DDX20: A Multifunctional Complex Protein
Abstract
:1. Introduction
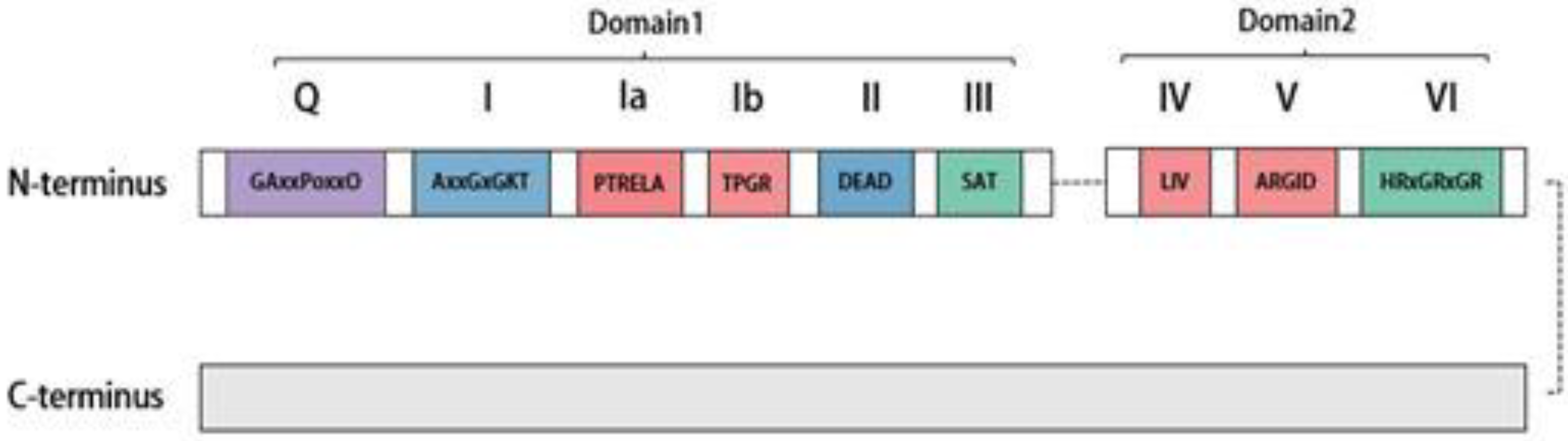
2. Distribution, Structure, and Subcellular Localization of DDX20
3. Splicing Features of DDX20
4. DDX20 Represses Transcription by Inhibiting Transcription Factors
5. Biogenesis of DDX20 and miRNA
6. Functions of DDX20 in the Innate Immune Signaling Pathway
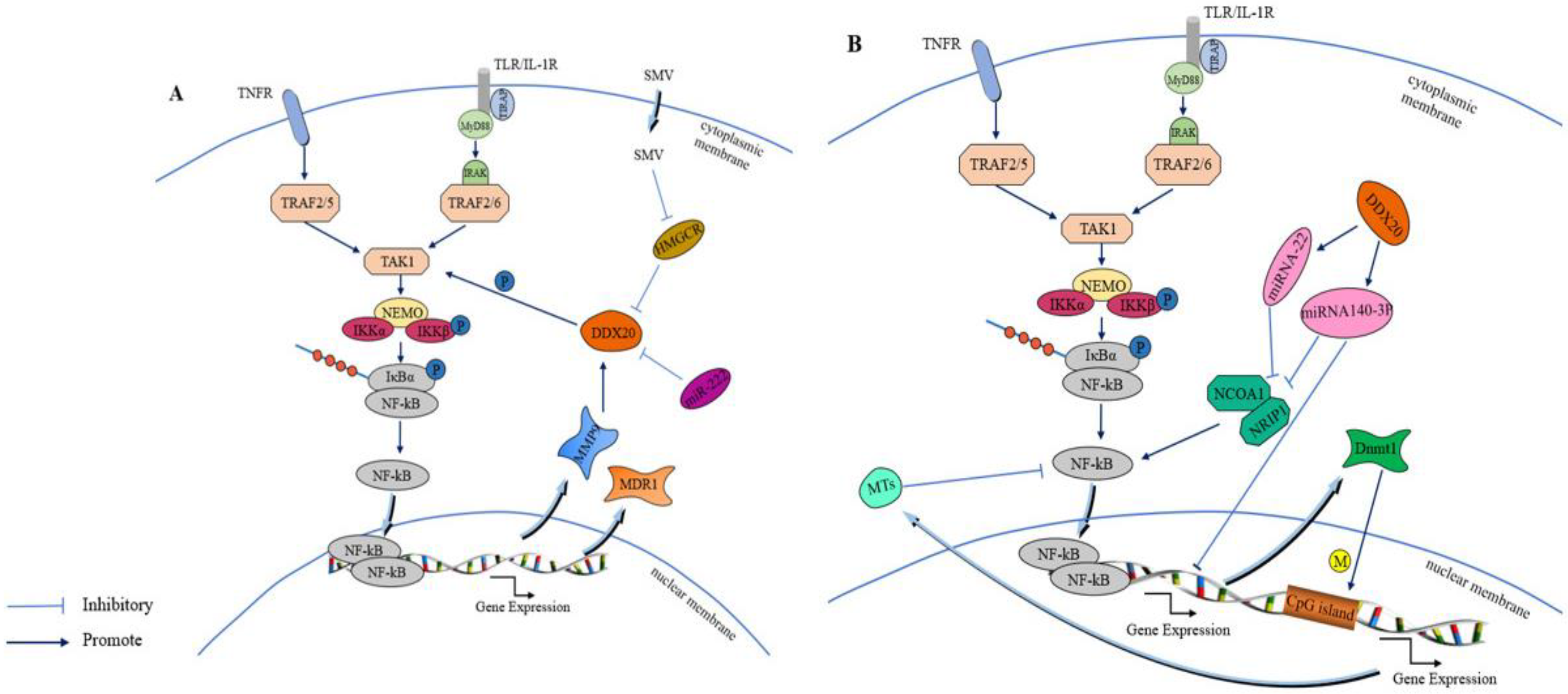
6.1. Effect of DDX20 on the NF-κB Signaling Pathway
6.2. DDX20 Affects p53 Signaling Pathway Conduction
7. DDX20 Plays Different Roles in Cancers through the NF-κB Signaling Pathway
8. Functions in Viral Infection
9. Concluding Remarks
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curmi, F.; Cauchi, R.J. The multiple lives of DEAD-box RNA helicase DP103/DDX20/Gemin3. Biochem. Soc. Trans. 2018, 46, 329–341. [Google Scholar] [CrossRef]
- Wang, X. The Role and Mechanisms of DEAD-Box RNA Helicase DDX20 on Antiviral Innate Immunity. Master’s Thesis, Soochow University, Suzhou, China, 2019. [Google Scholar]
- Mouillet, J.-F.; Yan, X.; Ou, Q.; Jin, L.; Muglia, L.J.; Crawford, P.A.; Sadovsky, Y. DEAD-box protein-103 (DP103, Ddx20) is essential for early embryonic development and modulates ovarian morphology and function. Endocrinology 2008, 149, 2168–2175. [Google Scholar] [CrossRef]
- Bizen, N.; Bepari, A.K.; Zhou, L.; Abe, M.; Sakimura, K.; Ono, K.; Takebayashi, H. Ddx20, an Olig2 binding factor, governs the survival of neural and oligodendrocyte progenitor cells via proper Mdm2 splicing and p53 suppression. Cell Death Differ. 2022, 29, 1028–1041. [Google Scholar] [CrossRef]
- Simankova, A.; Bizen, N.; Saitoh, S.; Shibata, S.; Ohno, N.; Abe, M.; Sakimura, K.; Takebayashi, H. Ddx20, DEAD box helicase 20, is essential for the differentiation of oligodendrocyte and maintenance of myelin gene expression. Glia 2021, 69, 2559–2574. [Google Scholar] [CrossRef]
- Hobani, Y.H.; Almars, A.I.; Alelwani, W.; Toraih, E.A.; Nemr, N.A.; Shaalan, A.A.M.; Fawzy, M.S.; Attallah, S.M. Genetic Variation in DEAD-Box Helicase 20 as a Putative Marker of Recurrence in Propensity-Matched Colon Cancer Patients. Genes 2022, 13, 1404. [Google Scholar] [CrossRef]
- Minasaki, R.; Puoti, A.; Streit, A. The DEAD-box protein MEL-46 is required in the germ line of the nematode Caenorhabditis elegans. BMC Dev. Biol. 2009, 9, 35. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Kojima, K.; Yoshikawa, T.; Kishikawa, T.; Ijichi, H.; Hirata, Y.; Tateishi, K.; Yoshida, H.; Omata, M.; et al. Abstract 3978: DDX20, a suppressor of hepatocarcinogenesis, controls NF-κB activity through regulating the function of miRNA-22 and miRNA-140-3p targeting transcriptional coactivators. Cancer Res. 2011, 71, 3978. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Hikiba, Y.; Obi, S.; Goto, T.; Kang, Y.J.; Maeda, S.; Yoshida, H.; et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 2013, 57, 162–170. [Google Scholar] [CrossRef]
- Ou, Q.; Mouillet, J.F.; Yan, X.; Dorn, C.; Crawford, P.A.; Sadovsky, Y. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 2001, 15, 69–79. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, P.; Li, X. High expression of DDX20 enhances the proliferation and metastatic potential of prostate cancer cells through the NF-κB pathway. Int. J. Mol. Med. 2016, 37, 1551–1557. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, M.; Pang, H.; Qiu, Y.; Sun, T.; Wang, T.; Shen, S.; Wang, W. A Macrophage Differentiation-Mediated Gene: DDX20 as a Molecular Biomarker Encompassing the Tumor Microenvironment, Disease Staging, and Prognoses in Hepatocellular Carcinoma. Oxidative Med. Cell. Longev. 2022, 2022, 9971776. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, Y.; Lin, R.; Weng, S.; Cai, F.; Zou, M.; Niu, H.; Ge, L.; Lin, Y. Analysis of the expression, function, prognosis and co-expression genes of DDX20 in gastric cancer. Comput. Struct. Biotechnol. J. 2020, 18, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Svobodova Kovarikova, A.; Grolich, T.; Prochazka, V.; Slaba, K.; Machackova, T.; Halamkova, J.; Svoboda, M.; Kala, Z.; Kiss, I.; et al. MicroRNA Biogenesis Pathway Genes Are Deregulated in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4460. [Google Scholar] [CrossRef] [PubMed]
- Kroiss, M.; Schultz, J.; Wiesner, J.; Chari, A.; Sickmann, A.; Fischer, U. Evolution of an RNP assembly system: A minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2008, 105, 10045–10050. [Google Scholar] [CrossRef]
- Lai, X.; Wang, T.; Tan, T.Z.; Casey, P.J.; Hong, W.; Sudol, M.; Tergaonkar, V.; Lee, S.C.; Kumar, A.P. Abstract A28: DEAD-box RNA helicase DP103 enhances YAP sumoylation for YAP-TEAD dependence and statin sensitivity in triple-negative breast cancer. Mol. Cancer Res. 2020, 18, A28. [Google Scholar] [CrossRef]
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352. [Google Scholar] [CrossRef]
- Schütz, P.; Karlberg, T.; van den Berg, S.; Collins, R.; Lehtiö, L.; Högbom, M.; Holmberg-Schiavone, L.; Tempel, W.; Park, H.W.; Hammarström, M.; et al. Comparative structural analysis of human DEAD-box RNA helicases. PLoS ONE 2010, 5, e12791. [Google Scholar] [CrossRef]
- Ali, M.A.M. The DEAD-box protein family of RNA helicases: Sentinels for a myriad of cellular functions with emerging roles in tumorigenesis. Int. J. Clin. Oncol. 2021, 26, 795–825. [Google Scholar] [CrossRef]
- Yan, X.; Mouillet, J.F.; Ou, Q.; Sadovsky, Y. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell Biol. 2003, 23, 414–423. [Google Scholar] [CrossRef]
- Coady, T.H.; Lorson, C.L. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip. Rev. RNA 2011, 2, 546–564. [Google Scholar] [CrossRef]
- Burlet, P.; Huber, C.; Bertrandy, S.; Ludosky, M.A.; Zwaenepoel, I.; Clermont, O.; Roume, J.; Delezoide, A.L.; Cartaud, J.; Munnich, A.; et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum. Mol. Genet. 1998, 7, 1927–1933. [Google Scholar] [CrossRef] [PubMed]
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell. Biol. 1999, 147, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Pellizzoni, L.; Yong, J.; Dreyfuss, G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002, 298, 1775–1779. [Google Scholar] [CrossRef] [PubMed]
- Lemm, I.; Girard, C.; Kuhn, A.N.; Watkins, N.J.; Schneider, M.; Bordonné, R.; Lührmann, R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell 2006, 17, 3221–3231. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L.; Boulon, S.; Lam, Y.W.; Urcia, R.; Boisvert, F.M.; Vandermoere, F.; Morrice, N.A.; Swift, S.; Rothbauer, U.; Leonhardt, H.; et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008, 183, 223–239. [Google Scholar] [CrossRef]
- Stanek, D.; Neugebauer, K.M. The Cajal body: A meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 2006, 115, 343–354. [Google Scholar] [CrossRef]
- Husedzinovic, A.; Oppermann, F.; Draeger-Meurer, S.; Chari, A.; Fischer, U.; Daub, H.; Gruss, O.J. Phosphoregulation of the human SMN complex. Eur. J. Cell Biol. 2014, 93, 106–117. [Google Scholar] [CrossRef]
- Riboldi, G.M.; Faravelli, I.; Kuwajima, T.; Delestrée, N.; Dermentzaki, G.; De Planell-Saguer, M.; Rinchetti, P.; Hao, L.T.; Beattie, C.C.; Corti, S.; et al. Sumoylation regulates the assembly and activity of the SMN complex. Nat. Commun. 2021, 12, 5040. [Google Scholar] [CrossRef]
- Lafarga, V.; Tapia, O.; Sharma, S.; Bengoechea, R.; Stoecklin, G.; Lafarga, M.; Berciano, M.T. CBP-mediated SMN acetylation modulates Cajal body biogenesis and the cytoplasmic targeting of SMN. Cell. Mol. Life Sci. CMLS 2018, 75, 527–546. [Google Scholar] [CrossRef]
- Meier, I.D.; Walker, M.P.; Matera, A.G. Gemin4 is an essential gene in mice, and its overexpression in human cells causes relocalization of the SMN complex to the nucleoplasm. Biol. Open 2018, 7, bio032409. [Google Scholar]
- Fallini, C.; Bassell, G.J.; Rossoll, W. Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res. 2012, 1462, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Patterson, W.L., 3rd; Georgel, P.T. Breaking the cycle: The role of omega-3 polyunsaturated fatty acids in inflammation-driven cancers. Biochem. Cell Biol. Biochim. Biol. Cell. 2014, 92, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Bühler, D.; Pillai, R.; Lottspeich, F.; Fischer, U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001, 3, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Battle, D.J.; Kasim, M.; Wang, J.; Dreyfuss, G. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 2007, 282, 27953–27959. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Lührmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001, 13, 290–301. [Google Scholar] [CrossRef]
- Massenet, S.; Pellizzoni, L.; Paushkin, S.; Mattaj, I.W.; Dreyfuss, G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 2002, 22, 6533–6541. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Embarc-Buh, A.; Francisco-Velilla, R. Emerging Roles of Gemin5: From snRNPs Assembly to Translation Control. Int. J. Mol. Sci. 2020, 21, 3868. [Google Scholar] [CrossRef]
- Li, D.K.; Tisdale, S.; Lotti, F.; Pellizzoni, L. SMN control of RNP assembly: From post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 2014, 32, 22–29. [Google Scholar] [CrossRef]
- Mouaikel, J.; Narayanan, U.; Verheggen, C.; Matera, A.G.; Bertrand, E.; Tazi, J.; Bordonné, R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003, 4, 616–622. [Google Scholar] [CrossRef]
- Otter, S.; Grimmler, M.; Neuenkirchen, N.; Chari, A.; Sickmann, A.; Fischer, U. A Comprehensive Interaction Map of the Human Survival of Motor Neuron (SMN) Complex. J. Biol. Chem. 2007, 282, 5825–5833. [Google Scholar] [CrossRef] [PubMed]
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Yong, J.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000, 148, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Cauchi, R.J. SMN and Gemins: ‘we are family’ … or are we?: Insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. BioEssays News Rev. Mol. Cell. Dev. Biol. 2010, 32, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Gubitz, A.K.; Wan, L.; Battle, D.J.; Dostie, J.; Golembe, T.J.; Dreyfuss, G. Gemins modulate the expression and activity of the SMN complex. Hum. Mol. Genet. 2005, 14, 1605–1611. [Google Scholar] [CrossRef] [PubMed]
- Cauchi, R.J.; Davies, K.E.; Liu, J.L. A motor function for the DEAD-box RNA helicase, Gemin3, in Drosophila. PLoS Genet. 2008, 4, e1000265. [Google Scholar] [CrossRef] [PubMed]
- Almstead, L.L.; Sarnow, P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes. Dev. 2007, 21, 1086–1097. [Google Scholar] [CrossRef]
- Shpargel, K.B.; Matera, A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2005, 102, 17372–17377. [Google Scholar] [CrossRef]
- Borg, R.M.; Fenech Salerno, B.; Vassallo, N.; Bordonne, R.; Cauchi, R.J. Disruption of snRNP biogenesis factors Tgs1 and pICln induces phenotypes that mirror aspects of SMN-Gemins complex perturbation in Drosophila, providing new insights into spinal muscular atrophy. Neurobiol. Dis. 2016, 94, 245–258. [Google Scholar] [CrossRef]
- Cacciottolo, R.; Ciantar, J.; Lanfranco, M.; Borg, R.M.; Vassallo, N.; Bordonné, R.; Cauchi, R.J. SMN complex member Gemin3 self-interacts and has a functional relationship with ALS-linked proteins TDP-43, FUS and Sod1. Sci. Rep. 2019, 9, 18666. [Google Scholar] [CrossRef]
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006, 34, 4206–4215. [Google Scholar] [CrossRef]
- Parker, K.L.; Schimmer, B.P. Steroidogenic factor 1: A key determinant of endocrine development and function. Endocr. Rev. 1997, 18, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Kim, K.W.; Ikeda, Y.; Anderson, K.K.; Beck, L.; Chase, S.; Tobet, S.A.; Parker, K.L. Central nervous system-specific knockout of steroidogenic factor 1 results in increased anxiety-like behavior. Mol. Endocrinol. 2008, 22, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yang, D.J.; Lee, S.; Hammer, G.D.; Kim, K.W.; Elmquist, J.K. Nutritional conditions regulate transcriptional activity of SF-1 by controlling sumoylation and ubiquitination. Sci. Rep. 2016, 6, 19143. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.B.; Lebedeva, L.A.; Suzawa, M.; Wadekar, S.A.; Desclozeaux, M.; Ingraham, H.A. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 2005, 25, 1879–1890. [Google Scholar] [CrossRef]
- Treuter, E.; Venteclef, N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim. Biophys. Acta 2011, 1812, 909–918. [Google Scholar] [CrossRef]
- Hoivik, E.A.; Lewis, A.E.; Aumo, L.; Bakke, M. Molecular aspects of steroidogenic factor 1 (SF-1). Mol. Cell. Endocrinol. 2010, 315, 27–39. [Google Scholar] [CrossRef]
- Yao, C.; Sun, Y.; Zhang, Z.; Jia, X.; Zou, P.; Wang, Y. Integration of RNAi and RNA-seq uncovers the regulation mechanism of DDX20 on vitellogenin expression in Scylla paramamosain. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 44, 101028. [Google Scholar] [CrossRef]
- Li, Q.; Xie, J.; He, L.; Wang, Y.; Yang, H.; Duan, Z.; Wang, Q. FOXL2 down-regulates vitellogenin expression at mature stage in Eriocheir sinensis. Biosci. Rep. 2015, 35, e00278. [Google Scholar] [CrossRef]
- Pisarska, M.D.; Bae, J.; Klein, C.; Hsueh, A.J. Forkhead l2 is expressed in the ovary and represses the promoter activity of the steroidogenic acute regulatory gene. Endocrinology 2004, 145, 3424–3433. [Google Scholar] [CrossRef]
- Lee, K.; Pisarska, M.D.; Ko, J.J.; Kang, Y.; Yoon, S.; Ryou, S.M.; Cha, K.Y.; Bae, J. Transcriptional factor FOXL2 interacts with DP103 and induces apoptosis. Biochem. Biophys. Res. Commun. 2005, 336, 876–881. [Google Scholar] [CrossRef]
- Sawka-Verhelle, D.; Escoubet-Lozach, L.; Fong, A.L.; Hester, K.D.; Herzig, S.; Lebrun, P.; Glass, C.K. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J. Biol. Chem. 2004, 279, 17772–17784. [Google Scholar] [CrossRef] [PubMed]
- Klappacher, G.W.; Lunyak, V.V.; Sykes, D.B.; Sawka-Verhelle, D.; Sage, J.; Brard, G.; Ngo, S.D.; Gangadharan, D.; Jacks, T.; Kamps, M.P.; et al. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 2002, 109, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Hester, K.D.; Verhelle, D.; Escoubet-Lozach, L.; Luna, R.; Rose, D.W.; Glass, C.K. Differential repression of c-myc and cdc2 gene expression by ERF and PE-1/METS. Cell Cycle 2007, 6, 1594–1604. [Google Scholar] [CrossRef] [PubMed]
- Taefehshokr, S.; Key, Y.A.; Khakpour, M.; Dadebighlu, P.; Oveisi, A. Early growth response 2 and Egr3 are unique regulators in immune system. Cent. Eur. J. Immunol. 2017, 42, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Borg, R.M.; Bordonne, R.; Vassallo, N.; Cauchi, R.J. Genetic Interactions between the Members of the SMN-Gemins Complex in Drosophila. PLoS ONE 2015, 10, e0130974. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Guo, Y.; Xiao, B.; Banerjee, S.; Saha, A.; Lu, J.; Glisovic, T.; Robertson, E.S. Epstein-Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis. PLoS Pathog. 2011, 7, e1002418. [Google Scholar] [CrossRef]
- Grundhoff, A.T.; Kremmer, E.; Türeci, O.; Glieden, A.; Gindorf, C.; Atz, J.; Mueller-Lantzsch, N.; Schubach, W.H.; Grässer, F.A. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 1999, 274, 19136–19144. [Google Scholar] [CrossRef]
- Shin, E.M.; Hay, H.S.; Lee, M.H.; Goh, J.N.; Tan, T.Z.; Sen, Y.P.; Lim, S.W.; Yousef, E.M.; Ong, H.T.; Thike, A.A.; et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J. Clin. Investig. 2014, 124, 3807–3824. [Google Scholar] [CrossRef]
- Gillian, A.L.; Svaren, J. The Ddx20/DP103 dead box protein represses transcriptional activation by Egr2/Krox-20. J. Biol. Chem. 2004, 279, 9056–9063. [Google Scholar] [CrossRef]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes. Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef]
- Murashov, A.K.; Chintalgattu, V.; Islamov, R.R.; Lever, T.E.; Pak, E.S.; Sierpinski, P.L.; Katwa, L.C.; Van Scott, M.R. RNAi pathway is functional in peripheral nerve axons. FASEB J. 2007, 21, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Lührmann, R.; Tuschl, T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Fontaine, J.M.; Hoppe, A.D.; Carra, S.; DeGuzman, C.; Martin, J.L.; Simon, S.; Vicart, P.; Welsh, M.J.; Landry, J.; et al. Abnormal interaction of motor neuropathy-associated mutant HspB8 (Hsp22) forms with the RNA helicase Ddx20 (gemin3). Cell Stress. Chaperones 2010, 15, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, D.; Shi, X.; Shen, C.; Hao, Y.; Zhang, T.; Yang, J.; Yuan, X.; Chen, X.; Zhao, D.; et al. Construction, Identification and Analysis of the Interaction Network of African Swine Fever Virus MGF360-9L with Host Proteins. Viruses 2021, 13, 1804. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV–human protein complexes. Nature 2012, 481, 365–370. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Dostie, J.; Mourelatos, Z.; Yang, M.; Sharma, A.; Dreyfuss, G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 2003, 9, 180–186. [Google Scholar] [CrossRef]
- Nelson, P.T.; Hatzigeorgiou, A.G.; Mourelatos, Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 2004, 10, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes. Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Bollrath, J.; Greten, F.R. IKK/NF-kappaB and STAT3 pathways: Central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009, 10, 1314–1319. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Gilmore, T.D. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef]
- Guttridge, D.C.; Albanese, C.; Reuther, J.Y.; Pestell, R.G.; Baldwin, A.S., Jr. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999, 19, 5785–5799. [Google Scholar] [CrossRef]
- Patel, M.; Horgan, P.G.; McMillan, D.C.; Edwards, J. NF-κB pathways in the development and progression of colorectal cancer. Transl. Res. J. Lab. Clin. Med. 2018, 197, 43–56. [Google Scholar] [CrossRef]
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Motolani, A.; Martin, M.; Sun, M.; Lu, T. 6.19—NF-κB and Cancer Therapy Drugs. In Comprehensive Pharmacology; Kenakin, T., Ed.; Elsevier: Oxford, UK, 2022; pp. 351–363. [Google Scholar]
- Tao, Y.; Zhou, J.; Wang, Z.; Tao, H.; Bai, J.; Ge, G.; Li, W.; Zhang, W.; Hao, Y.; Yang, X.; et al. Human bone mesenchymal stem cells-derived exosomal miRNA-361-5p alleviates osteoarthritis by downregulating DDX20 and inactivating the NF-κB signaling pathway. Bioorganic Chem. 2021, 113, 104978. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Wang, C.; Goh, J.N.; Loo, S.Y.; Yap, C.T.; Kumar, A.P. DEAD-box RNA Helicases: The microRNA managers of breast cancer. RNA Dis. 2015, 2, e846. [Google Scholar]
- Takata, A.; Otsuka, M.; Kojima, K.; Yoshikawa, T.; Kishikawa, T.; Yoshida, H.; Koike, K. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem. Biophys. Res. Commun. 2011, 411, 826–831. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Kudo, Y.; Kojima, K.; Maeda, S.; Tateishi, K.; Ikenoue, T.; Ijichi, H.; Hirata, Y.; Yoshida, H.; et al. Abstract 2065: DDX20 deficiency enhances NF-κB by impairing NF-κB suppressive-microRNA function and leads to hepatocarcinogenesis. Cancer Res. 2010, 70, 2065. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, C.; Li, K.; Ye, Y.; Shen, A.; Guo, L.; Chen, P.; Meng, C.; Wang, Q.; Yang, X.; et al. Death-associated protein kinase 1 suppresses hepatocellular carcinoma cell migration and invasion by upregulation of DEAD-box helicase 20. Cancer Sci. 2020, 111, 2803–2813. [Google Scholar] [CrossRef]
- Shi, D.; Jiang, P. A Different Facet of p53 Function: Regulation of Immunity and Inflammation During Tumor Development. Front. Cell Dev. Biol. 2021, 9, 762651. [Google Scholar] [CrossRef]
- Bordon, Y. Cell death: Tumour suppressor p53 helps phagocytes clean up. Nat. Rev. Immunol. 2015, 15, 525. [Google Scholar] [CrossRef]
- Aylon, Y.; Oren, M. Living with p53, dying of p53. Cell 2007, 130, 597–600. [Google Scholar] [CrossRef]
- Cooks, T.; Harris, C.C.; Oren, M. Caught in the cross fire: p53 in inflammation. Carcinogenesis 2014, 35, 1680–1690. [Google Scholar] [CrossRef]
- Casey, P.; Yap, C. RNA helicase DDX20 as a surrogate marker of statin activity in invasive breast cancer. Chin. J. Pharmacol. Toxicol. 2015, S1, 84. [Google Scholar]
- Zender, L.; Xue, W.; Zuber, J.; Semighini, C.P.; Krasnitz, A.; Ma, B.; Zender, P.; Kubicka, S.; Luk, J.M.; Schirmacher, P.; et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 2008, 135, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef]
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Seyed Saleh, S.H.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Ramadass, V.; Vaiyapuri, T.; Tergaonkar, V. Small Molecule NF-κB Pathway Inhibitors in Clinic. Int. J. Mol. Sci. 2020, 21, 5164. [Google Scholar] [CrossRef]
- Cai, W.; Xiong Chen, Z.; Rane, G.; Satendra Singh, S.; Choo, Z.; Wang, C.; Yuan, Y.; Zea Tan, T.; Arfuso, F.; Yap, C.T.; et al. Wanted DEAD/H or Alive: Helicases Winding Up in Cancers. J. Natl. Cancer Inst. 2017, 109, djw278. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.M.; Osato, M.; Kumar, A.P.; Tergaonkar, V. RNA helicase DP103 and TAK1: A new connection in cancer. Mol. Cell Oncol. 2015, 2, e985911. [Google Scholar] [CrossRef]
- Bentires-Alj, M.; Barbu, V.; Fillet, M.; Chariot, A.; Relic, B.; Jacobs, N.; Gielen, J.; Merville, M.P.; Bours, V. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 2003, 22, 90–97. [Google Scholar] [CrossRef]
- Nee, G.J. Characterization of Statins-Induced DDX20 Silencing in Invasive Breast Cancers. Master’s Thesis, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 2013. [Google Scholar]
- Chao, W. Interplay between Mevalonate and Hippo Pathways Regulates Ddx20 Transciption via Yao-Tead Complex in Triple Negative Breast Cancers. Ph.D. Thesis, National University of Singapore, Singapore, 2017. [Google Scholar]
- Pohl, S.Ö.-G. Mediation of Triple-Negative Breast Cancer Cell Fate via Cellular Redox and Wnt Signalling. Ph.D. Thesis, Curtin University, Bentley, WA, Australia, 2018. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes. Dev. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Kudo, Y.; Goto, T.; Yoshida, H.; Koike, K. A miRNA machinery component DDX20 controls NF-κB via microRNA-140 function. Biochem. Biophys. Res. Commun. 2012, 420, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Spitz, M.R.; Yang, H.; Wu, X. Abstract 1174: Genetic variations in microRNA biogenesis pathway genes as susceptibility loci for lung cancer risk. Cancer Res. 2011, 71, 1174. [Google Scholar] [CrossRef]
- Bourova-Flin, E.; Derakhshan, S.; Goudarzi, A.; Wang, T.; Vitte, A.L.; Chuffart, F.; Khochbin, S.; Rousseaux, S.; Aminishakib, P. The combined detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 expression predicts aggressive forms of oral squamous cell carcinoma. Br. J. Cancer 2021, 125, 1122–1134. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. Epstein-Barr virus: Exploiting the immune system. Nat. Rev. Immunol. 2001, 1, 75–82. [Google Scholar] [CrossRef]
- Kawa, K. Epstein-Barr virus-associated diseases in humans. Int. J. Hematol. 2000, 71, 108–117. [Google Scholar] [PubMed]
- Hille, A.; Badu-Antwi, A.; Holzer, D.; Grässer, F.A. Lysine residues of Epstein-Barr virus-encoded nuclear antigen 2 do not confer secondary modifications via ubiquitin or SUMO-like proteins but modulate transcriptional activation. J. Gen. Virol. 2002, 83, 1037–1042. [Google Scholar] [CrossRef]
- Strobl, L.J.; Höfelmayr, H.; Stein, C.; Marschall, G.; Brielmeier, M.; Laux, G.; Bornkamm, G.W.; Zimber-Strobl, U. Both Epstein-Barr Viral Nuclear Antigen 2 (EBNA2) and Activated Notch1 Transactivate Genes by Interacting with the Cellular Protein RBP-Jκ. Immunobiology 1997, 198, 299–306. [Google Scholar] [CrossRef]
- Ling, P.D.; Rawlins, D.R.; Hayward, S.D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 9237–9241. [Google Scholar] [CrossRef]
- Harada, S.; Kieff, E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 1997, 71, 6611–6618. [Google Scholar] [CrossRef]
- Voss, M.D.; Hille, A.; Barth, S.; Spurk, A.; Hennrich, F.; Holzer, D.; Mueller-Lantzsch, N.; Kremmer, E.; Grässer, F.A. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J. Virol. 2001, 75, 11781–11790. [Google Scholar] [CrossRef]
- West, M.J. Structure and function of the Epstein-Barr virus transcription factor, EBNA 3C. Curr. Protein Pept. Sci. 2006, 7, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, C.; Knight, J.S.; Robertson, E.S. The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front. Biosci. J. Virtual Libr. 2002, 7, d704–d716. [Google Scholar] [CrossRef] [PubMed]
- Styles, C.T.; Paschos, K.; White, R.E.; Farrell, P.J. The Cooperative Functions of the EBNA3 Proteins Are Central to EBV Persistence and Latency. Pathogens 2018, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Pelletier, J. Perturbations of RNA helicases in cancer. Wiley Interdiscip. Rev. RNA 2013, 4, 333–349. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, J.; Wang, J.; Lu, X.; Jin, C.; Xie, T.; Wu, N. Identify Potential Regulators in HIV-1 Latency by Joint microRNA and mRNA Analysis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 36, 569–584. [Google Scholar] [CrossRef]
- Greenwood, E.J.D.; Williamson, J.C.; Sienkiewicz, A.; Naamati, A.; Matheson, N.J.; Lehner, P.J. Promiscuous Targeting of Cellular Proteins by Vpr Drives Systems-Level Proteomic Remodeling in HIV-1 Infection. Cell Rep. 2019, 27, 1579–1596.e7. [Google Scholar] [CrossRef]



| Interacting Protein | Key Function | Interaction Technique | Refs |
|---|---|---|---|
| SMN | RNP assembly function | GST-pulldown, Y2H, co-IP, genetic interaction | [23,42] |
| Gemin2 | RNP assembly function | co-IP, genetic interaction | [42,66] |
| Gemin4 | SMN complex member | Y2H, co-IP, genetic interaction | [42,43] |
| Gemin5 | RNP assembly function | co-IP, genetic interaction | [42,66] |
| Sm B | snRNP component | GST-pulldown | [23] |
| Sm D2 | snRNP component | GST-pulldown | [23] |
| Sm D3 | snRNP component | Y2H, co-IP | [23] |
| pICln | RNP assembly function | Y2H, genetic interaction | [49] |
| Tgs1 | snRNP cap | Y2H, genetic interaction | [49] |
| EBNA2 | Epstein–Barr virus-encoded nuclear antigens | Y2H, co-IP | [67,68] |
| EBNA3C | Epstein–Barr virus-encoded nuclear antigens | Y2H, co-IP, GST-pulldown | [67,68] |
| p53 | Tumor suppressor | GST-pulldown | [4,67] |
| TAK1 | Regulator of NF-κB signaling pathway | co-IP | [69] |
| SF-1 | Transcription factor | M2H, His-pulldown | [10,20] |
| METS/PE1 | Transcription factor | Y2H, GST-pulldown | [63,64] |
| FOXL2 | Transcription factor | Y2H, co-IP | [59,61] |
| Egr2 / Krox-20 | Transcription factor | Y2H | [70] |
| AGO2/eIF2C2 | RNA silencing | co-IP, GST-pulldown | [71,72,73] |
| HspB8/Hsp22 | Heat shock protein | Y2H, co-IP | [74] |
| Olig2 | Transcription factor | Y2H | [4] |
| FTZ-F1 | Transcription factor | co-IP | [59] |
| MGF360-9L | Type I interferon inhibitors | LC-MS, co-IP | [75] |
| IRF3 | Transcription factor | co-IP | [2] |
| Vpr | Accessory factors | AP-MS | [76] |
| Cancer Type | DDX20 Expression | DDX20 Function | Mode of Action | Refs |
|---|---|---|---|---|
| Gastric cancer | Upward revision | In vitro promotion, high expression inhibition | Immune activation | [13] |
| Prostate cancer | Upward revision | Enhances the proliferation and metastasis of cancer cells | Activating the NF-κB signaling pathway | [11] |
| Oral squamous cell carcinoma | —— | Predicts OSCC invasion forms | —— | [118] |
| Colorectal cancer | Genetic variation | Presumptive markers of relapse | —— | [6] |
| Hepatocellular carcinoma | Lower | Promotes hepatocellular carcinoma | Impaired miRNA140-3P function | [9,96] |
| Invasive breast cancer | Upward revision | Biomarker and metastasis-driving oncogene | DDX20–NF-κB–MMP9 axis Reduced miR-222 | [69,111] |
| Triple-negative breast cancer | Upward revision | Biomarker and metastasis-driving oncogene | DDX20-NF-κB-MMP9 axis Reduced miR-222 TCF 4 and Wnt/beta-catenin signaling | [69,111,113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Yang, J.; Hao, Y.; Yang, X.; Shi, X.; Zhang, D.; Zhao, D.; Yan, W.; Bie, X.; Chen, L.; et al. DDX20: A Multifunctional Complex Protein. Molecules 2023, 28, 7198. https://doi.org/10.3390/molecules28207198
He L, Yang J, Hao Y, Yang X, Shi X, Zhang D, Zhao D, Yan W, Bie X, Chen L, et al. DDX20: A Multifunctional Complex Protein. Molecules. 2023; 28(20):7198. https://doi.org/10.3390/molecules28207198
Chicago/Turabian StyleHe, Lu, Jinke Yang, Yu Hao, Xing Yang, Xijuan Shi, Dajun Zhang, Dengshuai Zhao, Wenqian Yan, Xintian Bie, Lingling Chen, and et al. 2023. "DDX20: A Multifunctional Complex Protein" Molecules 28, no. 20: 7198. https://doi.org/10.3390/molecules28207198






