Toward Accurate yet Effective Computations of Rotational Spectroscopy Parameters for Biomolecule Building Blocks
Abstract
1. Introduction
- Unsupervised perception of the molecular system to identify hard and soft degrees of freedom [15];
- Exploration of the PES governed by soft degrees of freedom using the same semi-empirical method of the previous step, guided by a purposely tailored evolutionary algorithm with the aim of finding other low-lying minima [10];
- Refinement of the most stable structures by hybrid and then double-hybrid density functionals [14];
- Analysis of relaxation paths between pairs of adjacent energy minima [13];
2. Results and Discussion
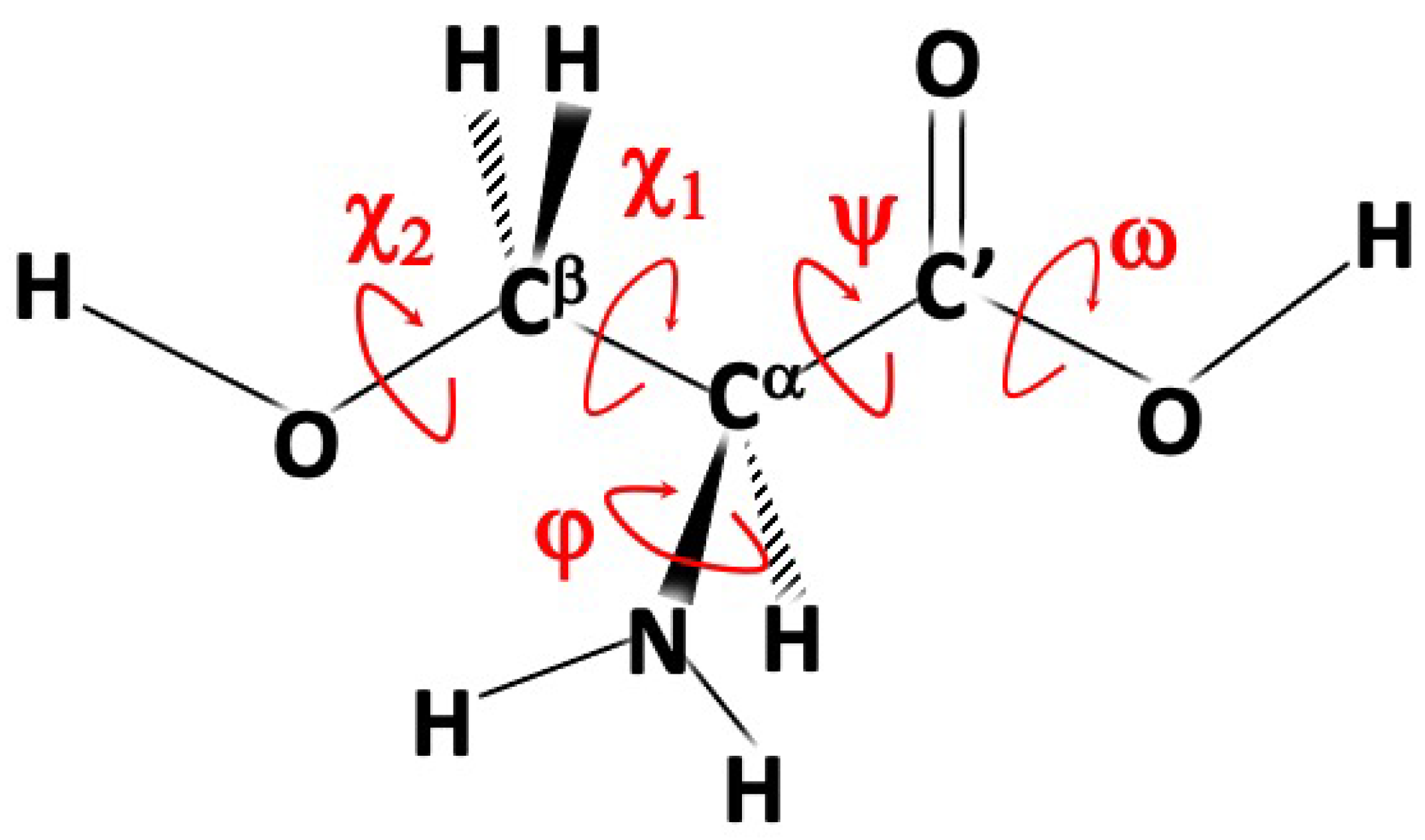
2.1. The Methodologic Approach
2.2. The Validation Step
2.3. Amino Acids
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schols, G. Atomic and Molecular Beam Methods; Oxford University Press: Oxford, UK, 1988. [Google Scholar]
- Alonso, J.L.; López, J.C. Microwave spectroscopy of biomolecular building blocks. In Gas-Phase IR Spectroscopy and Structure of Biological Molecules; Springer: Berlin/Heidelberg, Germany, 2015; pp. 335–401. [Google Scholar]
- Godfrey, P.D.; Rodgers, F.M.; Brown, R.D. Theory versus Experiment in Jet Spectroscopy: Glycolic Acid. J. Am. Chem. Soc. 1997, 119, 2232–2239. [Google Scholar] [CrossRef]
- Florio, G.M.; Christie, R.A.; Jordan, K.D.; Zwier, T.S. Conformational Preferences of Jet-Cooled Melatonin: Probing trans- and cis-Amide Regions of the Potential Energy Surface. J. Am. Chem. Soc. 2002, 124, 10236–10247. [Google Scholar] [CrossRef]
- Helgaker, T.; Klopper, W.; Tew, D.P. Quantitative quantum chemistry. Mol. Phys. 2008, 106, 2107–2143. [Google Scholar] [CrossRef]
- Karton, A. A computational chemist’s guide to accurate thermochemistry for organic molecules. WIRES Comp. Mol. Sci. 2016, 6, 292–310. [Google Scholar] [CrossRef]
- Puzzarini, C.; Bloino, J.; Tasinato, N.; Barone, V. Accuracy and interpretability: The devil and the holy grail. New routes across old boundaries in computational spectroscopy. Chem. Rev. 2019, 119, 8131–8191. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, M.K.; Karton, A.; Martin, J.M. Benchmark ab initio conformational energies for the proteinogenic amino acids through explicitly correlated methods. Assessment of density functionale methods. J. Chem. Theory Comput. 2016, 12, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Shu, C.; Ye, H.; Biczysko, M. Structural and energetic properties of amino acids and peptides benchmarked by accurate theoretical and experimental data. J. Phys. Chem. A 2021, 125, 9826–9837. [Google Scholar] [CrossRef]
- Mancini, G.; Fusè, M.; Lazzari, F.; Chandramouli, B.; Barone, V. Unsupervised search of low-lying conformers with spectroscopic accuracy: A two-step algorithm rooted into the island model evolutionary algorithm. J. Chem. Phys. 2020, 153, 124110. [Google Scholar] [CrossRef]
- Ferro-Costas, D.; Mosquera-Lois, I.; Fernandez-Ramos, A. Torsiflex: An automatic generator of torsional conformers. application to the twenty proteinogenic amino acids. J. Cheminf. 2021, 13, 100. [Google Scholar] [CrossRef]
- Barone, V.; Puzzarini, C.; Mancini, G. Integration of theory, simulation, artificial intelligence and virtual reality: A four-pillar approach for reconciling accuracy and interpretability in computational spectroscopy. Phys. Chem. Chem. Phys. 2021, 23, 17079–17096. [Google Scholar] [CrossRef]
- León, I.; Fusè, M.; Alonso, E.R.; Mata, S.; Mancini, G.; Puzzarini, C.; Alonso, J.L.; Barone, V. Unbiased disentanglement of conformational baths with the help of microwave spectroscopy, quantum chemistry and artificial intelligence: The puzzling case of homocysteine. J. Chem. Phys. 2022, 157, 074107. [Google Scholar] [CrossRef]
- Mancini, G.; Fusè, M.; Lazzari, F.; Barone, V. Fast exploration of potential energy surfaces with a joint venture of quantum chemistry, evolutionary algorithms and unsupervised learning. Digit. Discov. 2022, 1, 10539–10547. [Google Scholar] [CrossRef]
- Lazzari, F.; Salvadori, A.; Mancini, G.; Barone, V. Molecular Perception for Visualization and Computation: The Proxima Library. J. Chem. Inf. Model. 2020, 60, 2668–2672. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, C.; Ehlert, S.; Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 2019, 15, 1652–1671. [Google Scholar] [CrossRef]
- Puzzarini, C.; Biczysko, M.; Barone, V.; Largo, L.; Pena, I.; Cabezas, C.; Alonso, J.L. Accurate characterization of the peptide linkage in the gas phase: A joint quantum-chemistry and rotational spectroscopy study of the glycine dipeptide analogue. J. Phys. Chem. Lett. 2014, 5, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, S.; Barone, V.; Puzzarini, C. Extension of the “Cheap” Composite Approach to Noncovalent Interactions: The jun-ChS Scheme. J. Chem. Theory Comput. 2020, 16, 988–1006. [Google Scholar] [CrossRef] [PubMed]
- Lupi, J.; Alessandrini, S.; Barone, V.; Puzzarini, C. junChS and junChS-F12 Models: Parameter-free Efficient yet Accurate Composite Schemes for Energies and Structures of Noncovalent Complexes. J. Chem. Theory Comput. 2021, 17, 6974–6992. [Google Scholar] [CrossRef] [PubMed]
- Barone, V. Anharmonic vibrational properties by a fully automated second order perturbative approach. J. Chem. Phys. 2005, 122, 014108. [Google Scholar] [CrossRef]
- Rosnik, A.M.; Polik, W.F. VPT2+K spectroscopic constants and matrix elements of the transformed vibrational Hamiltonian of a polyatomic molecule with resonances using Van Vleck perturbation theory. Mol. Phys. 2014, 112, 261–300. [Google Scholar] [CrossRef]
- Franke, P.R.; Stanton, J.F.; Douberly, G.E. How to VPT2: Accurate and Intuitive Simulations of CH Stretching Infrared Spectra Using VPT2+ K with Large Effective Hamiltonian Resonance Treatments. J. Phys. Chem. A 2021, 125, 1301–1324. [Google Scholar] [CrossRef]
- Mendolicchio, M.; Bloino, J.; Barone, V. Perturb-then-diagonalize vibrational engine exploiting curvilinear internal coordinates. J. Chem. Theory Comput. 2022, 18, 7603–7619. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Supramolecular Binding Thermodynamics by Dispersion-Corrected Density Functional Theory. Chem. A Eur. J. 2012, 18, 9955–9964. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Spada, L.; Alessandrini, S.; Zheng, Y.; Lengsfels, K.G.; Grabow, J.U.; Feng, G.; Puzzarini, C.; Barone, V. Stacked but not Stuck: Unveiling the role of π–π* Interactions with the Help of the Benzofuran-Formaldehyde Complex. Angew. Chem. Int. Ed. Engl. 2022, 61, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Alonso, E.R.; Fusé, M.; León, I.; Puzzarini, C.; Alonso, J.L.; Barone, V. Exploring the Maze of Cycloserine Conformers in the Gas Phase Guided by Microwave Spectroscopy and Quantum Chemistry. J. Phys. Chem. A 2021, 125, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.E.; Blanco, S.; López, J.C.; Alonso, J.L. Rotational Probes of Six Conformers of Neutral Cysteine. Angew. Chem. Int. Ed. 2008, 47, 6216–6220. [Google Scholar] [CrossRef] [PubMed]
- Lovas, F.J.; Kawashima, Y.; Grabow, J.U.; Suenram, R.D.; Fraser, G.T.; Hirota, E. Microwave Spectra, Hyperfine Structure, and Electric Dipole Moments for Conformers I and II of Glycine. Astrophys. J. 1995, 455, L201–L204. [Google Scholar] [CrossRef]
- Blanco, S.; Lesarri, A.; López, J.C.; Alonso, J.L. The gas-phase structure of alanine. J. Am. Chem. Soc. 2004, 126, 11675–11683. [Google Scholar] [CrossRef]
- Lesarri, E.J.; Cocinero, J.C.; López, J.C.; Alonso, J.L. The Shape of Neutral Valine. Angew. Chem. Int. Ed. Engl. 2004, 43, 605–610. [Google Scholar] [CrossRef]
- Lesarri, E.J.; Sanchez, R.; Cocinero, E.J.; López, J.C.; Alonso, J.L. Coded Amino Acids in Gas Phase: The Shape of Isoleucine. J. Am. Chem. Soc. 2005, 127, 12952–12956. [Google Scholar] [CrossRef]
- Cocinero, J.C.; Lesarri, E.J.; Grabow, J.U.; López, J.C.; Alonso, J.L. The Shape of Leucine in the Gas Phase. Chem. Phys. Chem. 2007, 8, 599–604. [Google Scholar] [CrossRef]
- Blanco, S.; Sanz, M.E.; López, J.C.; Alonso, J.L. Revealing the multiple structures of serine. Proc. Natl. Acad. Sci. USA 2007, 104, 20183–20188. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.L.; Peña, I.; López, J.C.; Vaquero, V. Rotational spectra signatures of four tautomers of guanine. Angew. Chem. Int. Ed. 2009, 48, 6141–6143. [Google Scholar] [CrossRef] [PubMed]
- Cabezas, C.; Varela, M.; Peña, I.; Mata, S.; López, J.C.; Alonso, J.L. The conformational locking of asparagine. Chem. Commun. 2012, 48, 5934–5936. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.E.; Cabezas, C.; Mata, S.; Alonso, J.L. Rotational spectrum of tryptophan. J. Chem. Phys. 2014, 140, 204318. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.; Mata, S.; Blanca, S.; Lopez, J.C.; Alonso, J.L. Jet-cooled rotational spectrum of laser-ablated phenylalanine. J. Phys. Chem. A 2011, 115, 9653–9657. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.; Mata, S.; Cabezas, C.; Lopez, J.C.; Alonso, J.L. The rotational spectrum of tyrosine. J. Phys. Chem. A 2015, 119, 3731–3735. [Google Scholar] [CrossRef]
- Godfrey, P.D.; Brown, R.D. Proportions of Species Observed in Jet Spectroscopy-Vibrational Energy Effects: Histamine Tautomers and Conformers. J. Am. Chem. Soc. 1998, 120, 10724–10732. [Google Scholar] [CrossRef]
- Alonso, E.R.; León, I.; Alonso, J.L. Intra- and Intermolecular Interactions Between Non-Covalently Bonded Species; Elsevier: Amsterdam, The Netherlands, 2020; pp. 93–141. [Google Scholar]
- Nguyen, H.V.L.; Kleiner, I. Understanding (coupled) large amplitude motions: The interplay of microwave spectroscopy, spectral modeling, and quantum chemistry. Phys. Sci. Rev. 2020, 20200037. [Google Scholar] [CrossRef]
- Santra, G.; Sylvetsky, N.; Martin, J.M. Minimally empirical double-hybrid functionals trained against the GMTKN55 database: RevDSD-PBEP86-D4, revDOD-PBE-D4, and DOD-SCAN-D4. J. Phys. Chem. A 2019, 123, 5129–5143. [Google Scholar] [CrossRef]
- Papajak, E.; Zheng, J.; Xu, X.; Leverentz, H.R.; Truhlar, D.G. Perspectives on Basis Sets Beautiful: Seasonal Plantings of Diffuse Basis Functions. J. Chem. Theory Comput. 2011, 7, 3027–3034. [Google Scholar] [CrossRef]
- Lupi, J.; Puzzarini, C.; Cavallotti, C.; Barone, V. State-of-the-Art Quantum Chemistry Meets Variable Reaction Coordinate Transition State Theory to Solve the Puzzling Case of the H2S + Cl System. J. Chem. Theory Comput. 2020, 16, 5090–5104. [Google Scholar] [CrossRef] [PubMed]
- García de la Concepción, J.; Puzzarini, C.; Barone, V.; Jiménez-Serra, I.; Roncero, O. Formation of phosphorus monoxide (PO) in the interstellar medium: Insights from quantum-chemical and kinetic calculations. Astrophys. J. Lett. 2021, 922, 169. [Google Scholar] [CrossRef]
- Barone, V.; Lupi, J.; Salta, Z.; Tasinato, N. Development and Validation of a Parameter-Free Model Chemistry for the Computation of Reliable Reaction Rates. J. Chem. Theory Comput. 2021, 17, 4913–4928. [Google Scholar] [CrossRef] [PubMed]
- Shavitt, I.; Bartlett, R.J. Many-Body Methods in Chemistry and Physics: MBPT and Coupled-Cluster Theory; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Tajti, A.; Szalay, P.G.; Császár, A.G.; Kállay, M.; Gauss, J.; Valeev, E.F.; Flowers, B.A.; Vázquez, J.; Stanton, J.F. HEAT: High accuracy extrapolated ab initio thermochemistry. J. Chem. Phys. 2004, 121, 11599–11613. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Biczysko, M.; Bloino, J.; Puzzarini, C. Accurate structure, thermochemistry and spectroscopic parameters from CC and CC/DFT schemes: The challenge of the conformational equilibrium in glycine. Phys. Chem. Chem. Phys. 2013, 15, 10094–10111. [Google Scholar] [CrossRef]
- Puzzarini, C.; Barone, V. The challenging playground of astrochemistry: An integrated rotational spectroscopy—Quantum chemistry strategy. Phys. Chem. Chem. Phys. 2020, 22, 6507–6523. [Google Scholar] [CrossRef]
- Puzzarini, C.; Barone, V. Extending the molecular size in accurate quantum-chemical calculations: The equilibrium structure and spectroscopic properties of uracil. Phys. Chem. Chem. Phys. 2011, 13, 7189–7197. [Google Scholar] [CrossRef] [PubMed]
- Møller, C.; Plesset, M.S. Note on an Approximation Treatment for Many-Electron Systems. Phys. Rev. 1934, 46, 618–622. [Google Scholar] [CrossRef]
- Raghavachari, K.; Trucks, G.W.; Pople, J.A.; Head-Gordon, M. A fifth-order perturbation comparison of electron correlation theories. Chem. Phys. Lett. 1989, 157, 479–483. [Google Scholar] [CrossRef]
- Helgaker, T.; Klopper, W.; Koch, H.; Noga, J. Basis-set convergence of correlated calculations on water. J. Chem. Phys. 1997, 106, 9639–9646. [Google Scholar] [CrossRef]
- Woon, D.E.; Dunning, T.H., Jr. Gaussian Basis Sets for Use in Correlated Molecular Calculations. V. Core-Valence Basis Sets for Boron through Neon. J. Chem. Phys. 1995, 103, 4572. [Google Scholar] [CrossRef]
- Peterson, K.A.; Dunning, T.H., Jr. Accurate correlation consistent basis sets for molecular core-valence correlation effects: The second row atoms Al-Ar, and the first row atoms B-Ne revisited. J. Chem. Phys. 2002, 117, 10548–10560. [Google Scholar] [CrossRef]
- Dunning, T.H., Jr. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. JChPh 1989, 90, 1007–1023. [Google Scholar] [CrossRef]
- Dunning, T.H.; Peterson, K.A.; Wilson, A.K. Gaussian basis sets for use in correlated molecular calculations. X. The atoms aluminum through argon revisited. J. Chem. Phys. 2001, 114, 9244–9253. [Google Scholar] [CrossRef]
- Knizia, G.; Adler, T.B.; Werner, H.J. Simplified CCSD(T)-F12 methods: Theory and benchmarks. J. Chem. Phys. 2009, 130, 054104. [Google Scholar] [CrossRef] [PubMed]
- Werner, H.-J.; Adler, T.B.; Manby, F.R. General orbital invariant MP2-F12 theory. J. Chem. Phys. 2007, 126, 164102. [Google Scholar] [CrossRef]
- Feller, D.; Peterson, K. An expanded calibration study of the explicitly correlated CCSD(T)-F12b method using large basis set standard CCSD(T) atomization energies. J. Chem. Phys. 2013, 139, 084110. [Google Scholar] [CrossRef]
- Puzzarini, C.; Biczysko, M.; Barone, V.; Peña, I.; Cabezas, C.; Alonso, J.L. Accurate Molecular Structure and Spectroscopic Properties of Nucleobases: A Combined Computational-Microwave Investigation of 2-Thiouracil as a Case Study. Phys. Chem. Chem. Phys. 2013, 15, 16965–16975. [Google Scholar] [CrossRef]
- Melli, A.; Melosso, M.; Lengsfeld, K.G.; Bizzocchi, L.; Rivilla, V.M.; Dore, L.; Barone, V.; Grabow, J.U.; Puzzarini, C. Spectroscopic Characterization of 3-Aminoisoxazole, a Prebiotic Precursor of Ribonucleotides. Molecules 2022, 27, 3278. [Google Scholar] [CrossRef]
- Barone, V.; Ceselin, G.; Fusè, M.; Tasinato, N. Accuracy meets interpretability for computational spectroscopy by means of hybrid and double-hybrid functionals. Front. Chem. 2020, 8, 584203. [Google Scholar] [CrossRef]
- Biczysko, M.; Panek, P.; Scalmani, G.; Bloino, J.; Barone, V. Harmonic and Anharmonic Vibrational Frequency Calculations with the Double-Hybrid B2PLYP Method: Analytic Second Derivatives and Benchmark Studies. J. Chem. Theory Comput. 2010, 6, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.M.L.; Kesharwani, M.K. Assessment of CCSD(T)-F12 approximations and basis sets for harmonic vibrational frequencies. J. Chem. Theory Comput. 2014, 10, 2085–2090. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Biczysko, M.; Bloino, J. Fully anharmonic IR and Raman spectra of medium-size molecular systems: Accuracy and interpretation. Phys. Chem. Chem. Phys. 2014, 16, 1759–1787. [Google Scholar] [CrossRef]
- Luchini, G.; Alegre-Requena, J.; Funes-Ardoiz, I.; Paton, R. GoodVibes: Automated thermochemistry for heterogeneous computational chemistry data [version 1; peer review: 2 approved with reservations]. F1000Research 2020, 9, 291. [Google Scholar] [CrossRef]
- Puzzarini, C.; Stanton, J.F. Connections between the accuracy of rotational constants and equilibrium molecular structures. Phys. Chem. Chem. Phys. 2023, 25. [Google Scholar] [CrossRef]
- Puzzarini, C.; Heckert, M.; Gauss, J. The accuracy of rotational constants predicted by high-level quantum-chemical calculations. I. Molecules containing first-row atoms. J. Chem. Phys. 2008, 128, 194108. [Google Scholar] [CrossRef] [PubMed]
- Piccardo, M.; Penocchio, E.; Puzzarini, C.; Biczysko, M.; Barone, V. Semi-experimental equilibrium structure determinations by employing B3LYP/SNSD anharmonic force fields: Validation and application to semirigid organic molecules. J. Phys. Chem. A 2015, 119, 2058–2082. [Google Scholar] [CrossRef]
- Penocchio, E.; Piccardo, M.; Barone, V. Semiexperimental Equilibrium Structures for Building Blocks of Organic and Biological Molecules: The B2PLYP Route. J. Chem. Theory Comput. 2015, 11, 4689–4707. [Google Scholar] [CrossRef]
- Ceselin, G.; Barone, V.; Tasinato, N. Accurate biomolecular structures by the nano-LEGO approach: Pick the bricks and build your geometry. J. Chem Theory Comput. 2021, 17, 7290–7311. [Google Scholar] [CrossRef]
- Melli, A.; Tonolo, F.; Barone, V.; Puzzarini, C. Extending the Applicability of the Semi-experimental Approach by Means of “Template Molecule” and “Linear Regression” Models on Top of DFT Computations. J. Phys. Chem. A 2021, 125, 9904–9916. [Google Scholar] [CrossRef]
- Puzzarini, C.; Stanton, J.F.; Gauss, J. Quantum-chemical calculation of spectroscopic parameters for rotational spectroscopy. Int. Rev. Phys. Chem. 2010, 29, 273–367. [Google Scholar] [CrossRef]
- Gordy, W.; Cook, R.L. Microwave Molecular Spectra; Wiley: Hoboken, NJ, USA, 1984. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Stanton, J.F.; Gauss, J.; Harding, M.E.; Szalay, P.G. CFOUR. A Quantum Chemical Program Package. 2016. Available online: http://www.cfour.de (accessed on 19 December 2022).
- Werner, H.J.; Knowles, P.J.; Manby, F.R.; Black, J.A.; Doll, K.; Hesselmann, A.; Kats, D.; Köhn, A.; Korona, T.; Kreplin, D.A.; et al. The Molpro quantum chemistry package. J. Chem. Phys. 2020, 152, 144107. [Google Scholar] [CrossRef]
- Puzzarini, C.; Spada, L.; Alessandrini, S.; Barone, V. The challenge of non-covalent interactions: Theory meets experiment for reconciling accuracy and interpretation. J. Phys. Condens. Matter 2020, 32, 343002. [Google Scholar] [CrossRef]
- Řezáč, J.; Hobza, P. Describing Noncovalent Interactions beyond the Common Approximations: How Accurate Is the “Gold Standard,” CCSD(T) at the Complete Basis Set Limit? J. Chem. Theory Comput. 2013, 9, 2151–2155. [Google Scholar] [CrossRef] [PubMed]
- Řezáč, J.; Dubecký, M.; Jurečka, P.; Hobza, P. Extensions and applications of the A24 data set of accurate interaction energies. Phys. Chem. Chem. Phys. 2015, 17, 19268–19277. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Puzzarini, C. Gas-Phase Computational Spectroscopy: The Challenge of the Molecular Bricks of Life. Ann. Rev. Phys. Chem. 2023, 74. [Google Scholar] [CrossRef] [PubMed]
- Benedict, W.S.; Gailar, N.; Plyler, E.K. Rotation-Vibration Spectra of Deuterated Water Vapor. J. Chem. Phys. 1956, 24, 1139. [Google Scholar] [CrossRef]
- Smith, A.M.; Coy, S.L.; Klemperer, W.; Lehmann, K.K. Fourier Transform Spectra of Overtone Bands of HCN from 5400 to 15100 cm−1. J. Mol. Spectrosc. 1989, 134, 134–153. [Google Scholar] [CrossRef]
- Teffo, J.L.; Sulakshina, O.N.; Perevalov, V.I. Effective Hamiltonian for Rovibrational Energies and Line Intensities of Carbon Dioxide]. J. Mol. Spectrosc. 1992, 156, 48–64. [Google Scholar] [CrossRef]
- Strey, G.; Mills, I.M. Anharmonic Force Field of Acetylene]. J. Mol. Spectrosc. 1976, 59, 103–115. [Google Scholar] [CrossRef]
- Yachmenev, A.; Yurchenko, S.N.; Jensen, P.; Thiel, W. A New Spectroscopic Potential Energy Surface for Formaldehyde in its Ground Electronic State. J. Chem. Phys. 2011, 134, 244307. [Google Scholar] [CrossRef] [PubMed]
- East, A.L.L.; Allen, W.D.; Klippenstein, S.J. [The Anharmonic Force Field and Equilibrium Molecular Structure of Ketene]. J. Chem. Phys. 1995, 102, 8506–8532. [Google Scholar] [CrossRef]
- Huang, X.; Schwenke, D.W.; Lee, T.J. Rovibrational Spectra of Ammonia. I. Unprecedented Accuracy of a Potential Energy Surface Used with Nonadiabatic Corrections. J. Chem. Phys. 2011, 134, 044320. [Google Scholar] [CrossRef]
- Wang, D.; Shi, Q.; Zhu, Q.S. An ab Initio Quartic Force Field of PH3. J. Chem. Phys. 2000, 112, 9624. [Google Scholar] [CrossRef]
- Shu, C.; Jiang, Z.; Biczysko, M. Toward accurate prediction of amino acid derivatives structure and energetics from DFT: Glycine conformers and their interconversions. J. Mol. Model. 2020, 26, 129. [Google Scholar] [CrossRef]
- Nacsa, A.B.; Csako, G. Benchmark ab initio proton affinity of glycine. Phys. Chem. Chem. Phys. 2021, 23, 9663–9671. [Google Scholar] [CrossRef] [PubMed]
- Puzzarini, C.; Barone, V. Diving for accurate structures in the ocean of molecular systems with the help of spectroscopy and quantum chemistry. Acc. Chem. Res. 2018, 51, 548–556. [Google Scholar] [CrossRef]
- Sheng, M.; Silvestrini, F.; Biczysko, M.; Puzzarini, C. Structural and vibrational properties of amino acids from composite schemes and double-hybrid DFT: Hydrogen bonding in serine as a test case. J. Phys. Chem. A 2021, 125, 9099–9114. [Google Scholar] [CrossRef] [PubMed]
| junChS | junChSF12 | junCCF12+CV | Best | |
|---|---|---|---|---|
| H2O···H2O | −21.10 | −21.00 | −21.03 | −21.07 |
| NH3···NH3 | −13.30 | −13.20 | −12.99 | −13.26 |
| HF···HF | −19.45 | −19.25 | −19.25 | −19.18 |
| CH2O···CH2O | −19.23 | −18.96 | −18.59 | −18.89 |
| HCN···HCN | −19.88 | −19.84 | −19.83 | −19.95 |
| C2H4···C2H4 | −4.75 | −4.66 | −4.36 | −4.64 |
| CH4···CH4 | −2.25 | −2.20 | −1.99 | −2.27 |
| H2O···NH3 | −27.57 | −27.47 | −27.34 | −27.39 |
| H2O···C2H4 | −10.86 | −10.77 | −10.55 | −10.82 |
| C2H4···CH2O | −6.94 | −6.83 | −6.56 | −6.84 |
| NH3···C2H4 | −5.89 | −5.82 | −5.61 | −5.84 |
| HF···CH4 | −7.13 | −7.04 | −6.85 | −6.95 |
| H2O···CH4 | −2.78 | −2.77 | −2.68 | −2.85 |
| NH3···CH4 | −3.24 | −3.23 | −3.11 | −3.26 |
| MAX | 0.34 | 0.11 | 0.30 | |
| MUE | 0.11 | 0.06 | 0.19 | |
| RMSD | 0.14 | 0.07 | 0.21 |
| junChS | DZCCF12 | DZCCF12+CV | TZCCF12+CV | junCCF12+CV | |
|---|---|---|---|---|---|
| 2nd-row (17 molecules) | |||||
| MAX(r) | 0.0042 | 0.0044 | 0.0027 | 0.0039 | 0.0044 |
| MAX(, ) | 1.52 | 0.61 | 0.62 | 0.79 | 0.62 |
| MUE(r) | 0.0011 | 0.0022 | 0.0009 | 0.0008 | 0.0007 |
| MUE(, ) | 0.19 | 0.19 | 0.16 | 0.16 | 0.15 |
| 3rd-row (7 molecules) | |||||
| MAX(r) | 0.0012 | 0.0046 | 0.0040 | 0.0021 | 0.0014 |
| MAX(, ) | 0.10 | 0.21 | 0.24 | 0.09 | 0.17 |
| MUE(r) | 0.0005 | 0.0018 | 0.0010 | 0.0004 | 0.0005 |
| MUE(, ) | 0.05 | 0.10 | 0.09 | 0.02 | 0.06 |
| DZCCF12 | junCCF12 | augCCF12 | rDSD | |
|---|---|---|---|---|
| MAX | 13.7 | 12.8 | 18.4 | 18.2 |
| MUE | 4.1 | 4.0 | 4.6 | 5.6 |
| RMSD | 5.0 | 5.2 | 6.1 | 7.1 |
| Parameter | Exp. | ChS | rDSD | rDSD-LRA | MP2/cc-pVTZ | B3LYP/SNSD | |
|---|---|---|---|---|---|---|---|
| Glycine (I) | A | 10,418.2 | 10,396.6 | 10,334.8 | 10,390.3 | 10,328.0 | 10,283.1 |
| B | 3906.9 | 3901.1 | 3879.9 | 3897.6 | 3905.0 | 3831.1 | |
| C | 2934.4 | 2930.4 | 2913.5 | 2927.4 | 2926.2 | 2882.9 | |
| −1.208(9) | −1.278 | −1.336 | |||||
| −0.343(8) | −0.464 | −0.448 | |||||
| 1.552(10) | 1.742 | 1.785 | |||||
| Glycine (II) | A | 10,144.5 | 10,205.3 | 10,139.3 | 10,193.8 | 10,178.7 | 10,135.0 |
| B | 4094.5 | 4095.6 | 4078.7 | 4097.2 | 4104.7 | 4043.4 | |
| C | 3024.7 | 3030.6 | 3021.3 | 3035.8 | 3041.0 | 2993.1 | |
| 1.773(2) | 1.876 | 1.922 | |||||
| −3.194(4) | −3.286 | −3.344 | |||||
| 1.421(4) | 1.413 | 1.422 | |||||
| E | 201.5 | 214.8 | 157.2 | 237.8 | |||
| Serine (Igg) | A | 4528.1 | 4499.4 | 4487.0 | 4510.4 | 4494.9 | 4485.2 |
| B | 1838.8 | 1841.3 | 1822.7 | 1831.6 | 1830.8 | 1809.1 | |
| C | 1460.9 | 1460.0 | 1451.8 | 1459.0 | 1462.7 | 1433.3 | |
| −4.302(3) | −4.416 | −4.554 | |||||
| 2.8236(6) | 2.852 | 2.868 | |||||
| 1.479(5) | 1.565 | 1.685 | |||||
| Serine (IIgg) | A | 3585.9 | 3560.2 | 3559.3 | 3578.1 | 3554.8 | 3547.2 |
| B | 2412.7 | 2410.2 | 2393.0 | 2404.7 | 2414.0 | 2342.0 | |
| C | 1754.4 | 1757.6 | 1739.7 | 1748.2 | 1757.9 | 1713.6 | |
| −3.462(2) | −3.530 | −3.670 | |||||
| 2.0797(9) | 2.149 | 2.134 | |||||
| 1.382(5) | 1.381 | 1.536 | |||||
| E | −167.2 | −161.8 | −185.5 | −100.8 |
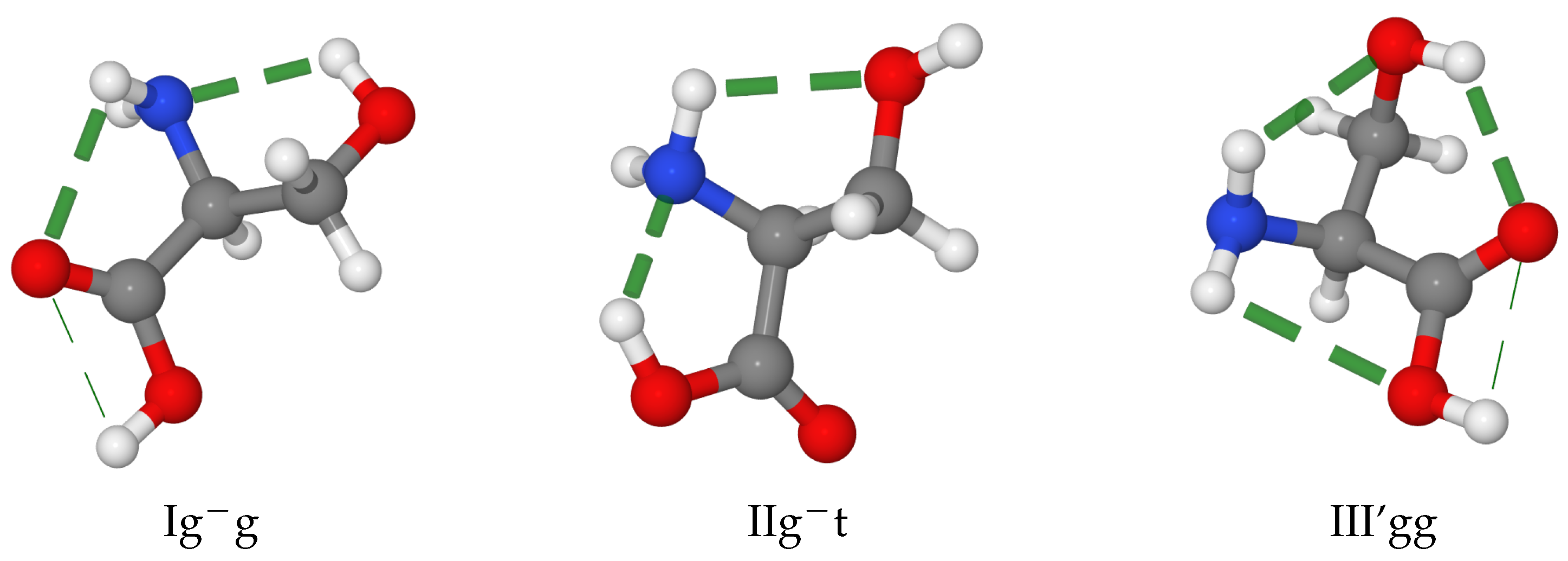
| Igg | IIgg | I’gg | IItg | III’gg | IIgt | III’tg | |
|---|---|---|---|---|---|---|---|
| Calc. | |||||||
| A | 4461.34 | 3549.33 | 3505.74 | 3630.86 | 3950.32 | 4508.13 | 3464.84 |
| B | 1823.01 | 2372.38 | 2305.21 | 2382.52 | 2222.91 | 1843.00 | 2304.68 |
| C | 1441.95 | 1734.67 | 1803.62 | 1515.28 | 1657.03 | 1462.05 | 1604.74 |
| −4.5535 | −3.6696 | −0.9235 | −3.8114 | −0.6094 | −0.3660 | −1.0975 | |
| 2.8681 | 2.1341 | 2.5528 | 2.1268 | −0.6702 | 2.0569 | −0.6582 | |
| 1.6854 | 1.5355 | −1.6293 | 1.6847 | 1.2796 | −1.6909 | 1.7557 | |
| G | 0.0 | 44.1 | 222.4 | 295.6 | 481.2 | 522.7 | 620.6 |
| Exp. | |||||||
| A | 4479.0320(12) | 3557.20088(35) | 3524.38806(41) | 3638.05784(38) | 3931.7548(76) | 4517.473(17) | 3510.4015(35) |
| B | 830.16170(25) | 2380.37208(40) | 2307.76826(70) | 2387.89651(99) | 2242.76701(70) | 1846.99360(30) | 2321.90829(24) |
| C | 1443.79545(28) | 1740.92458(10) | 1805.20788(60) | 1519.18716(36) | 1664.53012(57) | 1463.79646(31) | 1584.38608(32) |
| −4.3023(27) | −3.4616(19) | −1.1343(35) | −3.6257(57) | −0.6733(67) | −0.6066(55) | −1.0486(55) | |
| 2.82359(63) | 2.07974(93) | 2.5043(50) | 2.06213(26) | −0.456(16) | 2.0723(82) | −0.5637(53) | |
| 1.4788(46) | 1.3819(47) | −1.3701(50) | 1.5906(50) | 1.129(16) | −1.466(30) | 1.612(21) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barone, V.; Di Grande, S.; Puzzarini, C. Toward Accurate yet Effective Computations of Rotational Spectroscopy Parameters for Biomolecule Building Blocks. Molecules 2023, 28, 913. https://doi.org/10.3390/molecules28020913
Barone V, Di Grande S, Puzzarini C. Toward Accurate yet Effective Computations of Rotational Spectroscopy Parameters for Biomolecule Building Blocks. Molecules. 2023; 28(2):913. https://doi.org/10.3390/molecules28020913
Chicago/Turabian StyleBarone, Vincenzo, Silvia Di Grande, and Cristina Puzzarini. 2023. "Toward Accurate yet Effective Computations of Rotational Spectroscopy Parameters for Biomolecule Building Blocks" Molecules 28, no. 2: 913. https://doi.org/10.3390/molecules28020913
APA StyleBarone, V., Di Grande, S., & Puzzarini, C. (2023). Toward Accurate yet Effective Computations of Rotational Spectroscopy Parameters for Biomolecule Building Blocks. Molecules, 28(2), 913. https://doi.org/10.3390/molecules28020913







