Bioprotection Potential of Lacticaseibacillus rhamnosus LRH01 and Lactiplantibacillus plantarum LP01 against Spoilage-Associated Penicillium Strains in Yoghurt
Abstract
:1. Introduction
2. Results
2.1. Antifungal Potential Evaluation
2.2. Minimal Inoculum Levels of LAB Cultures to Achieve Fungal Inhibition
2.3. Antifungal Activity of Selected LAB Cultures in Yoghurt Serum
2.3.1. Interaction of Penicillium Strains and C-Fermentates with Live Cells
2.3.2. CF-Fermentates
2.3.3. Contribution of Volatiles to the Antifungal Activity
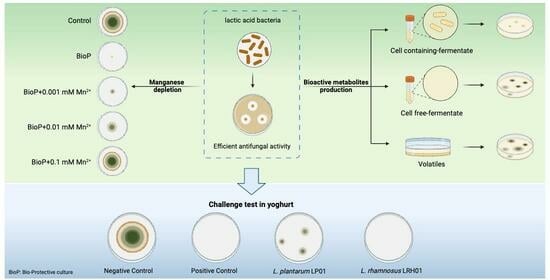
2.4. Effect of Manganese on the Antifungal Activity
2.5. Challenge Test in Yoghurt
3. Discussion
4. Materials and Methods
4.1. Organisms and Chemicals
4.2. Antifungal Activity of LAB Cultures
4.3. Determination of Minimal Inoculum Levels of LAB Cultures to Achieve Fungal Inhibition
4.4. Sensitivity to LAB in Yoghurt Serum
4.4.1. Interaction with C-Fermentates
4.4.2. Interaction with CF-Fermentates
4.4.3. Contribution of Volatiles to the Antifungal Activity
4.5. Manganese Depletion Test
4.6. A Challenge Test-Yoghurt Production and Biopreservation
4.6.1. Yoghurt Production
4.6.2. Yoghurt Contamination and Biopreservation Test
4.7. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garnier, L.; Valence, F.; Mounier, J. Diversity and Control of Spoilage Fungi in Dairy Products: An Update. Microorganisms 2017, 5, 42. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: New York, NY, USA, 2009; ISBN 9780387922065. [Google Scholar]
- Pitt, J.I.; Hocking, A.D.; Pitt, J.I.; Hocking, A.D. Spoilage of Stored, Processed and Preserved Foods. In Fungi and Food Spoilage; Springer: New York, NY, USA, 1997; pp. 489–507. [Google Scholar]
- Buehler, A.J.; Evanowski, R.L.; Martin, N.H.; Boor, K.J.; Wiedmann, M. Internal Transcribed Spacer (ITS) Sequencing Reveals Considerable Fungal Diversity in Dairy Products. J. Dairy Sci. 2017, 100, 8814–8825. [Google Scholar] [CrossRef] [PubMed]
- Novais, C.; Molina, A.K.; Abreu, R.M.V.; Santo-Buelga, C.; Ferreira, I.C.F.R.; Pereira, C.; Barros, L. Natural Food Colorants and Preservatives: A Review, a Demand, and a Challenge. J. Agric. Food Chem. 2022, 70, 2789–2805. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Maktabdar, M. Lactic Acid Bacteria as Biopreservation Against Spoilage Molds in Dairy Products—A Review. Front. Microbiol. 2022, 12, 819684. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Saleh, H.A.; El Ahmady, S.; Elmassry, M.M. Dissecting Yogurt: The Impact of Milk Types, Probiotics, and Selected Additives on Yogurt Quality. Food Rev. Int. 2022, 38, 643–650. [Google Scholar] [CrossRef]
- Center for Food Safety and Applied Nutrition Food Additives & Ingredients—Food Additive Status List. Available online: http://www.fda.gov/food/ingredientspackaginglabeling/foodadditivesingredients/ucm091048.htm (accessed on 26 September 2023).
- Li, Q.Q.; Zeng, X.Q.; Fu, H.L.; Wang, X.M.; Guo, X.J.; Wang, M. Lactiplantibacillus plantarum: A Comprehensive Review of its Antifungal and Anti-Mycotoxic Effects. Trends Food Sci. Technol. 2023, 136, 224–238. [Google Scholar] [CrossRef]
- Stefańska, I.; Kwiecień, E.; Jóźwiak-Piasecka, K.; Garbowska, M.; Binek, M.; Rzewuska, M. Antimicrobial Susceptibility of Lactic Acid Bacteria Strains of Potential Use as Feed Additives—The Basic Safety and Usefulness Criterion. Front. Vet. Sci. 2021, 8, 687071. [Google Scholar] [CrossRef]
- Xu, R.; Sa, R.; Jia, J.; Li, L.; Wang, X.; Liu, G. Screening of Antifungal Lactic Acid Bacteria as Bioprotective Cultures in Yogurt and a Whey Beverage. J. Food Prot. 2021, 84, 953–961. [Google Scholar] [CrossRef]
- Luana, V.S.; Raiane, R.S.; Andressa, F.; Andressa, F.; Evandro, M.; Cinzia, C.; Cinzia, L.R.; Antonio, F.C. Evaluation of Antifungal Activity of Lactic Acid Bacteria against Fungi in Simulated Cheese Matrix. LWT 2023, 182, 114773. [Google Scholar]
- Garnier, L.; Mounier, J.; Lê, S.; Pawtowski, A.; Pinon, N.; Camier, B.; Chatel, M.; Garric, G.; Thierry, A.; Coton, E.; et al. Development of Antifungal Ingredients for Dairy Products: From in vitro Screening to Pilot Scale Application. Food Microbiol. 2019, 81, 97–107. [Google Scholar] [CrossRef]
- Aunsbjerg, S.D.; Honoré, A.H.; Marcussen, J.; Ebrahimi, P.; Vogensen, F.K.; Benfeldt, C.; Skov, T.; Knøchel, S. Contribution of Volatiles to the Antifungal Effect of Lactobacillus Paracasei in Defined Medium and Yogurt. Int. J. Food Microbiol. 2015, 194, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective Mechanisms of Lactic Acid Bacteria against Fungal Spoilage of Food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Rau, M.H.; Bidstrup, S.; Vento, J.M.; Aunsbjerg, S.D.; Bosma, E.F.; Mcnair, L.M.; Beisel, C.L.; Neves, A.R. Competitive Exclusion Is a Major Bioprotective Mechanism of Lactobacilli against Fungal Spoilage in Fermented Milk Products. Appl. Environ. Microbiol. 2020, 86, e02312–e02319. [Google Scholar] [CrossRef]
- Shi, C.; Knøchel, S. Sensitivity of Molds from Spoiled Dairy Products Towards Bioprotective Lactic Acid Bacteria Cultures. Front. Microbiol. 2021, 12, 631730. [Google Scholar] [CrossRef]
- Liang, N.; Zhao, Z.; Curtis, J.M.; Gänzle, M.G. Antifungal Cultures and Metabolites of Lactic Acid Bacteria for Use in Dairy Fermentations. Int. J. Food Microbiol. 2022, 383, 109938. [Google Scholar] [CrossRef]
- Abouloifa, H.; Gaamouche, S.; Rokni, Y.; Hasnaoui, I.; Bellaouchi, R.; Ghabbour, N.; Karboune, S.; Brasca, M.; D’Hallewin, G.; Ben Salah, R.; et al. Antifungal Activity of Probiotic Lactobacillus Strains Isolated from Natural Fermented Green Olives and Their Application as Food Bio-Preservative. Biol. Control 2021, 152, 104450. [Google Scholar] [CrossRef]
- Guimarães, A.; Venancio, A.; Abrunhosa, L. Antifungal Effect of Organic Acids from Lactic Acid Bacteria on Penicillium nordicum. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2018, 35, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Leyva Salas, M.; Mounier, J.; Valence, F.; Coton, M.; Thierry, A.; Coton, E. Antifungal Microbial Agents for Food Biopreservation—A Review. Microorganisms 2017, 5, 37. [Google Scholar] [CrossRef]
- Leila, B.F.; Neyssene, A.; Raquel, T.; Carlos, L.; Giuseppe, M.; Ferid, A. Correlation Between Metabolites of Lactic Acid Bacteria Isolated from Dairy Traditional Fermented Tunisian Products and Antifungal and Antioxidant Activities. J. Appl. Microbiol. 2022, 133, 3069–3082. [Google Scholar]
- Mani-López, E.; Arrioja-Bretón, D.; López-Malo, A. The Impacts of Antimicrobial and Antifungal Activity of Cell-Free Supernatants from Lactic Acid Bacteria in vitro and Foods. Compr. Rev. Food Sci. Food Saf. 2022, 21, 604–641. [Google Scholar] [CrossRef]
- Luz, C.; D’Opazo, V.; Quiles, J.M.; Romano, R.; Mañes, J.; Meca, G. Biopreservation of Tomatoes Using Fermented Media by Lactic Acid Bacteria. LWT 2020, 130, 109618. [Google Scholar] [CrossRef]
- Shi, C.; Knøchel, S. Susceptibility of Dairy Associated Molds towards Microbial Metabolites with Focus on the Response to Diacetyl. Food Control 2021, 121, 107573. [Google Scholar] [CrossRef]
- Bin, L.; Zhirong, W.; Gang, Y.; Shan, H.; Shenglan, L.; Kewei, C.; Muying, D.; Zsolt, Z.; Ferenc, H.; Jianquan, K. Biocontrol Potential of 1-Pentanal Emitted from Lactic Acid Bacteria Strains against Aspergillus Flavus in Red Pepper (Capsicum Annuum L.). Food Control 2022, 142, 109261. [Google Scholar]
- Schmidt, S.B.; Jensen, P.E.; Husted, S. Manganese Deficiency in Plants: The Impact on Photosystem II. Trends Plant Sci. 2016, 21, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Jensen, A.N.; Jensen, L.T. Manganese Transport, Trafficking and Function in Invertebrates. In Manganese in Health and Disease; Issues in Toxicology; Royal Society of Chemistry: Cambridge, UK, 2014; pp. 1–33. [Google Scholar]
- Bosma, E.F.; Rau, M.H.; van Gijtenbeek, L.A.; Siedler, S. Regulation and Distinct Physiological Roles of Manganese in Bacteria. FEMS Microbiol. Rev. 2021, 45, fuab028. [Google Scholar] [CrossRef] [PubMed]
- Porcheron, G.; Garénaux, A.; Proulx, J.; Sabri, M.; Dozois, C.M. Iron, Copper, Zinc, and Manganese Transport and Regulation in Pathogenic Enterobacteria: Correlations between Strains, Site of Infection and the Relative Importance of the Different Metal Transport Systems for Virulence. Front. Cell Infect. Microbiol. 2013, 3, 90. [Google Scholar] [CrossRef]
- Tong, Y.; Wang, G.; Zhang, Q.; Tian, F.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Systematic Understanding of the Potential Manganese-Adsorption Components of a Screened: Lactobacillus plantarum CCFM436. RSC Adv. 2016, 6, 102804–102813. [Google Scholar] [CrossRef]
- Ebrahimi, P.; van den Berg, F.; Aunsbjerg, S.D.; Honoré, A.; Benfeldt, C.; Jensen, H.M.; Engelsen, S.B. Quantitative Determination of Mold Growth and Inhibition by Multispectral Imaging. Food Control 2015, 55, 82–89. [Google Scholar] [CrossRef]
- Inglin, R.C.; Stevens, M.J.A.; Meile, L.; Lacroix, C.; Meile, L. High-Throughput Screening Assays for Antibacterial and Antifungal Activities of Lactobacillus Species. J. Microbiol. Methods 2015, 114, 26–29. [Google Scholar] [CrossRef]
- Ouiddir, M.; Bettache, G.; Leyva Salas, M.; Pawtowski, A.; Donot, C.; Brahimi, S.; Mabrouk, K.; Coton, E.; Mounier, J. Selection of Algerian Lactic Acid Bacteria for Use as Antifungal Bioprotective Cultures and Application in Dairy and Bakery Products. Food Microbiol. 2019, 82, 160–170. [Google Scholar] [CrossRef]
- Salas, M.L.; Thierry, A.; Lemaître, M.; Garric, G.; Harel-Oger, M.; Chatel, M.; Lê, S.; Mounier, J.; Valence, F.; Coton, E. Antifungal Activity of Lactic Acid Bacteria Combinations in Dairy Mimicking Models and Their Potential as Bioprotective Cultures in Pilot Scale Applications. Front. Microbiol. 2018, 9, 1787. [Google Scholar]





| Organism | Abbreviation | Source | Provider |
|---|---|---|---|
| Bacteria | |||
| Lacticaseibacillus rhamnosus LRH01 | L. rhamnosus LRH01 | Dairy | SACCO |
| Lacticaseibacillus rhamnosus LRH05 | L. rhamnosus LRH05 | Dairy | SACCO |
| Lacticaseibacillus rhamnosus LRH14 | L. rhamnosus LRH14 | Dairy | SACCO |
| Lacticaseibacillus rhamnosus LRH16 | L. rhamnosus LRH16 | Cereals | SACCO |
| Lacticaseibacillus rhamnosus LRH43 | L. rhamnosus LRH43 | Dairy | SACCO |
| Lactiplantibacillus plantarum LP01 | L. plantarum LP01 | Dairy | SACCO |
| Lactiplantibacillus plantarum LP37 | L. plantarum LP37 | Cereals | SACCO |
| Lactiplantibacillus plantarum LP48 | L. plantarum LP48 | Meat | SACCO |
| Lacticaseibacillus paracasei LPC44 | L. paracasei LPC44 | Dairy | SACCO |
| Lacticaseibacillus paracasei LPC46 | L. paracasei LPC46 | Dairy | SACCO |
| Mold | |||
| Penicillium crustosum 01180001 | P. crustosum 01180001 | Yoghurt/Skyr | Arla |
| Penicillium commune ISI2 | P. commune ISI2 | Greek yoghurt | ISI |
| Penicillium glabrum ISI3 | P. glabrum ISI3 | Crème fraiche 18% | ISI |
| Penicillium solitum ISI5 | P. solitum ISI5 | Crème fraiche 30% | ISI |
| Penicillium palitans PPa01 | P. palitans PPa01 | Rahka (Finnish quark) | SACCO |
| LAB cultures/Molds | P. crustosum 01180001 | P. commune ISI2 | P. glabrum ISI3 | P. solitum ISI5 | P. palitans PPa01 |
|---|---|---|---|---|---|
| L. rhamnosus LRH01 | 105 | 105 | 104 | 104 | 104 |
| L. rhamnosus LRH05 | 105 | 106 | 105 | 105 | 105 |
| L. rhamnosus LRH14 | 106 | 107 | 106 | 106 | 105 |
| L. rhamnosus LRH16 | 106 | 107 | 105 | 105 | 105 |
| L. rhamnosus LRH43 | 106 | 107 | 106 | 106 | 105 |
| L. plantarum LP01 | 104 | 106 | 105 | 104 | 104 |
| L. plantarum LP37 | 105 | 107 | 106 | 105 | 105 |
| L. plantarum LP48 | 107 | 107 | 107 | 107 | 107 |
| L. paracasei LPC44 | 107 | 106 | 106 | 106 | 107 |
| L. paracasei LPC46 | 107 | 107 | 107 | 107 | 107 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Knøchel, S. Bioprotection Potential of Lacticaseibacillus rhamnosus LRH01 and Lactiplantibacillus plantarum LP01 against Spoilage-Associated Penicillium Strains in Yoghurt. Molecules 2023, 28, 7397. https://doi.org/10.3390/molecules28217397
Shi C, Knøchel S. Bioprotection Potential of Lacticaseibacillus rhamnosus LRH01 and Lactiplantibacillus plantarum LP01 against Spoilage-Associated Penicillium Strains in Yoghurt. Molecules. 2023; 28(21):7397. https://doi.org/10.3390/molecules28217397
Chicago/Turabian StyleShi, Ce, and Susanne Knøchel. 2023. "Bioprotection Potential of Lacticaseibacillus rhamnosus LRH01 and Lactiplantibacillus plantarum LP01 against Spoilage-Associated Penicillium Strains in Yoghurt" Molecules 28, no. 21: 7397. https://doi.org/10.3390/molecules28217397
APA StyleShi, C., & Knøchel, S. (2023). Bioprotection Potential of Lacticaseibacillus rhamnosus LRH01 and Lactiplantibacillus plantarum LP01 against Spoilage-Associated Penicillium Strains in Yoghurt. Molecules, 28(21), 7397. https://doi.org/10.3390/molecules28217397