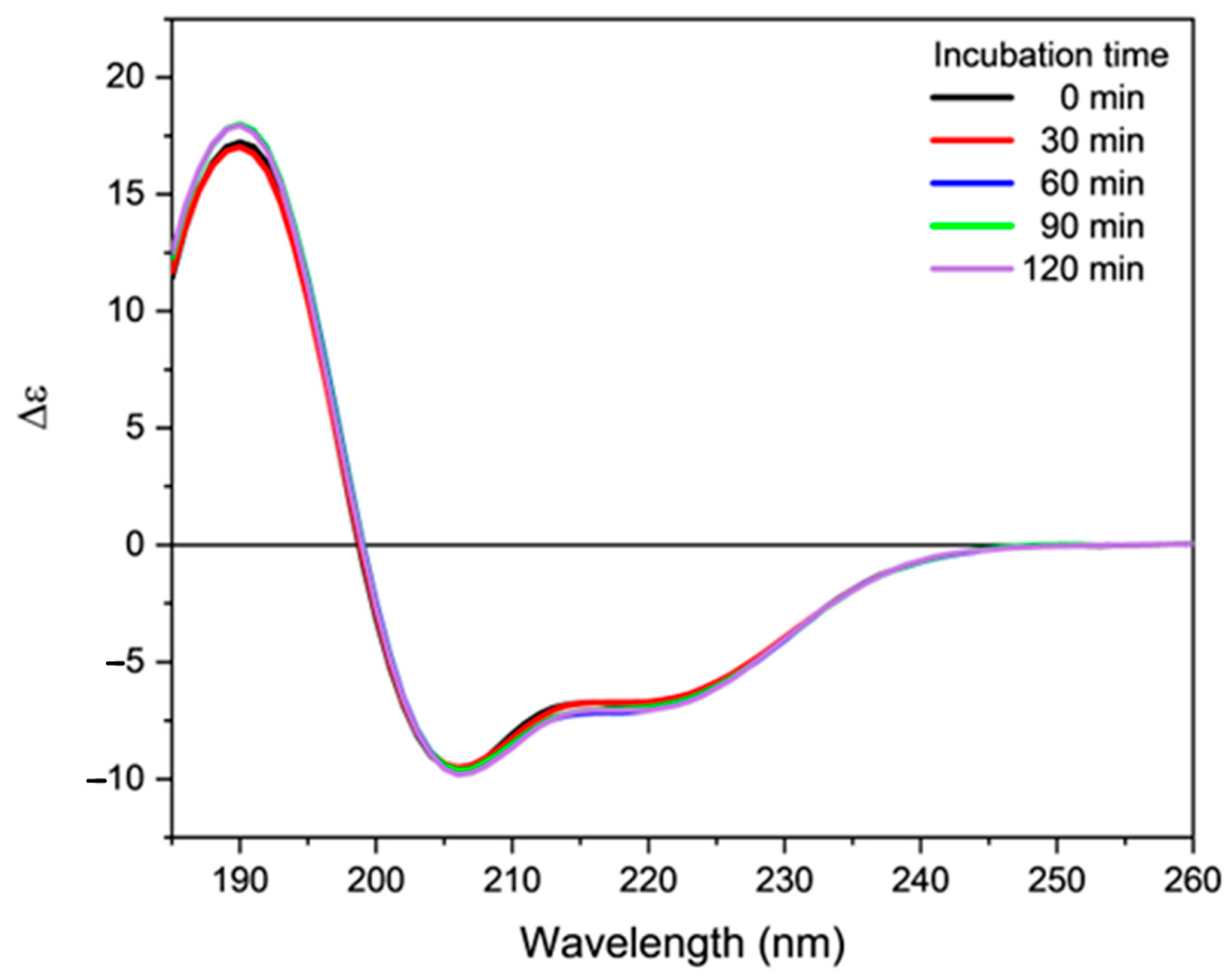
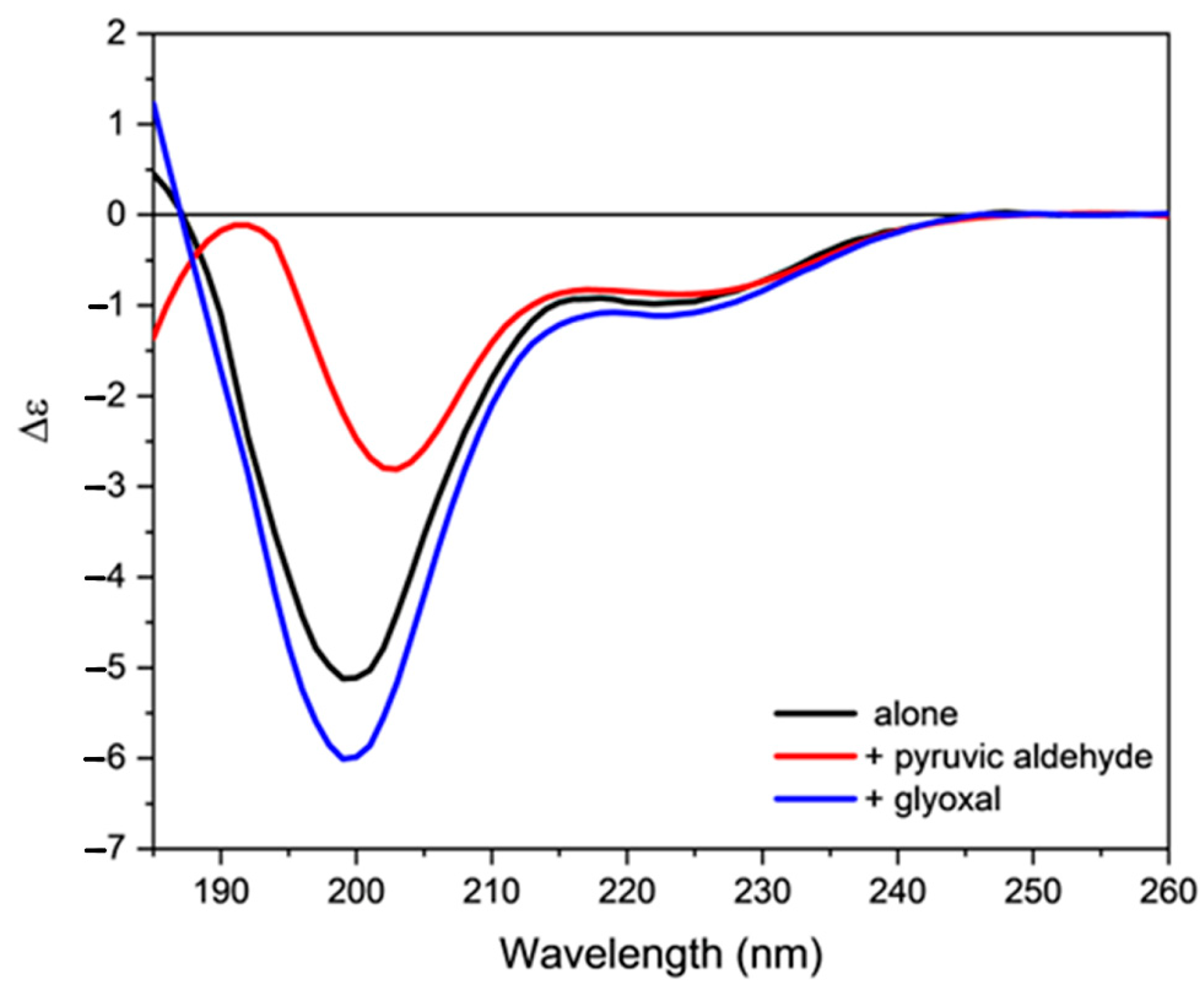
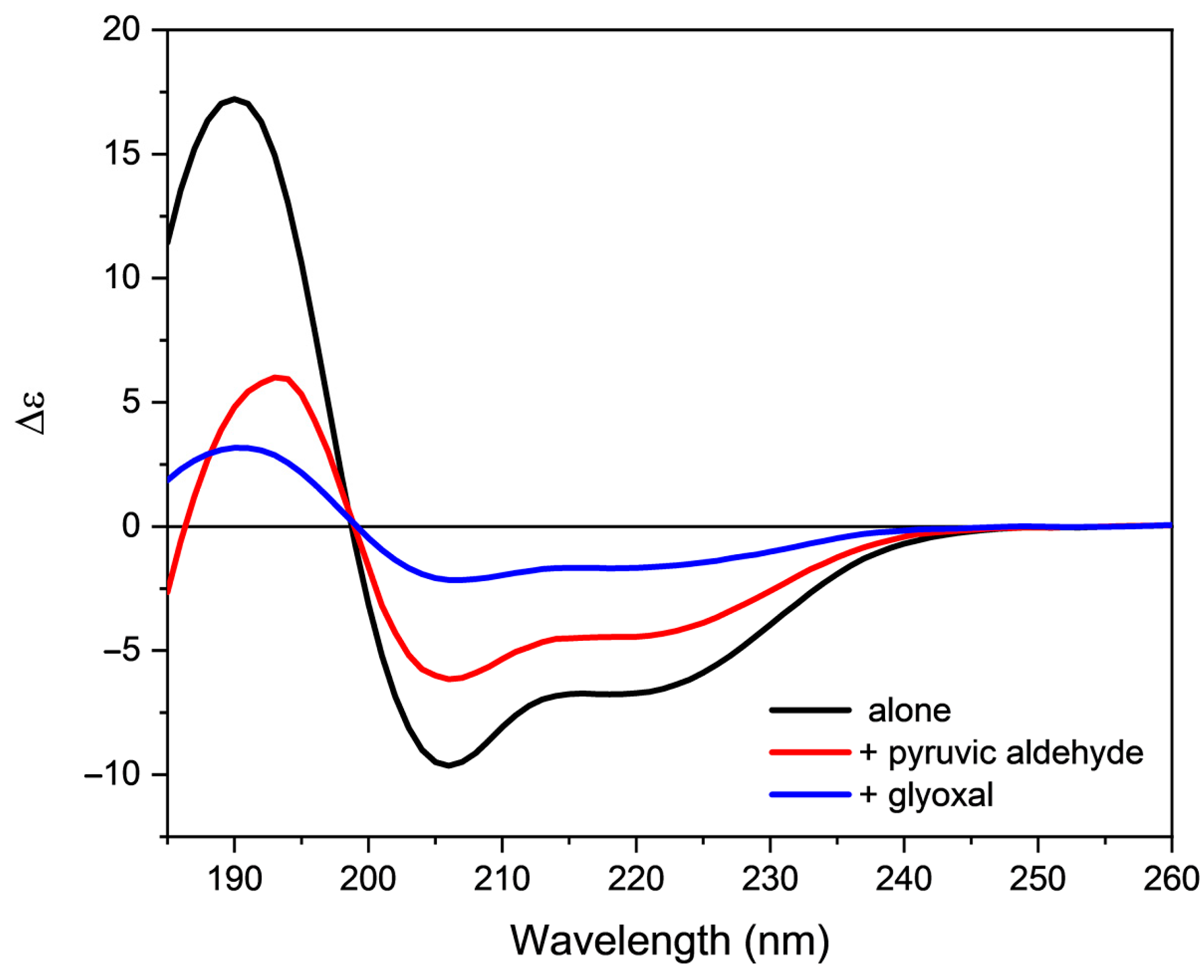
2.3. Effect of Oxidative Stress on the Secondary Structure of Orexin-B
3. Conclusions
4. Material and Methods
4.1. Peptide Synthesis
4.2. Circular Dichroism Studies
4.3. Induction of Oxidative Stress
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sakurai, T.; Amemiya, A.; Ishii, M.; Matsuzaki, I.; Chemelli, R.M.; Tanaka, H.; Williams, S.C.; Richardson, J.A.; Kozlowski, G.P.; Wilson, S.; et al. Orexins and Orexin Receptors: A Family of Hypothalamic Neuropeptides and G Protein-Coupled Receptors that Regulate Feeding Behavior. Cell 1998, 92, 573–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Lecea, L.; Kilduff, T.; Peyron, C.; Gao, X.B.; Foye, P.E.; Danielson, P.E.; Fukahara, C.; Battenberg, E.L.F.; Gautvik, V.T.; Barlett, F.S.; et al. The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. USA 1998, 95, 322–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takai, T.; Takaya, T.; Nakano, M.; Akutsu, H.; Nakagawa, A.; Aimoto, S.; Nagai, K.; Ikegami, T. Orexin-A is composed of a highly conserved C-terminal and a specific, hydrophilic N-terminal region, revealing the structural basis of specific recognition by the Orexin-1 receptor. J. Pept. Sci. 2006, 12, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Soll, R.M.; Durrenberger, F.; Dautzenberg, F.M.; Beck-Sickinger, A.G. Structure—Activity Studies of Orexin A and Orexin B at the Human Orexin 1 and Orexin 2 Receptors Led to Orexin 2 Receptor Selective and Orexin 1 Receptor Preferring Ligands. J. Med. Chem. 2004, 47, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- German, A.N.; Decker, N.A.; Gilmor, B.P.; Thomas, B.F.; Zhang, Y. Truncated Orexin Peptides: Structure-Activity Relationship Studies. Med. Chem. Lett. 2013, 4, 1224–1227. [Google Scholar] [CrossRef]
- Lee, J.H.; Bang, E.; Chae, K.J.; Kim, J.Y.; Lee, D.W.; Lee, W. Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/Orexin-B. Eur. J. Biochem. 1999, 266, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Miskolzie, M.; Lucyk, S.; Kotovych, G. NMR conformational studies of micelle-bound Orexin-B: A neuropeptide involved in the sleep/awake cycle and feeding regulation. J. Biomol. Struct. Dyn. 2003, 21, 341–351. [Google Scholar] [CrossRef]
- Voisin, T.; Rouet-Benzineb, P.; Reuter, N.; Laburthe, M. Orexins and their receptors: Structural aspects and role in peripheral tissues. Cell Mol. Life Sci. 2003, 60, 72–87. [Google Scholar] [CrossRef]
- Chemelli, R.M.; Willie, J.T.; Sinton, C.M.; Elmquist, J.K.; Scammell, T.; Lee, C. Narcolepsy in Orexin knockout mice: Molecular genetics of sleep regulation. Cell 1999, 98, 437–451. [Google Scholar] [CrossRef] [Green Version]
- Monda, M.; Viggiano, A.; Viggiano, A.; Mondola, R.; Viggiano, E.; Messina, G. Olanzapine blocks the sympathetic and hyperthermic reactions due to cerebral injection of Orexin A. Peptides 2008, 29, 120–126. [Google Scholar] [CrossRef]
- Lubkin, M.; Stricker-Krongrad, A. Independent Feeding and Metabolic Actions of Orexins in Mice. Biochem. Biophys. Res. Commun. 1998, 253, 241–245. [Google Scholar] [CrossRef]
- Martin-Fardon, R.; Cauvi, G.; Kerr, T.M.; Weiss, F. Differential role of hypothalamic Orexin/hypocretin neurons in reward seeking motivated by cocaine versus palatable food. Addict. Biol. 2018, 23, 6–15. [Google Scholar] [CrossRef]
- Yang, L.; Zou, B.; Xiong, X.; Pascual, C.; Xie, J.; Malik, A. Hypocretin/Orexin Neurons Contribute to Hippocampus-Dependent Social Memory and Synaptic Plasticity in Mice. J. Neurosci. 2013, 33, 5275–5284. [Google Scholar] [CrossRef] [Green Version]
- Arendt, D.H.; Ronan, P.J.; Oliver, K.D.; Callahan, L.B.; Summers, T.R.; Summers, C.H. Depressive behavior and activation of the Orexin/hypocretin system. Behav. Neurosci. 2013, 127, 86–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B. Antioxidants and human disease: A general introduction. Nutr. Rev. 1997, 55, 44–52. [Google Scholar] [CrossRef]
- Gutteridge, J.M.C. Lipid peroxidation and antioxidants as biomarkers of tissue damage. Clin. Chem. 1995, 41, 1819–1828. [Google Scholar] [CrossRef]
- Cross, C.E.; Halliwell, B.; Borish, E.T.; Pryor, W.A.; Ames, B.A.; Saul, R.L.; McCord, J.M.; Harman, D. Oxygen Radicals and Human Disease. Ann. Int. Med. 1987, 107, 526–545. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [Green Version]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 3rd ed.; Oxford Science Publications: Oxford, UK, 1999. [Google Scholar]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitocco, D.; Zaccardi, F.; Di Stasio, E. Oxidative Stress, Nitric Oxide, and Diabetes. Rev. Diabet. Stud. 2010, 7, 15–25. [Google Scholar] [CrossRef]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Internatl. Microbiol. 2000, 3, 3–8. [Google Scholar]
- Davies, M.J. Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 2004, 3, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Stadtman, E.R. Oxidative modification of proteins during aging. Exp. Gerontol. 2001, 36, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Scaloni, A.; Giustarini, D. Proteins as biomarkers of oxidative/nitrosative stress in diseases: The contribution of redox proteomics. Mass Spectr. Rev. 2005, 24, 55–99. [Google Scholar] [CrossRef]
- Aldini, G.; Domingues, M.R.; Spickett, C.M.; Domingues, P.; Altomare, A.; Sanchez-Gomez, F.J. Protein lipoxidation: Detection strategies and challenges. Redox Biol. 2015, 5, 253–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Peptide Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Carpino, L.A.; Imazumi, H.; El-Faham, A.; Ferrer, F.J.; Zhang, C.; Lee, Y.; Foxman, B.M.; Henklein, P.; Hanay, C.; Mügge, C.; et al. The Uronium/Guanidinium Peptide Coupling Reagents: Finally the True Uronium Salts. Angew. Chem. Int. Ed. 2002, 41, 441–445. [Google Scholar] [CrossRef]
- Van Stokkum, I.H.M.; Spoelder, H.J.W.; Bloemendal, M.; Van Grondelle, R.; Groen, F.C.A. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 1990, 191, 110–118. [Google Scholar] [CrossRef]
- Hussain, R.; Benning, K.; Javorfi, T.; Longo, E.; Rudd, T.R.; Pulford, B.; Siligardi, G. CDApps: Integrated software for experimental planning and data processing at beamline B23, Diamond Light Source. J. Synchrotron Radiat. 2015, 22, 465–468. [Google Scholar] [CrossRef] [Green Version]
- Sónnichsen, F.D.; Van Eyk, J.E.; Hodges, R.S.; Sykes, B.D. Effect of Trifluoroethanol on Protein Secondary Structure: An NMR and CD Study Using a Synthetic Actin Peptide. Biochemistry 1992, 31, 8790–8798. [Google Scholar] [CrossRef]
- Nelson, J.W.; Kallenbach, N.R. Stabilization of the ribonuclease S-peptide alpha-helix by trifluoroethanol. Proteins 1986, 1, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Tanford, C. Protein denaturation. Adv. Protein Chem. 1968, 23, 121–282. [Google Scholar] [CrossRef] [PubMed]
- Kallenbach, N.R.; Lyu, P.; Zhou, H. CD Spectroscopy and the Helix-Coil Transition in Peptides and Polypeptides. In Circular Dichroism and the Conformational Analysis of Biomolecules; Fasman, G.D., Ed.; Springer: Boston, MA, USA, 1996. [Google Scholar] [CrossRef]
- Ruzza, P.; Honisch, C.; Hussain, R.; Siligardi, G. Free Radical Generation in Far-UV Synchrotron Radiation Circular Dichroism Assays—Protein and Buffer Composition Contribution. Int. J. Mol. Sci. 2021, 22, 6512. [Google Scholar] [CrossRef] [PubMed]
- Honisch, C.; Hussain, R.; Siligardi, G.; Ruzza, P. Influence of small molecules on the photo-stability of watersoluble porcine lens proteins. Chirality 2020, 32, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nakayoshi, T.; Kurimoto, E.; Oda, A. Mechanisms of Deamidation of Asparagine Residues and Effects of Main-Chain Conformation on Activation Energy. Int. J. Mol. Sci. 2020, 21, 7035. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotondo, M.; Honisch, C.; Tartaggia, S.; Ruzza, P. Circular Dichroism Study of Orexin B under Oxidative Stress Conditions. Molecules 2023, 28, 484. https://doi.org/10.3390/molecules28020484
Rotondo M, Honisch C, Tartaggia S, Ruzza P. Circular Dichroism Study of Orexin B under Oxidative Stress Conditions. Molecules. 2023; 28(2):484. https://doi.org/10.3390/molecules28020484
Chicago/Turabian StyleRotondo, Martina, Claudia Honisch, Stefano Tartaggia, and Paolo Ruzza. 2023. "Circular Dichroism Study of Orexin B under Oxidative Stress Conditions" Molecules 28, no. 2: 484. https://doi.org/10.3390/molecules28020484





