Preparation, Purification and Characterization of Antibacterial and ACE Inhibitory Peptides from Head Protein Hydrolysate of Kuruma Shrimp, Marsupenaeus japonicus
Abstract
:1. Introduction
2. Results and Discussions
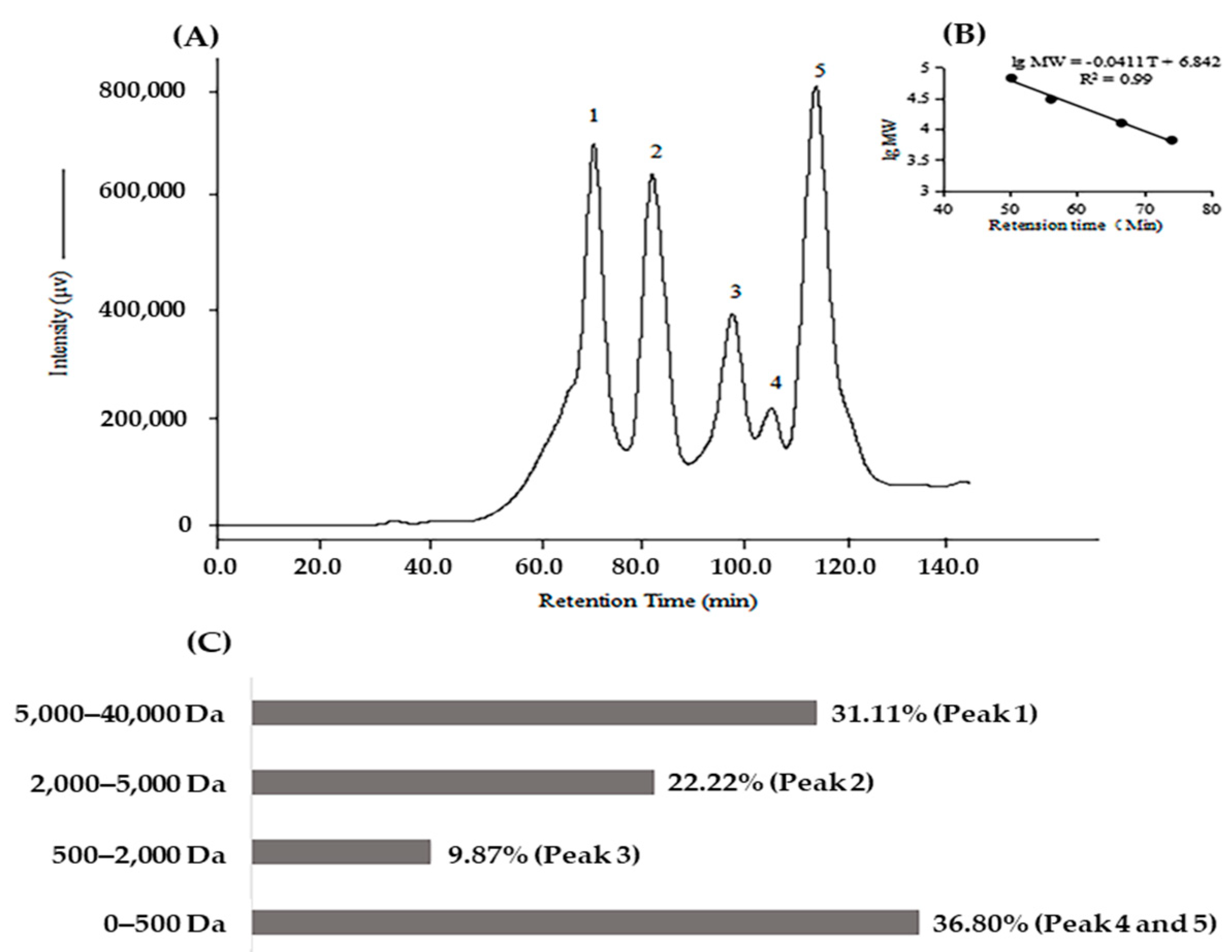
2.1. Biochemical Characteristics of Protein Hydrolysate of Kuruma Shrimp Heads (KSHPH)
2.2. Purification of Antibacterial and ACE Inhibitory Peptides
2.3. Amino Acid Sequence of the Purified Peptide
2.4. ACE Inhibition Pattern of the ACE Inhibitory Peptide
2.5. Molecular Mechanism between the ACE Inhibitory Peptide and ACE
2.6. Hemolytic Activity
3. Materials and Methods
3.1. Materials
3.2. Preparation of Protein Hydrolysate of Kuruma Shrimp Heads (KSHPH)
3.3. Determination of Molecular Weight Distribution of KSHPH
3.4. Purification of Peptides
3.5. Antibacterial Activity
3.5.1. Disc Diffusion Assay
3.5.2. Determination of Minimum Inhibitory Concentration (MIC)
3.6. ACE Inhibitory Activity
3.7. Peptide Identification and Structural Analysis
3.8. Determination of ACE Inhibition Pattern
3.9. The Molecular Docking Simulation
3.10. Hemolytic Activity Assay
3.11. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mao, X.; Guo, N.; Sun, J.; Xue, C. Comprehensive utilization of shrimp waste based on biotechnological methods: A review. J. Clean. Prod. 2017, 143, 814–823. [Google Scholar] [CrossRef]
- Knorr, D. Recovery and utilization of chitin and chitosan in food processing waste management. Food Techcol. 1991, 45, 114–122. [Google Scholar]
- Mizani, M.; Aminlari, M.; Khodabandeh, M. An effective method for producing a nutritive protein extract powder from shrimp-head waste. Food Sci. Technol. Int. 2005, 11, 49–54. [Google Scholar] [CrossRef]
- Harnedy, P.A.; FitzGerald, R.J. Bioactive peptides from marine processing waste and shellfish: A review. J. Funct. Foods 2012, 4, 6–24. [Google Scholar] [CrossRef]
- Kim, S.K.; Wijesekara, I. Development and biological activities of marine-derived bioactive peptides: A review. J. Funct. Foods 2010, 2, 1–9. [Google Scholar] [CrossRef]
- Jemil, I.; Abdelhedi, O.; Nasri, R.; Mora, L.; Jridi, M.; Aristoy, M.C.; Toldrá, F.; Nasri, M. Novel bioactive peptides from enzymatic hydrolysate of Sardinelle (Sardinella aurita) muscle proteins hydrolysed by Bacillus subtilis A26 proteases. Food Res. Int. 2017, 100, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Cézard, C.; Silva-Pires, V.; Mullié, C.; Sonnet, P. Antibacterial peptides: A review. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; pp. 926–937. [Google Scholar]
- Wald, M.; Schwarz, K.; Rehbein, H.; Bußmann, B.; Beermann, C. Detection of antibacterial activity of an enzymatic hydrolysate generated by processing rainbow trout by-products with trout pepsin. Food Chem. 2016, 205, 221–228. [Google Scholar] [CrossRef]
- Yang, S.; Huang, H.; Wang, F.; Aweya, J.J.; Zheng, Z.; Zhang, Y. Prediction and characterization of a novel hemocyanin-derived antimicrobial peptide from shrimp Litopenaeus vannamei. Amino Acids 2018, 50, 995–1005. [Google Scholar] [CrossRef]
- Anbuchezian, R.; Ravichandran, S.; Rajan, D.K.; Tilivi, S.; Devi, S.P. Identification and functional characterization of antimicrobial peptide from the marine crab Dromia dehaani. Microb. Pathog. 2018, 125, 60–65. [Google Scholar] [CrossRef]
- Robert, M.; Zatylny-Gaudin, C.; Fournier, V.; Corre, E.; Le Corguillé, G.; Bernay, B.; Henry, J. Transcriptomic and peptidomic analysis of protein hydrolysates from the white shrimp (L. vannamei). J. Biotechnol. 2014, 186, 30–37. [Google Scholar] [CrossRef]
- Beaulieu, L.; Thibodeau, J.; Bonnet, C.; Bryl, P.; Carbonneau, M.É. Detection of antibacterial activity in an enzymatic hydrolysate fraction obtained from processing of Atlantic rock crab (Cancer irroratus) by-product. PharmaNutrition 2013, 1, 49–157. [Google Scholar] [CrossRef]
- Barman, J.; Baishya, A. Aetiologocal study of hypertension in ayurveda. Global J. Res. Analysis 2019, 8, 27–28. [Google Scholar]
- Barbana, C.; Boye, J.I. Angiotensin I-converting enzyme inhibitory properties of lentil protein hydrolysates: Determination of the kinetics of inhibition. Food Chem. 2011, 127, 94–101. [Google Scholar] [CrossRef]
- Jao, C.L.; Huang, S.L.; Hsu, K.C. Angiotensin I-converting enzyme inhibitory peptides: Inhibition mode, bioavailability, and antihypertensive effects. BioMedicine 2012, 2, 130–136. [Google Scholar] [CrossRef]
- Patchett, A.; Harris, A.E.; Tristram, E.W.; Wyvratt, M.J.; Wu, M.; Taub, T.D.; Peterson, E.R.; Ikeler, T.; Broeke, J.J.L.; Payne, G.; et al. A new class of angiotensin-converting enzyme inhibitors. Nature 1980, 5788, 280–283. [Google Scholar] [CrossRef]
- He, H.-L.; Chen, X.-L.; Sun, C.-Y.; Zhang, Y.-Z.; Zhou, B.-C. Analysis of novel angiotensin I-converting enzyme inhibitory peptides from protease-hydrolyzed marine shrimp Acetes chinensis. J. Pept. Sci. 2006, 12, 726–733. [Google Scholar]
- Nii, Y.; Fukuta, K.; Yoshimoto, R.; Sakai, K.; Ogawa, T. Determination of antihypertensive peptides from an izumi shrimp hydrolysate. Biosci. Biotechnol. Biochem. 2008, 72, 861–864. [Google Scholar] [CrossRef] [Green Version]
- Tsai, J.S.; Chen, J.L.; Pan, B.S. ACE-inhibitory peptides identified from the muscle protein hydrolysate of hard clam (Meretrix lusoria). Process Biochem. 2008, 43, 743–747. [Google Scholar] [CrossRef]
- Shiozaki, K.; Shiozaki, M.; Masuda, J.; Yamauchi, A.; Ohwada, S.; Nakano, T.; Yamaguchi, T.; Saito, T.; Muramoto, K.; Sato, M. Identification of oyster-derived hypotensive peptide acting as angiotensin I-converting enzyme inhibitor. Fish. Sci. 2010, 76, 865–872. [Google Scholar] [CrossRef]
- Balti, R.; Nedjar-Arroume, N.; Bougatef, A.; Guillochon, D.; Nasri, M. Three novel angiotensin I-converting enzyme (ACE) inhibitory peptides from cuttlefish (Sepia officinalis) using digestive proteases. Food Res. Int. 2010, 43, 1136–1143. [Google Scholar] [CrossRef]
- Martínez-Cruz, O.; Cabrera-Chávez, F.; Paredes-López, O. Biochemical characteristics, and nutraceutical and technological uses of amaranth globulins. In Globulins: Biochemistry, Production and Role in Immunity; Milford, S.D., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2014; pp. 41–70. [Google Scholar]
- Zarei, M.; Forghani, B.; Ebrahimpour, A.; Abdul-Hamid, A.; Anwar, F.; Saari, N. In vitro and in vivo antihypertensive activity of palm kernel cake protein hydrolysates: Sequencing and characterization of potent bioactive peptides. Ind. Crops Prod. 2015, 76, 112–120. [Google Scholar] [CrossRef]
- He, H.L.; Chen, X.; Wu, L.H.; Sun, C.Y.Y.; Zhang, Z.B.; Zhou, C. High throughput and rapid screening of marine protein hydrolysates enriched in peptides with angiotensin-I-converting enzyme inhibitory activity by capillary electrophoresis. Bioresour. Technol. 2007, 98, 3499–3505. [Google Scholar] [CrossRef]
- Abdel-Hamid, M.; Goda, H.A.; De Gobba, C.; Jenssen, H.; Osman, A. Antibacterial activity of papain hydrolysed camel whey and its fractions. Int. Dairy J. 2016, 61, 91–98. [Google Scholar] [CrossRef]
- Kobbi, S.; Balti, R.; Bougatef, A.; Le Flem, G.; Firdaous, L.; Bigan, M.; Chataigné, G.; Chaabouni, S.; Dhulster, P.; Nedjar, N. Antibacterial activity of novel peptides isolated from protein hydrolysates of RuBisCO purified from green juice alfalfa. J. Funct. Foods 2015, 18, 703–713. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Y.; Cheserek, M.J.; Su, G.; Le, G. Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2013, 97, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Feng, Z.; Ren, T.; Sun, H.; Han, H.; Jin, W.; Dang, T.; Tao, Y. Purification, characterization and application of a novel antimicrobial peptide from Andrias davidianus blood. Lett. Appl. Microbiol. 2018, 66, 8–43. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Muraoka, T.; Cheetham, A.G.; Stupp, S.I. Self-assembly of giant peptide nanobelts. Nano Lett. 2009, 9, 945–951. [Google Scholar] [CrossRef] [Green Version]
- Bougherra, F.; Dilmi-Bouras, A.; Balti, R.; Przybylski, R.; Adoui, F.; Elhameur, H.; Chevalier, M.; Flahaut, C.; Dhulster, P.; Naima, N. Antibacterial activity of new peptide from bovine casein hydrolyzed by a serine metalloprotease of Lactococcus lactis subsp BR16. J. Funct. Foods 2017, 32, 112–122. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 2003, 6922, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Kohmura, M.; Nio, N.; Kubo, K.; Minoshima, Y.; Munekata, E.; Ariyoshi, Y. Inhibition of angiotensin-converting enzyme by synthetic peptides of human β-casein. Agric. Biol. Chem. 1989, 53, 2107–2114. [Google Scholar]
- Wu, J.R.; Aluko, E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure−activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Asoodeh, A.; Homayouni-Tabrizi, M.; Shabestarian, H.; Emtenani, S.; Emtenani, S. Biochemical characterization of a novel antioxidant and angiotensin I-converting enzyme inhibitory peptide from Struthio camelus egg white protein hydrolysis. J. Food Drug Anal. 2016, 24, 332–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Ruiz, J.Á.; Ramos, M.; Recio, I. Identification of novel angiotensin- converting enzyme-inhibitory peptides from ovine milk proteins by CE-MS and chromatographic techniques. Electrophoresis 2007, 28, 4202–4211. [Google Scholar] [CrossRef] [PubMed]
- Khueychai, S.; Jangpromma, N.; Choowongkomon, K.; Joompang, A.; Daduang, S.; Vesaratchavest, M.; Payoungkiattikun, W.; Tachibana, S.; Klaynongsruang, S. A novel ACE inhibitory peptide derived from alkaline hydrolysis of ostrich (Struthio camelus) egg white ovalbumin. Process Biochem. 2018, 73, 235–245. [Google Scholar] [CrossRef]
- Wu, Q.; Jia, J.; Yan, H.; Du, J.; Gui, Z. A novel angiotensin-I converting enzyme (ACE) inhibitory peptide from gastrointestinal protease hydrolysate of silkworm pupa (Bombyx mori) protein: Biochemical characterization and molecular docking study. Peptides 2015, 68, 17–24. [Google Scholar] [CrossRef]
- Pan, D.; Guo, H.; Zhao, B.; Cao, J. The molecular mechanisms of interactions between bioactive peptides and angiotensin-converting enzyme. Bioorganic Med. Chem. Lett. 2011, 21, 3898–3904. [Google Scholar] [CrossRef]
- Fan, H.; Liao, W.; Wu, J. Molecular interactions, bioavailability, and cellular mechanisms of angiotensin-converting enzyme inhibitory peptides. Bioorganic Med. Chem. Lett. 2019, 43, 12572. [Google Scholar]
- Pina, A.S.; Roque, A.C.A. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J. Mol. Recognit. 2009, 22, 162–168. [Google Scholar] [CrossRef]
- Jung, W.K.; Mendis, E.; Je, J.Y.; Park, P.J.B.; Son, W.; Kim, H.C.; Choi, Y.K.; Kim, S.K. Angiotensin I-converting enzyme inhibitory peptide from yellowfin sole (Limanda aspera) frame protein and its antihypertensive effect in spontaneously hypertensive rats. Food Chem. 2006, 94, 26–32. [Google Scholar] [CrossRef]
- Elavarasan, K.; Shamasundar, B.A. Proteolysis of water washed meat from Catla catla using bromelain and papain: Optimization of hydrolysis parameters. J. Fish. Life Sci. 2016, 16, 1–7. [Google Scholar]
- Indu, M.N.A.; Hatha, A.M.; Abirosh, C.; Harsha, U.; Vivekanandan, G. Antimicrobial activity of some of the south-Indian spices against serotypes of Escherichia coli, Salmonella, Listeria monocytogenes and Aeromonas hydrophila. Braz. J. Microbiol. 2006, 37, 53–158. [Google Scholar] [CrossRef] [Green Version]
- Cushman, D.W.; Cheung, H.S. Spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung. Biochem. Pharmacol. 1971, 20, 637–1648. [Google Scholar] [CrossRef] [PubMed]







| Bacterial Strains | Zone of Inhibition (mm) | ||
|---|---|---|---|
| A | B | C | |
| S. aureus NBRC 102135 | + | ++ | +++ |
| M. luteus NBRC 3066 | – | + | ++ |
| E. coli Y1090 | – | – | – |
| S. putrefaciens IAM 1509 | – | – | – |
| Purification Step | IC50 Value (mg/mL) a,b | Purification Fold |
|---|---|---|
| KSHPH | 1.90 ± 0.03 | 1.00 |
| KSHPH-F9 | 0.97 ± 0.04 | 1.96 |
| KSHPH-F9-III | 0.045 ± 0.005 | 42.22 |
| Bacterial Strains | MIC Values (mg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| KSHPH−F9 | KSHPH−F9-I | Tetracycline | |||||
| mg/mL | mM | Mg/mL | mM | mg/mL | mM | ||
| Gram (+) | S. aureus NBRC 102135 | 5.00 a | NA | 0.67 a | 1.62 a | 0.0017 a | 0.0038 a |
| M. luteus NBRC 3066 | 5.00 a | NA | 0.83 a | 2.00 a | 0.0017 a | 0.0038 a | |
| Gram (−) | E. coli Y1090 | 16.67 b | NA | 3.33 b | 8.03 b | 0.3333 b | 0.7499 b |
| S. putrefaciens IAM 1509 | 6.67 a | NA | 1.00 a | 2.41 a | 0.0025 a | 0.0056 a | |
| Hydrogen Bond Obtained from Molecular Docking | ||||
|---|---|---|---|---|
| ARL | Distance (Å) | ARI | Distance (Å) | |
| Glu162: O(E2) | × | √ | 2.17 | |
| Gln281: N(E2) | √ | 2.21 | √ | 1.93 |
| Ala354: O | √ | 2.13 | √ | 2.22 |
| Ala354: O | √ | 2.59 | ||
| His383: N(E2) | √ | 3.35 | √ | 3.35 |
| Glu384: O(E2) | √ | 1.78 | √ | 1.90 |
| Glu384: O(E2) | √ | 2.08 | √ | 2.27 |
| Glu384: O(E2) | √ | 2.18 | ||
| Lys511: N(Z) | √ | 1.69 | √ | 1.70 |
| His513: N(E2) | √ | 2.20 | √ | 2.22 |
| Tyr520: O(H) | √ | 2.14 | √ | 1.92 |
| Tyr523: O(H) | √ | 1.81 | √ | 1.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Han, Q.; Koyama, T.; Ishizaki, S. Preparation, Purification and Characterization of Antibacterial and ACE Inhibitory Peptides from Head Protein Hydrolysate of Kuruma Shrimp, Marsupenaeus japonicus. Molecules 2023, 28, 894. https://doi.org/10.3390/molecules28020894
Zhou J, Han Q, Koyama T, Ishizaki S. Preparation, Purification and Characterization of Antibacterial and ACE Inhibitory Peptides from Head Protein Hydrolysate of Kuruma Shrimp, Marsupenaeus japonicus. Molecules. 2023; 28(2):894. https://doi.org/10.3390/molecules28020894
Chicago/Turabian StyleZhou, Jie, Qiuyu Han, Tomoyuki Koyama, and Shoichiro Ishizaki. 2023. "Preparation, Purification and Characterization of Antibacterial and ACE Inhibitory Peptides from Head Protein Hydrolysate of Kuruma Shrimp, Marsupenaeus japonicus" Molecules 28, no. 2: 894. https://doi.org/10.3390/molecules28020894
APA StyleZhou, J., Han, Q., Koyama, T., & Ishizaki, S. (2023). Preparation, Purification and Characterization of Antibacterial and ACE Inhibitory Peptides from Head Protein Hydrolysate of Kuruma Shrimp, Marsupenaeus japonicus. Molecules, 28(2), 894. https://doi.org/10.3390/molecules28020894







