The Synergistic Activity of Rhamnolipid Combined with Linezolid against Linezolid-Resistant Enterococcus faecium
Abstract
:1. Introduction
2. Results
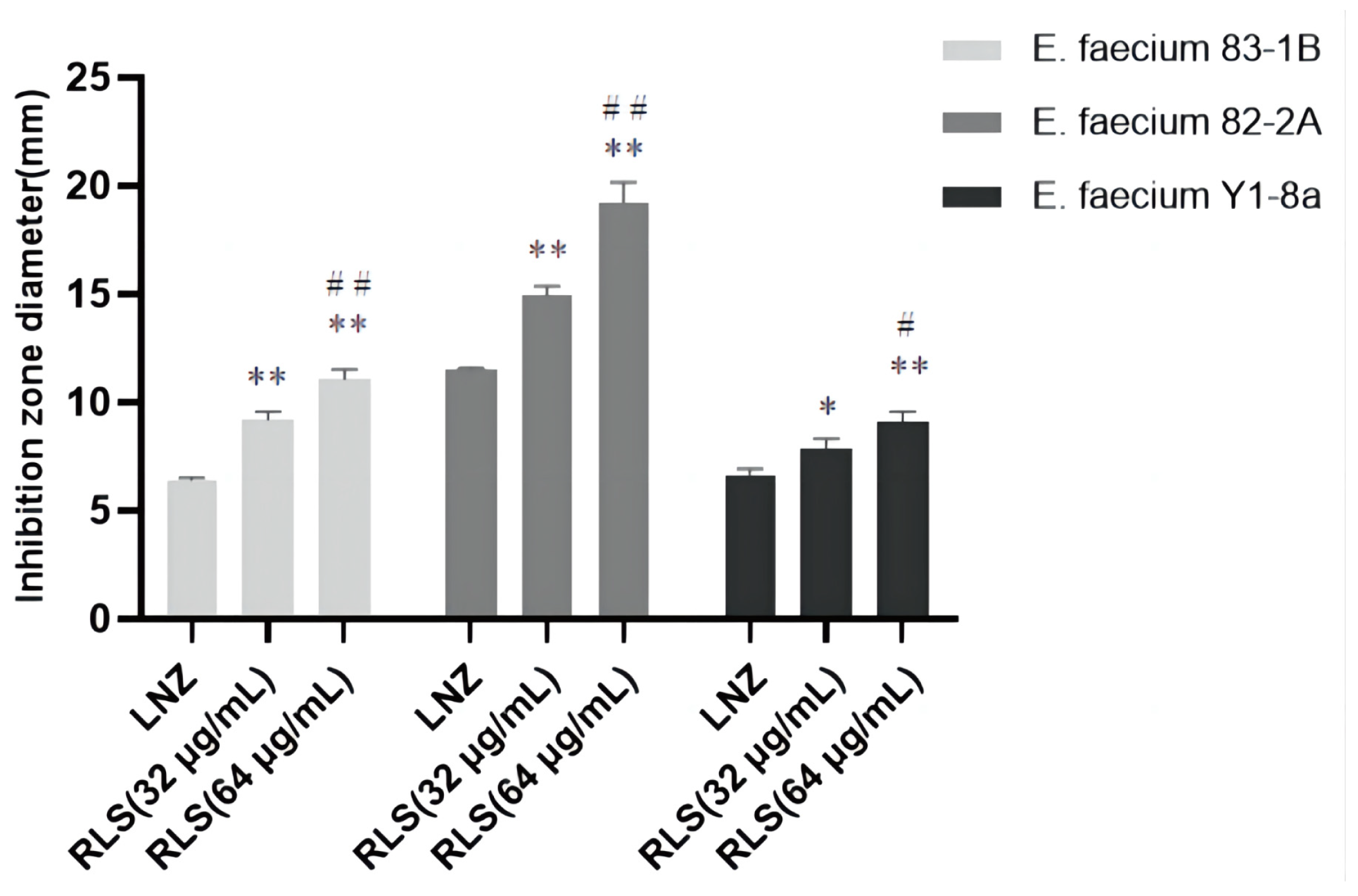
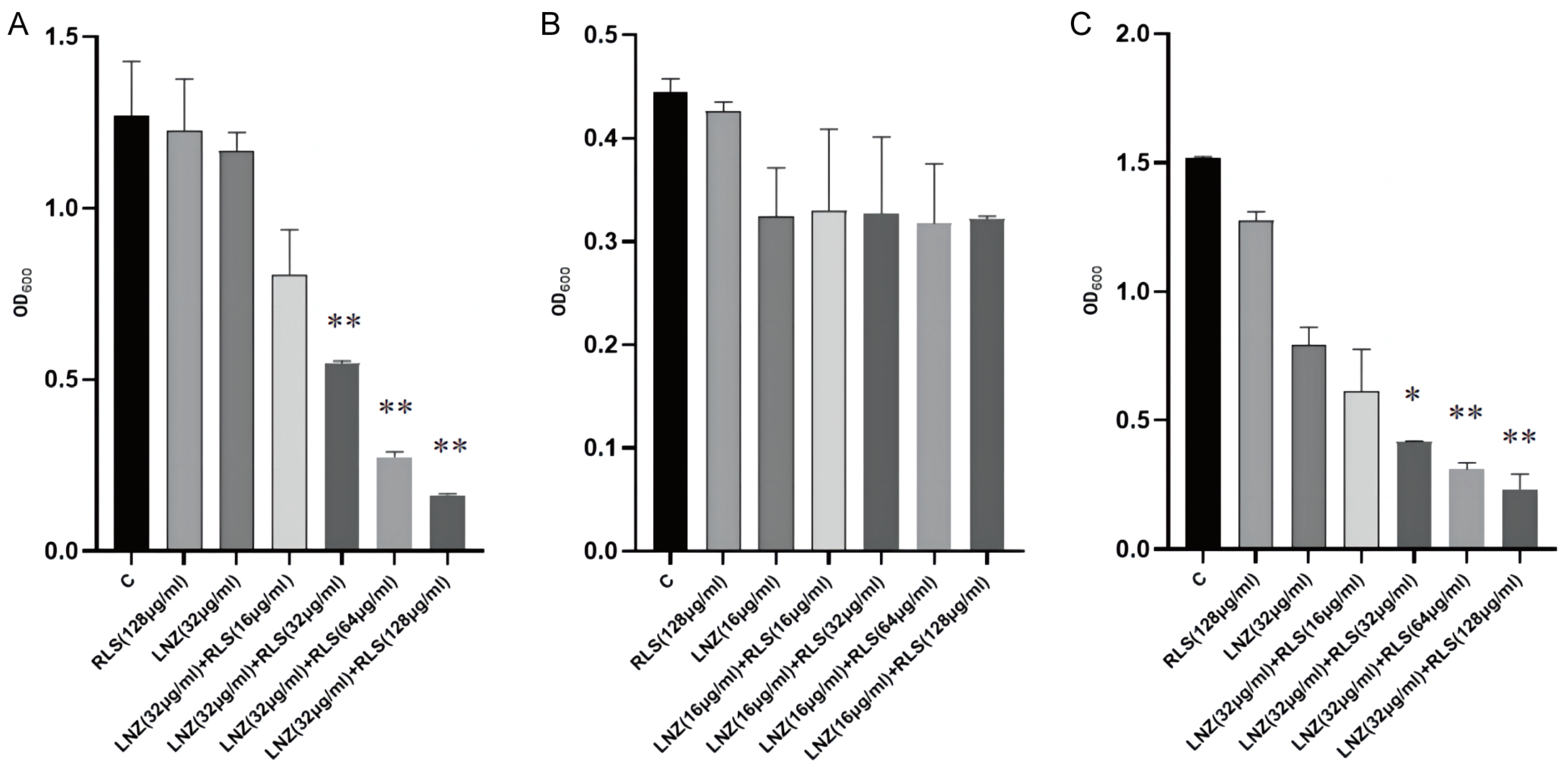
2.1. Rhamnolipid Restores the Sensitivity of Linezolid-Resistant Enterococci to Linezolid In Vitro
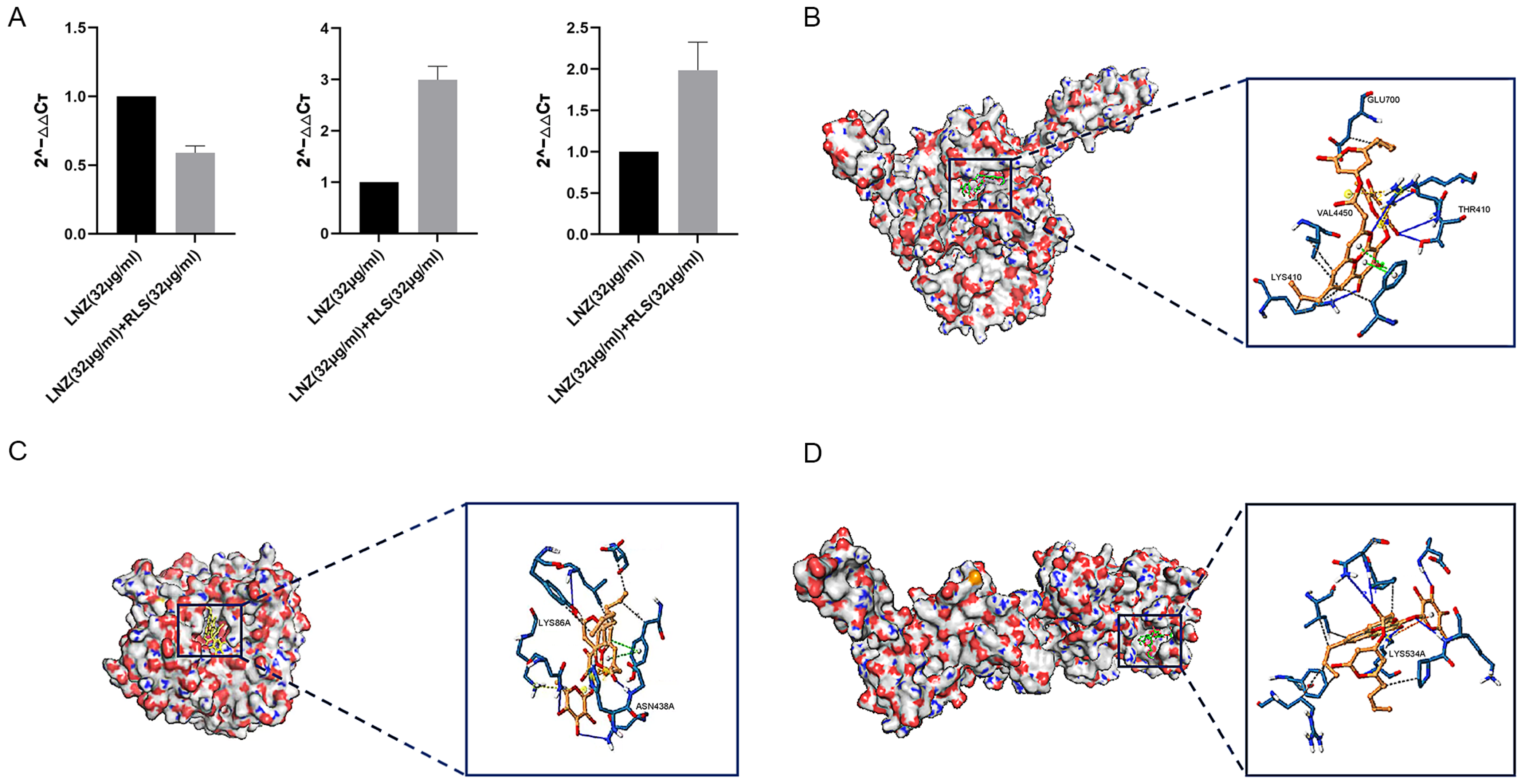
2.2. Rhamnolipid Is Directly Involved in the Inhibitory Activity of poxtA
2.3. Rhamnolipid with Linezolid Showed a Synergistic Impact In Vivo Compared to Monotherapy
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Chemicals
4.2. Checkerboard Assays
4.3. Growth Curves
4.4. Time–Killing Assays
4.5. Combined Disk Test
4.6. Biofilm Eradication Test
4.7. Molecular Docking
4.8. qPCR
4.9. Animal Studies
4.10. Mouse Systemic Infection Model
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of Animal Origin. Microbiol. Spectr. 2018, 6, 1–41. [Google Scholar] [CrossRef] [PubMed]
- van Hal, S.J.; Willems, R.J.L.; Gouliouris, T.; Ballard, S.A.; Coque, T.M.; Hammerum, A.M.; Hegstad, K.; Westh, H.T.; Howden, B.P.; Malhotra-Kumar, S.; et al. The Global Dissemination of Hospital Clones of Enterococcus faecium. Genome Med. 2021, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.P.; Buss, P.W.; Marlow, N.; Brown, N.M.; Millar, M.R. Enterococcus faecium Meningitis. Case Rep. 1994, 70, F78–F79. [Google Scholar] [CrossRef]
- Chou, Y.-Y.; Lin, T.-Y.; Lin, J.-C.; Wang, N.-C.; Peng, M.-Y.; Chang, F.-Y. Vancomycin-Resistant Enterococcal Bacteremia: Comparison of Clinical Features and Outcome between Enterococcus faecium and Enterococcus faecalis. J. Microbiol. Immunol. Infect. Wei Mian Yu Gan Ran Za Zhi 2008, 41, 124–129. [Google Scholar]
- Ch’ng, J.-H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-Associated Infection by Enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef]
- Woksepp, H.; Ryberg, A.; Billström, H.; Hällgren, A.; Nilsson, L.E.; Marklund, B.-I.; Olsson-Liljequist, B.; Schön, T. Evaluation of High-Resolution Melting Curve Analysis of Ligation-Mediated Real-Time PCR, a Rapid Method for Epidemiological Typing of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter Species) Pathogens. J. Clin. Microbiol. 2014, 52, 4339–4342. [Google Scholar] [CrossRef]
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.T.; Dunny, G.M. Multiple Roles for Enterococcus faecalis Glycosyltransferases in Biofilm-Associated Antibiotic Resistance, Cell Envelope Integrity, and Conjugative Transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105. [Google Scholar] [CrossRef]
- Mercuro, N.J.; Davis, S.L.; Zervos, M.J.; Herc, E.S. Combatting Resistant Enterococcal Infections: A Pharmacotherapy Review. Expert Opin. Pharmacother. 2018, 19, 979–992. [Google Scholar] [CrossRef]
- Cattoir, V.; Giard, J.-C. Antibiotic Resistance in Enterococcus faecium Clinical Isolates. Expert Rev. Anti-Infect. Ther. 2014, 12, 239–248. [Google Scholar] [CrossRef]
- Dresser, L.D.; Rybak, M.J. The Pharmacologic and Bacteriologic Properties of Oxazolidinones, a New Class of Synthetic Antimicrobials. Pharmacotherapy 1998, 18, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Gao, S.; Xu, H.; Zhang, Z.; Chen, F.; Shen, H.; Zhang, C. Distribution of the optrA Gene in Enterococcus Isolates at a Tertiary Care Hospital in China. J. Glob. Antimicrob. Resist. 2019, 17, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Platforms for Antibiotic Discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Ten Threats to Global Health in 2019. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 15 May 2023).
- Wt, L.; Ej, V.; Sa, B. Synergy between Essential Oil Components and Antibiotics: A Review. Crit. Rev. Microbiol. 2014, 40, 76–94. [Google Scholar] [CrossRef]
- Thakur, P.; Saini, N.K.; Thakur, V.K.; Gupta, V.K.; Saini, R.V.; Saini, A.K. Rhamnolipid the Glycolipid Biosurfactant: Emerging Trends and Promising Strategies in the Field of Biotechnology and Biomedicine. Microb. Cell Factories 2021, 20, 1. [Google Scholar] [CrossRef]
- Christova, N.; Tuleva, B.; Kril, A.; Georgieva, M.; Konstantinov, S.; Terziyski, I.; Nikolova, B.; Stoineva, I. Chemical Structure and In Vitro Antitumor Activity of Rhamnolipids from Pseudomonas aeruginosa BN10. Appl. Biochem. Biotechnol. 2013, 170, 676–689. [Google Scholar] [CrossRef]
- Vatsa, P.; Sanchez, L.; Clement, C.; Baillieul, F.; Dorey, S. Rhamnolipid Biosurfactants as New Players in Animal and Plant Defense against Microbes. Int. J. Mol. Sci. 2010, 11, 5095–5108. [Google Scholar] [CrossRef]
- Zezzi Do Valle Gomes, M.; Nitschke, M. Evaluation of Rhamnolipid and Surfactin to Reduce the Adhesion and Remove Biofilms of Individual and Mixed Cultures of Food Pathogenic Bacteria. Food Control 2012, 25, 441–447. [Google Scholar] [CrossRef]
- Sajid, M.; Ahmad Khan, M.S.; Singh Cameotra, S.; Safar Al-Thubiani, A. Biosurfactants: Potential Applications as Immunomodulator Drugs. Immunol. Lett. 2020, 223, 71–77. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, A.; Sharma, R.; Aurora, R.; Jain, S.K. Biosurfactants as a Novel Additive in Pharmaceutical Formulations: Current Trends and Future Implications. Curr. Drug Metab. 2020, 21, 885–901. [Google Scholar] [CrossRef]
- Pogue, J.M.; Kaye, K.S.; Cohen, D.A.; Marchaim, D. Appropriate Antimicrobial Therapy in the Era of Multidrug-Resistant Human Pathogens. Clin. Microbiol. Infect. 2015, 21, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Durack, D.T.; Beeson, P.B.; Petersdorf, R.G. Experimental Bacterial Endocarditis. 3. Production and Progress of the Disease in Rabbits. Br. J. Exp. Pathol. 1973, 54, 142–151. [Google Scholar] [PubMed]
- Borst, L.B.; Suyemoto, M.M.; Sarsour, A.H.; Harris, M.C.; Martin, M.P.; Strickland, J.D.; Oviedo, E.O.; Barnes, H.J. Pathogenesis of Enterococcal Spondylitis Caused by Enterococcus cecorum in Broiler Chickens. Vet. Pathol. 2017, 54, 61–73. [Google Scholar] [CrossRef]
- Santín, M.; Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in Domesticated and Wild Animals. Res. Vet. Sci. 2011, 90, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Tumpa, A.; Štritof, Z.; Pintarić, S. Prevalence and Antimicrobial Susceptibility of Enterococcus spp. from Urine of Dogs and Cats in Northwestern Croatia. Res. Vet. Sci. 2022, 151, 42–46. [Google Scholar] [CrossRef]
- Sulis, G.; Sayood, S.; Katukoori, S.; Bollam, N.; George, I.; Yaeger, L.H.; Chavez, M.A.; Tetteh, E.; Yarrabelli, S.; Pulcini, C.; et al. Exposure to World Health Organization’s AWaRe Antibiotics and Isolation of Multidrug Resistant Bacteria: A Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2022, 28, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Shikongo, A.; Owolabi, K.M. On the Hindering Evolution of Drug Resistance Due to Intraspecific Competition Arising during the Facilitation Survival for Non-Genetic Resistance with Fractal Fractional Derivative Order. Model. Earth Syst. Environ. 2023, 9, 2637–2650. [Google Scholar] [CrossRef]
- Bolhuis, M.S.; van der Laan, T.; Kosterink, J.G.W.; van der Werf, T.S.; van Soolingen, D.; Alffenaar, J.-W.C. In Vitro Synergy between Linezolid and Clarithromycin against Mycobacterium tuberculosis. Eur. Respir. J. 2014, 44, 808–811. [Google Scholar] [CrossRef]
- Kaskatepe, B.; Ozturk, S. Assessment of Synergistic Activity of Rhamnolipid and Linezolid against Methicillin-Resistant Staphylococcus aureus In-Vitro and In-Vivo with Galleria mellonella Larvae Model. Microb. Pathog. 2023, 174, 105945. [Google Scholar] [CrossRef]
- Sotirova, A.; Avramova, T.; Stoitsova, S.; Lazarkevich, I.; Lubenets, V.; Karpenko, E.; Galabova, D. The Importance of Rhamnolipid-Biosurfactant-Induced Changes in Bacterial Membrane Lipids of Bacillus subtilis for the Antimicrobial Activity of Thiosulfonates. Curr. Microbiol. 2012, 65, 534–541. [Google Scholar] [CrossRef]
- Zheng, J.; Bai, B.; Lin, Z.; Pu, Z.; Yao, W.; Chen, Z.; Li, D.; Deng, X.; Deng, Q.; Yu, Z. Characterization of Biofilm Formation by Enterococcus faecalis Isolates Derived from Urinary Tract Infections in China. J. Med. Microbiol. 2018, 67, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Jonas, D.; Huber, I.; Karygianni, L.; Wölber, J.; Hellwig, E.; Arweiler, N.; Vach, K.; Wittmer, A.; Al-Ahmad, A. Enterococcus faecalis from Food, Clinical Specimens, and Oral Sites: Prevalence of Virulence Factors in Association with Biofilm Formation. Front. Microbiol. 2016, 6, 1534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Li, X.; Yu, C.; Wang, Y. Promising Therapeutic Strategies Against Microbial Biofilm Challenges. Front. Cell. Infect. Microbiol. 2020, 10, 359. [Google Scholar] [CrossRef] [PubMed]
- Heikens, E.; Singh, K.V.; Jacques-Palaz, K.D.; van Luit-Asbroek, M.; Oostdijk, E.A.N.; Bonten, M.J.M.; Murray, B.E.; Willems, R.J.L. Contribution of the Enterococcal Surface Protein Esp to Pathogenesis of Enterococcus faecium Endocarditis. Microbes Infect. 2011, 13, 1185–1190. [Google Scholar] [CrossRef]
- Paganelli, F.L.; Willems, R.J.L.; Jansen, P.; Hendrickx, A.; Zhang, X.; Bonten, M.J.M.; Leavis, H.L. Enterococcus faecium Biofilm Formation: Identification of Major Autolysin AtlAEfm, Associated Acm Surface Localization, and AtlAEfm-Independent Extracellular DNA Release. mBio 2013, 4, e00154. [Google Scholar] [CrossRef]
- Pamp, S.J.; Frees, D.; Engelmann, S.; Hecker, M.; Ingmer, H. Spx Is a Global Effector Impacting Stress Tolerance and Biofilm Formation in Staphylococcus aureus. J. Bacteriol. 2006, 188, 4861–4870. [Google Scholar] [CrossRef]
- Schaffer, S.D.; Hutchison, C.A.; Rouchon, C.N.; Mdluli, N.V.; Weinstein, A.J.; McDaniel, D.; Frank, K.L. Diverse Enterococcus faecalis Strains Show Heterogeneity in Biofilm Properties. Res. Microbiol. 2023, 174, 103986. [Google Scholar] [CrossRef]
- Chandankere, R.; Ravikumar, Y.; Zabed, H.M.; Sabapathy, P.C.; Yun, J.; Zhang, G.; Qi, X. Conversion of Agroindustrial Wastes to Rhamnolipid by Enterobacter sp. UJS-RC and Its Role against Biofilm-Forming Foodborne Pathogens. J. Agric. Food Chem. 2020, 68, 15478–15489. [Google Scholar] [CrossRef]
- Ciandrini, E.; Campana, R.; Casettari, L.; Perinelli, D.R.; Fagioli, L.; Manti, A.; Palmieri, G.F.; Papa, S.; Baffone, W. Characterization of Biosurfactants Produced by Lactobacillus spp. and Their Activity against Oral Streptococci Biofilm. Appl. Microbiol. Biotechnol. 2016, 100, 6767–6777. [Google Scholar] [CrossRef]
- Firdose, A.; Chong, N.H.H.; Ramli, R.; Aqma, W.S. Antimicrobial, Antiadhesive, and Antibiofilm Actions of Rhamnolipids on ESKAPE Pathogens. Lett. Appl. Microbiol. 2023, 76, ovad013. [Google Scholar] [CrossRef]
- E Silva, S.S.; Carvalho, J.W.P.; Aires, C.P.; Nitschke, M. Disruption of Staphylococcus aureus Biofilms Using Rhamnolipid Biosurfactants. J. Dairy Sci. 2017, 100, 7864–7873. [Google Scholar] [CrossRef] [PubMed]
- Paladino, J.A. Linezolid: An Oxazolidinone Antimicrobial Agent. Am. J. Health-Syst. Pharm. 2002, 59, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Linezolid In Vitro: Mechanism and Antibacterial Spectrum. J. Antimicrob. Chemother. 2003, 51 (Suppl. S2), ii9–ii16. [Google Scholar] [CrossRef] [PubMed]
- Top, J.; Paganelli, F.L.; Zhang, X.; van Schaik, W.; Leavis, H.L.; van Luit-Asbroek, M.; van der Poll, T.; Leendertse, M.; Bonten, M.J.M.; Willems, R.J.L. The Enterococcus faecium Enterococcal Biofilm Regulator, EbrB, Regulates the esp Operon and Is Implicated in Biofilm Formation and Intestinal Colonization. PLoS ONE 2013, 8, e65224. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M.; Huh, H.J.; Song, D.J.; Shim, H.J.; Park, K.S.; Kang, C.-I.; Ki, C.-S.; Lee, N.Y. Resistance Mechanisms of Linezolid-Nonsusceptible Enterococci in Korea: Low Rate of 23S rRNA Mutations in Enterococcus faecium. J. Med. Microbiol. 2017, 66, 1730–1735. [Google Scholar] [CrossRef] [PubMed]
- Moure, Z.; Lara, N.; Marín, M.; Sola-Campoy, P.J.; Bautista, V.; Gómez-Bertomeu, F.; Gómez-Dominguez, C.; Pérez-Vázquez, M.; Aracil, B.; Campos, J.; et al. Interregional Spread in Spain of Linezolid-Resistant Enterococcus spp. Isolates Carrying the optrA and poxtA Genes. Int. J. Antimicrob. Agents 2020, 55, 105977. [Google Scholar] [CrossRef]
- Wang, Q.; Peng, K.; Liu, Z.; Li, Y.; Xiao, X.; Du, X.-D.; Li, R.; Wang, Z. Genomic Insights into Linezolid-Resistant Enterococci Revealed Its Evolutionary Diversity and poxtA Copy Number Heterogeneity. Int. J. Antimicrob. Agents 2023, 62, 106929. [Google Scholar] [CrossRef]
- Asadi, S.; Nayeri-Fasaei, B.; Zahraei-Salehi, T.; Yahya-Rayat, R.; Shams, N.; Sharifi, A. Antibacterial and Anti-Biofilm Properties of Carvacrol Alone and in Combination with Cefixime against Escherichia coli. BMC Microbiol. 2023, 23, 55. [Google Scholar] [CrossRef]
- Sotirova, A.; Spasova, D.; Vasileva-Tonkova, E.; Galabova, D. Effects of Rhamnolipid-Biosurfactant on Cell Surface of Pseudomonas aeruginosa. Microbiol. Res. 2009, 164, 297–303. [Google Scholar] [CrossRef]
- Saadati, F.; Shahryari, S.; Sani, N.M.; Farajzadeh, D.; Zahiri, H.S.; Vali, H.; Noghabi, K.A. Effect of MA01 Rhamnolipid on Cell Viability and Expression of Quorum-Sensing (QS) Genes Involved in Biofilm Formation by Methicillin-Resistant Staphylococcus aureus. Sci. Rep. 2022, 12, 14833. [Google Scholar] [CrossRef]
- Meyer-Hoffert, U.; Zimmermann, A.; Czapp, M.; Bartels, J.; Koblyakova, Y.; Gläser, R.; Schröder, J.-M.; Gerstel, U. Flagellin Delivery by Pseudomonas aeruginosa Rhamnolipids Induces the Antimicrobial Protein Psoriasin in Human Skin. PLoS ONE 2011, 6, e16433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Qin, S.; Wu, Y.; Zhang, R.; Xu, Y.; Yang, C. Rhamnolipids Regulate Lipid Metabolism, Immune Response, and Gut Microbiota in Rats. Front. Nutr. 2022, 9, 886256. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Q.; Hua, Y.; Chen, J.; Zhang, H.; Wang, H. Potential Applications of Biosurfactant Rhamnolipids in Agriculture and Biomedicine. Appl. Microbiol. Biotechnol. 2017, 101, 8309–8319. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.; Hill, H.J.; Elkhouly, G.E.; Bakkar, M.R.; Raya, N.R.; Stamataki, Z.; Abo-zeid, Y. Rhamnolipid Nano-Micelles Inhibit SARS-CoV-2 Infection and Have No Dermal or Eye Toxic Effects in Rabbits. Antibiotics 2022, 11, 1556. [Google Scholar] [CrossRef]
- Brennan-Krohn, T.; Grote, A.; Rodriguez, S.; Kirby, J.E.; Earl, A.M. Transcriptomics Reveals How Minocycline-Colistin Synergy Overcomes Antibiotic Resistance in Multidrug-Resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2022, 66, e0196921. [Google Scholar] [CrossRef]
- Guo, Y.; Lv, X.; Wang, Y.; Zhou, Y.; Lu, N.; Deng, X.; Wang, J. Honokiol Restores Polymyxin Susceptibility to MCR-1-Positive Pathogens Both In Vitro and In Vivo. Appl. Environ. Microbiol. 2020, 86, e02346-19. [Google Scholar] [CrossRef]
- Sun, W.; Liu, H.; Liu, J.; Jiang, Q.; Pan, Y.; Yang, Y.; Zhu, X.; Ge, J. Detection of optrA and poxtA Genes in Linezolid-Resistant Enterococcus Isolates from Fur Animals in China. Lett. Appl. Microbiol. 2022, 75, 1590–1595. [Google Scholar] [CrossRef]
- Ma, W. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; M7-A7; CLSI NCCLS: Wayne, PA, USA, 2006; Volume 26. [Google Scholar]
- Yarlagadda, V.; Sarkar, P.; Samaddar, S.; Manjunath, G.B.; Mitra, S.D.; Paramanandham, K.; Shome, B.R.; Haldar, J. Vancomycin Analogue Restores Meropenem Activity against NDM-1 Gram-Negative Pathogens. ACS Infect. Dis. 2018, 4, 1093–1101. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, T.; Guo, Y.; Liu, S.; Wang, J.; Shen, Y.; Tang, S.; Wang, Y.; Deng, X. In Vitro/Vivo Activity of Potential MCR-1 Inhibitor in Combination with Colistin Againsts Mcr-1-Positive Klebsiella pneumonia. Front. Microbiol. 2018, 9, 1615. [Google Scholar] [CrossRef]
- Levett, P.N. Time-Dependent Killing of Clostridium difficile by Metronidazole and Vancomycin. J. Antimicrob. Chemother. 1991, 27, 55–62. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, Y.; Yang, K.; Tong, Z.; Shi, J.; Li, R.; Xiao, X.; Ren, W.; Hardeland, R.; Reiter, R.J.; et al. Melatonin Overcomes MCR-Mediated Colistin Resistance in Gram-Negative Pathogens. Theranostics 2020, 10, 10697–10711. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Feng, L.; Xu, M.; Wen, H.; Yao, Z.; Shi, S.; Wu, Q.; Zhou, C.; Cao, J.; et al. In Vitro and in Vivo Synergistic Effect of Chrysin in Combination with Colistin against Acinetobacter baumannii. Front. Microbiol. 2022, 13, 961498. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.J.; Yu, H.H.; Kim, Y.J.; Lee, N.-K.; Paik, H.-D. Anti-Biofilm Activity of Grapefruit Seed Extract against Staphylococcus Aureus and Escherichia Coli. J. Microbiol. Biotechnol. 2019, 29, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Gu, S.; Shi, Y.; Cui, X.; Wen, S.; Ge, J. The Effect of Emodin on Staphylococcus aureus Strains in Planktonic Form and Biofilm Formation In Vitro. Arch. Microbiol. 2017, 199, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of Gut Microbiota during Probiotic-Mediated Attenuation of Metabolic Syndrome in High Fat Diet-Fed Mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes Clones’ Adaption to Mammalian Gut Accounts for Their Association with Dairy Products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]


| Gene | Primer | Primer Sequence (5′-3′) | References |
|---|---|---|---|
| 16SrRNA | 27F | AGAGTTTGATCMTGGCTCAG | [67] |
| 1492R | GGTTACCTTGTTACGACTT | ||
| gyrB | UP-1 | GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTYGA | [68] |
| UP-2r | AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGTCAT | ||
| Esp | Esp-F | CCACGAGTTAGCGGGAACAG | [22] |
| Esp-R | TTGGAGCCCCATCTTTTTCA | ||
| tufA | tufA-F | TACACGCCACTACGCTCAC | [22] |
| tufA-R | AGCTCCGTCCATTTGAGCAG | ||
| optrA | optrA-F | CACTGATTTGAGCAAGCTGTTGGTC | |
| optrA-R | TATGGATGGTGTGGCAGCATTGTC | ||
| poxtA | poxtA-F | TATGGATGGTGTGGCAGCATTGTC | |
| poxtA-R | GGTCGGTATTGTCGGCGTGAAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Q.; Chen, H.; Li, Y.; Li, H.; Yang, Z.; Zeng, J.; Zhang, P.; Ge, J.; Gao, M. The Synergistic Activity of Rhamnolipid Combined with Linezolid against Linezolid-Resistant Enterococcus faecium. Molecules 2023, 28, 7630. https://doi.org/10.3390/molecules28227630
Chang Q, Chen H, Li Y, Li H, Yang Z, Zeng J, Zhang P, Ge J, Gao M. The Synergistic Activity of Rhamnolipid Combined with Linezolid against Linezolid-Resistant Enterococcus faecium. Molecules. 2023; 28(22):7630. https://doi.org/10.3390/molecules28227630
Chicago/Turabian StyleChang, Qingru, Huinan Chen, Yifan Li, Hai Li, Zaixing Yang, Jiankai Zeng, Ping Zhang, Junwei Ge, and Mingchun Gao. 2023. "The Synergistic Activity of Rhamnolipid Combined with Linezolid against Linezolid-Resistant Enterococcus faecium" Molecules 28, no. 22: 7630. https://doi.org/10.3390/molecules28227630
APA StyleChang, Q., Chen, H., Li, Y., Li, H., Yang, Z., Zeng, J., Zhang, P., Ge, J., & Gao, M. (2023). The Synergistic Activity of Rhamnolipid Combined with Linezolid against Linezolid-Resistant Enterococcus faecium. Molecules, 28(22), 7630. https://doi.org/10.3390/molecules28227630





