Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins: Part 2
Abstract
1. Introduction
2. Results and Discussion
2.1. Salt2-HSA, Salt2-AGP, Salt2-HGG, Salt2-CNS Interaction Analysis
2.2. Salt2–Protein Binding Sites Assessment
3. Materials and Methods
3.1. Methods
3.1.1. Sample Preparation
3.1.2. Emission and Absorption Spectra Measurements
- Fcor and Fobs—corrected and observed fluorescence (after subtraction the solvent scattering spectrum), respectively,
- Aex and Aem—the absorbance at the excitation and emission wavelength, respectively.
- F, F0—the fluorescence intensities at the maximum wavelength of albumin in the presence and absence of a quencher, respectively,
- —bimolecular quenching rate constant in mol−1·L·s−1,
- [L]—ligand concentration in mol·L−1 ([L] = [Lb] + [Lf], where [Lb] and [Lf] are the bound and unbound (free) drug concentrations, respectively),
- KS-V—Stern–Volmer constant in mol−1·L.
- r—number of ligand moles bound to 1 mole of protein; , ,
- n—number of binding sites classes,
- Ka—association constant in mol−1·L,
- [Lf]—free ligand concentration in mol·L−1.
3.1.3. Circular Dichroism (CD) Measurements
- MRW—mean residue weight (MRWHSA = 113.7 Da; MRWAGP = 236.3 Da),
- θλ—observed ellipticity at wavelength λ in deg,
- l—optical path length in cm,
- c—protein concentration in g·cm−3.
3.2. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 30 June 2021).
- Owczarzy, A.; Zięba, A.; Pożycka, J.; Kulig, K.; Rogóż, W.; Szkudlarek, A.; Maciążek-Jurczyk, M. Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins. Part 1. Molecules 2021, 26, 4776. [Google Scholar] [CrossRef] [PubMed]
- Bojko, B.; Vuckovic, D.; Pawliszyn, J. Comparison of solid phase microextraction versus spectroscopic techniques for binding studies of carbamazepine. J. Pharm. Biomed. Anal. 2012, 66, 91–99. [Google Scholar] [CrossRef]
- Kragh-Hansen, U.; Chuang, V.T.G.; Otagiri, M. Practical Aspects of the Ligand-Binding and Enzymatic Properties of Human Serum Albumin. Biol. Pharm. Bull. 2002, 25, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, Y.; Liang, H. Interactive Association of Drugs Binding to Human Serum Albumin. Int. J. Mol. Sci. 2014, 15, 3580–3595. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef]
- Fanali, G.; Di Masi, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human serum albumin: From bench to bedside. Mol. Asp. Med. 2012, 33, 209–290. [Google Scholar] [CrossRef]
- Yamasaki, K.; Chuang, V.T.G.; Maruyama, T.; Otagiri, M. Albumin–drug interaction and its clinical implication. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 5435–5443. [Google Scholar] [CrossRef] [PubMed]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding speci-ficity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef]
- Dente, L.; Rüther, U.; Tripodi, M.; Wagner, E.F.; Cortese, R. Expression of human alpha 1-acid glycoprotein genes in cultured cells and in transgenic mice. Genes Dev. 1988, 2, 259–266. [Google Scholar] [CrossRef]
- Fitos, I.; Visy, J.; Zsila, F.; Bikádi, Z.; Mády, G.; Simonyi, M. Specific ligand binding on genetic variants of human α1-acid glycoprotein studied by circular dichroism spectroscopy. Biochem. Pharmacol. 2004, 67, 679–688. [Google Scholar] [CrossRef]
- Bteich, M. An overview of albumin and alpha-1-acid glycoprotein main characteristics: Highlighting the roles of amino acids in binding kinetics and molecular interactions. Heliyon 2019, 5, e02879. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.M.; Wilting, J.; Janssen, L.H. Drug binding to human alpha-1-acid glycoprotein in health and disease. Pharmacol. Rev. 1988, 40, 1–47. [Google Scholar] [PubMed]
- Fournier, T.; Medjoubi, N.N.; Porquet, D. Alpha-1-acid glycoprotein. BBA 2000, 1482, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Imre, T.; Schlosser, G.; Pocsfalvi, G.; Siciliano, R.; Molnár-Szöllösi, É.; Kremmer, T.; Malorni, A.; Vèkey, K. Glycosylation site analysis of human alpha-1-acid glycoprotein (AGP) by capillary liquid chromatography—electrospray mass spectrometry. J. Mass Spectrom. 2005, 40, 1483–1742. [Google Scholar] [CrossRef] [PubMed]
- Israili, Z.H.; Dayton, P.G. HUMAN ALPHA-1-GLYCOPROTEIN AND ITS INTERACTIONS WITH DRUGS†,‡. Drug Metab. Rev. 2001, 33, 161–235. [Google Scholar] [CrossRef] [PubMed]
- Azada, M.A.K.; Huangb, J.X.; Cooperb, M.A.; Robertsc, K.D.; Thompsonc, P.E.; Nationa, R.L.; Lia, J.; Velkova, T. Structure activity relationships for the binding of polymyxins with human α—1 acid glycoproteins. Biochem. Pharmacol. 2012, 84, 278–291. [Google Scholar] [CrossRef]
- Albani, J.R. Relation between the secondary structure of carbohydrate residues of α1-acid glycoprotein (orosomucoid) and the fluorescence of the protein. Carbohydr. Res. 2003, 338, 1097–1101. [Google Scholar] [CrossRef]
- Yongchun, L.; Wenying, H.; Wenhua, G.; Zhide, H.; Xingguo, C. Binding of wogonin to human gamma globulin. Int. J. Biol. Macromol. 2005, 37, 1–11. [Google Scholar]
- Zięba, A.; Sochanik, A.; Szurko, A.; Rams, M.; Mrozek, A.; Cmoch, P. Synthesis and in vitro antiproliferative activity of 5-alkyl-12(H)-quino[3,4-b] [1,4]benzothiazinium salts. Eur. J. Med. Chem. 2010, 45, 4733–4739. [Google Scholar] [CrossRef]
- Zięba, A.; Bober, K. Lipophilicity analysis of newly synthetized quinobenzothiaines by use of TLC. J. Liq. Chromatogr. Relat. 2016, 39, 104–109. [Google Scholar] [CrossRef]
- Smith, S.A.; Waters, N. Pharmacokinetic and Pharmacodynamic Considerations for Drugs Binding to Alpha-1-Acid Glyco-protein. Pharm. Res. 2019, 36, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Olins, D.E.; Edelman, M.D. The antigenic structure of the polypeptide chains of human ɣ-globulin. Exp. Med. 1962, 116, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Saeidifar, M.; Mansouri-Torshizi, H.; Saboury, A.A. Biophysical study on the interaction between two palladium(II) complexes and human serum albumin by Multispectroscopic methods. J. Lumin. 2015, 167, 391–398. [Google Scholar] [CrossRef]
- Peters, T. All about Albumin. In All About Albumin: Biochemistry, Genetics, and Medical Applications, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1995; pp. 1–40. [Google Scholar]
- Evans, T.W. Review article: Albumin as a drug-biological effects of albumin unrelated to osmotic pressure. Aliment. Pharmacol. Ther. 2002, 16, 6–11. [Google Scholar]
- Szkudlarek, A.; Wilk, M.; Maciążek-Jurczyk, M. In Vitro Investigations of Acetohexamide Binding to Glycated Serum Albumin in the Presence of Fatty Acid. Molecules 2020, 25, 2340. [Google Scholar] [CrossRef]
- Eftink, C.A.; Ghiron, C.A. Exposure of Tryptophanyl Residues in Proteins. Quantitative Determination by Fluorescence Quenching Studies. Biochemistry 1976, 15, 672–680. [Google Scholar] [CrossRef]
- Maciążek-Jurczyk, M.; Morak-Młodawska, B.; Jeleń, M.; Kopeć, W.; Szkudlarek, A.; Owczarzy, A.; Kulig, K.; Rogóż, W.; Pożycka, J. The Influence of Oxidative Stress on Serum Albumin Structure as a Carrier of Selected Diazaphenothiazine with Potential Anticancer Activity. Pharmaceuticals 2021, 14, 285. [Google Scholar] [CrossRef]
- Parkhomenko, T.V.; Klicenko, O.A.; Shavlovski, M.M.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Biophysical char-acterization of albumin preparations from blood serum of healthy donors and patients with renal diseases. Part I: Spectro-fluorometric analysis. Med. Sci. Monit. 2002, 8, BR261-5. [Google Scholar]
- Ichikawa, T.; Terada, H. Estimation of State amount of Phenylalanine residues in proteins by second derivative spectropho-tometry. BBA 1979, 580, 120–128. [Google Scholar]
- Levine, R.L.; Federici, M.M. Quantitation of aromatic Residues in Protein: Model Compound for Second-Derivative Spectrocopy. Biochemistry 1982, 21, 2600–2606. [Google Scholar] [CrossRef]
- Balestrieri, C.; Colinna, G.; Giovane, A.; Irace, G.; Servillo, L. Second—Derivative Spectroscopy of Proteins. A Method for the Quantitative Determination of Aroma Acids in Proteins. Eur. J. Biochem. 1978, 90, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Terada, H.; Inoue, Y.; Ichikawa, T. Second derivative spectral properties of tryptophan and tyrosine residues in proteins. Effects of guanidine hydrochloride and dodecyl sulfate on the residues in lysozyme, ribonuclease and serum albumin. Chem. Pharm. Bull. 1984, 32, 585–590. [Google Scholar] [CrossRef]
- Maciążek-Jurczyk, M. Phenylbutazone and ketoprofen binding to serum albumin. Fluorescence study. Pharmacol. Rep. 2014, 66, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; pp. 1–26, 277–330, 529–535. [Google Scholar]
- Van de Weert, M.; Stella, L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J. Mol. Struct. 2011, 998, 144–150. [Google Scholar] [CrossRef]
- Summerfield, S.G.; Yates, J.W.T.; Fairman, D.A. Free Drug Theory—No Longer Just a Hypothesis? Pharm. Res. 2022, 39, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, T.; Gan, L.-S. Plasma protein binding: From discovery to development. J. Pharm. Sci. 2013, 102, 2953–2994. [Google Scholar] [CrossRef] [PubMed]
- Pitekova, B.; Uhlikova, E.; Kupcova, V.; Durfinova, M.; Mojto, V.; Turecky, L. Can alpha-1-acid glycoprotein affect the outcome of treatment in a cancer patient? Bratisl. Med. J. 2019, 120, 9–14. [Google Scholar] [CrossRef]
- Bailey, D.N.; Briggs, J.R. The Binding of Selected Therapeutic Drugs to Human Serum α-1 Acid Glycoprotein and to Human Serum Albumin In Vitro. Ther. Drug Monit. 2004, 26, 40–43. [Google Scholar] [CrossRef]
- Pelton, J.T.; McLean, L.R. Spectroscopic Methods for Analysis of Protein Secondary Structure. Anal. Biochem. 2000, 277, 167–176. [Google Scholar] [CrossRef]
- Maciążek-Jurczyk, M.; Janas, K.; Pożycka, J.; Szkudlarek, A.; Rogóż, W.; Owczarzy, A.; Kulig, K. Human Serum Albumin Aggregation/Fibrillation and its Abilities to Drugs Binding. Molecules 2020, 25, 618. [Google Scholar] [CrossRef]
- Munro, A.W.; Lindsay, J.; Coggins, J.R.; Kelly, S.M.; Price, N.C. Structural and enzymological analysis of the interaction of isolated domains of cytochromeP-450 BM3. FEBS Lett. 1994, 343, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Zsila, F.; Bikádi, Z.; Simonyi, M. Induced circular dichroism spectra reveal binding of the antiinflammatory curcumin to human α1-acid glycoprotein. Bioorganic Med. Chem. 2004, 12, 3239–3245. [Google Scholar] [CrossRef]
- Valeur, B. Molecular Fluorescence, Principles and Applications; Wiley-VCH: Weinheim, Germany, 2009; pp. 34–123. [Google Scholar]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar] [PubMed]
- Kirby, E.P. Experimental Techniques, Chapter 2. In Excited States of Protein and Nucleic Acids; Steiner, R.F., Weinryb, I., Eds.; Plenum Press: New York, NY, USA, 1971; pp. 27–61. [Google Scholar]
- Zsila, F. Subdomain Ib Is the Third major Drug Binding Region of Human Serum Albumin: Toward the Tree—Sites Models. Mol. Pharm. 2013, 10, 1668–1682. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.J.; Ghuman, J.; Zunszain, P.A.; Chung, C.-W.; Curry, S. Structural basis of binding of fluorescent, site-specific dansylated amino acids to human serum albumin. J. Struct. Biol. 2011, 174, 84–91. [Google Scholar] [CrossRef]
- Schönfeld, D.L.; Ravelli, R.B.; Mueller, U.; Skerra, A. The 1.8-Å Crystal Structure of α1-Acid Glycoprotein (Orosomucoid) Solved by UV RIP Reveals the Broad Drug-Binding Activity of This Human Plasma Lipocalin. J. Mol. Biol. 2008, 384, 393–405. [Google Scholar] [CrossRef]
- Nishi, K.; Fukunaga, N.; Otagiri, M. Construction of expression system for human alpha—1—Acid glycoproteins in Pichia pastoris and evaluation of this drug-binding properties. Drug Metab. Dispos. 2004, 32, 1069–1074. [Google Scholar] [CrossRef]
- Klotz, I.M.; Hunston, D.L. Properties of graphical representations of multiple classes of binding sites. Biochemistry 1971, 10, 3065–3069. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Venyaminov, S.; Vassilenko, K. Determination of Protein Tertiary Structure Class from Circular Dichroism Spectra. Anal. Biochem. 1994, 222, 176–184. [Google Scholar] [CrossRef]
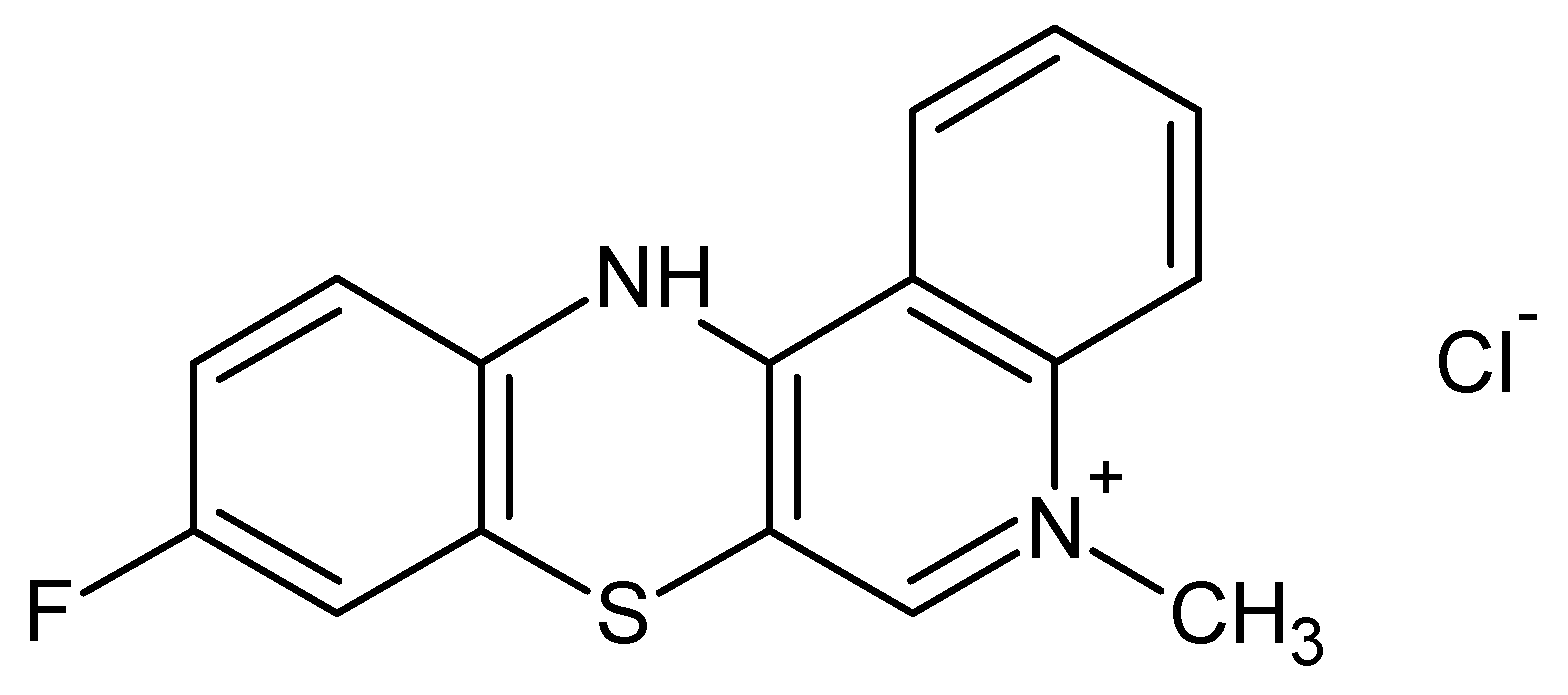




| Salt2:protein Molar Ratio | λex 275 nm | λex 295 nm | ||
|---|---|---|---|---|
| Molar Ratio | Percent of Fluorescence Quenching [%] | Molar Ratio | Percent of Fluorescence Quenching [%] | |
| Salt2:HSA | 0:1 ÷ 7:1 | 30.03 | 0:1 ÷ 6:1 | 29.02 |
| Salt2:AGP | 0:1 ÷ 8:1 | 66.07 | 0:1 ÷ 7:1 | 60.88 |
| Salt2:HGG | 0:1 ÷ 7:1 | 21.25 | 0:1 ÷ 6:1 | 25.07 |
| Salt2:CNS | 0:1 ÷ 7:1 | 28.79 | 0:1 ÷ 6:1 | 30.14 |
| Protein | 275 nm | 295 nm | ||||
|---|---|---|---|---|---|---|
| Salt2 [mol·L−1] | λmax [nm] | Parameter A | Salt2 [mol·L−1] | λmax [nm] | Parameter A | |
| HSA | 0 | 334 | 0.80 | 0 | 339 | 1.15 |
| 2.1 × 10−5 | 337 | 0.90 | 1.8 × 10−5 | 343 | 1.32 | |
| AGP | 0 | 331 | 0.61 | 0 | 333 | 0.64 |
| 2.4 × 10−5 | 333 | 0.67 | 2.1 × 10−5 | 337 | 0.77 | |
| HGG | 0 | 331 | 0.61 | 0 | 332 | 0.67 |
| 2.1 × 10−5 | 334 | 0.72 | 1.8 × 10−5 | 335 | 0.79 | |
| CNS | 0 | 331 | 0.60 | 0 | 335 | 0.83 |
| 2.4 × 10−5 | 334 | 0.76 | 2.1 × 10−5 | 337 | 0.93 | |
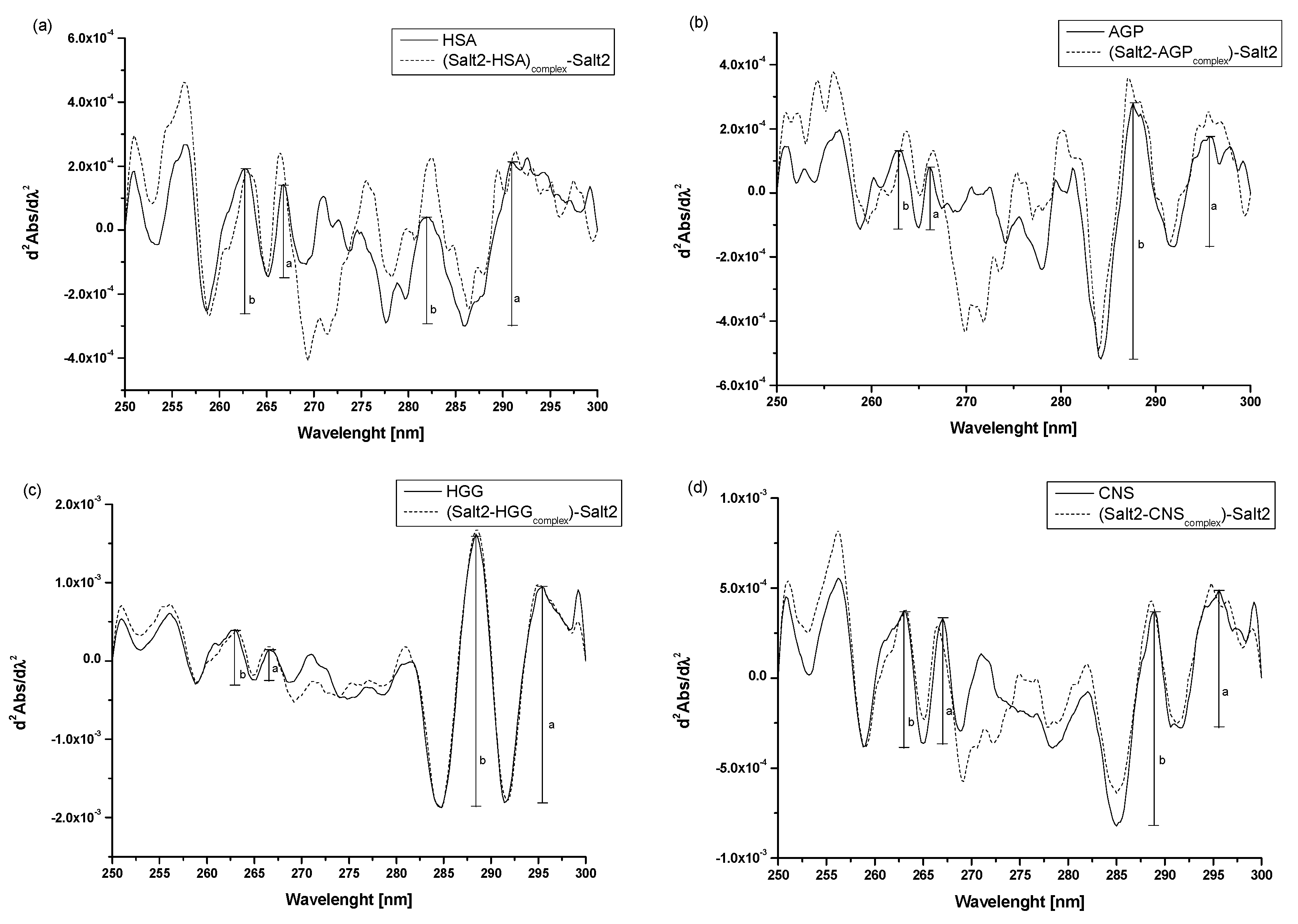
| r Values | |||
|---|---|---|---|
| λ 250–270 nm | λ > 270 nm | 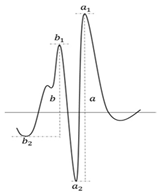 | |
| HSA | 0.65 | 2.00 | |
| Salt2-HSAcomplex | 0.84 | 2.24 | |
| AGP | 0.77 | 0.43 | |
| Salt2-AGPcomplex | 0.62 | 0.49 | |
| HGG | 1.00 | 0.80 | |
| Salt2-HGGcomplex | 0.55 | 0.77 | |
| CNS | 0.92 | 0.64 | |
| Salt2-CNScomplex | 0.67 | 0.72 | |
| λex 275 nm | λex 295 nm | |||
|---|---|---|---|---|
| (Ks-v104) ± SD * [mol·L−1] | (kq1012) ± SD * [mol−1·L·s−1] | (Ks-v104) ± SD * [mol·L−1] | (kq1012) ± SD * [mol−1·L·s−1] | |
| Salt2-HSAcomplex | 2.01 ± 0.04 | 3.35 ± 0.06 | 2.27 ± 0.02 | 3.79 ± 0.04 |
| Salt2-AGPcomplex | 8.13 ± 0.15 | 13.55 ± 2.43 | 7.28 ± 2.13 | 12.13 ± 3.54 |
| Salt2-HGGcomplex | 1.27 ± 0.02 | 2.08 ± 0.04 | 1.83 ± 0.03 | 3.00 ± 0.05 |
| Salt2-CNScomplex | 1.79 ± 0.03 | 2.84 ± 0.05 | 2.06 ± 0.07 | 3.43 ± 0.12 |
| λex 275 nm | λex 295 nm | |||
|---|---|---|---|---|
| (Ka104) ± SD * [mol·L−1] | n ± SD * | (Ka104) ± SD * [mol·L−1] | n ± SD * | |
| (Salt2-HSA)complex | 1.98 ± 0.07 | 0.97 ± 0.04 | 2.33 ± 0.18 | 0.97 ± 010 |
| (Salt2-AGP)complex | 5.04 ± 0.25 | 0.92 ± 0.07 | 6.28 ± 0.11 | 0.95 ± 0.04 |
| (Salt2-HGG)complex | 3.45 ± 0.29 | 0.97 ± 0.12 | 3.28 ± 0.10 | 0.96 ± 0.12 |
| (Salt2-CNS)complex | 0.94 ± 0.04 | 0.99 ± 0.05 | 0.91 ± 0.43 | 1.99 ± 1.19 |
| [Θ]MRW at 208.8 nm [mdeg·cm2·dmol−1] | [Θ]MRW at 222 nm [mdeg·cm2·dmol−1] | % α-helix | % β-sheet | % Turn | % Random | |
|---|---|---|---|---|---|---|
| HSA a | −21,690.171 | −20,413.846 | 37.0 | 10.3 | 21.5 | 31.2 |
| Salt2-HSAcomplex a | −21,794.444 | −21,690.171 | 37.4 | 10.1 | 21.7 | 30.9 |
| AGP b | - | −9130.429 | 16.5 | 83.5 | - | - |
| Salt2- AGPcomplex b | - | −9027.758 | 16.4 | 85.6 | - | - |
| CSalt2 [mol·L−1] | [HSA]:[dGly] Molar Ratio | [HSA]:[dPhe] Molar Ratio |
|---|---|---|
| 1:1 | ||
| Percentage of Displacement [%] | ||
| 0 | - | - |
| 3.3 × 10−5 | 65.55 | 46.11 |
| CSalt2 [mol·L−1] | [AGP]:[QR] 1:0.5 Molar Ratio | [AGP]:[QR] 1:1 Molar Ratio |
|---|---|---|
| Percentage of Displacement [%] | ||
| 0 | - | - |
| 4.8 × 10−5 | 66.5 | 69.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Owczarzy, A.; Rogóż, W.; Kulig, K.; Pożycka, J.; Zięba, A.; Maciążek-Jurczyk, M. Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins: Part 2. Molecules 2023, 28, 698. https://doi.org/10.3390/molecules28020698
Owczarzy A, Rogóż W, Kulig K, Pożycka J, Zięba A, Maciążek-Jurczyk M. Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins: Part 2. Molecules. 2023; 28(2):698. https://doi.org/10.3390/molecules28020698
Chicago/Turabian StyleOwczarzy, Aleksandra, Wojciech Rogóż, Karolina Kulig, Jadwiga Pożycka, Andrzej Zięba, and Małgorzata Maciążek-Jurczyk. 2023. "Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins: Part 2" Molecules 28, no. 2: 698. https://doi.org/10.3390/molecules28020698
APA StyleOwczarzy, A., Rogóż, W., Kulig, K., Pożycka, J., Zięba, A., & Maciążek-Jurczyk, M. (2023). Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins: Part 2. Molecules, 28(2), 698. https://doi.org/10.3390/molecules28020698







