HPLC-DAD-MS Characterization, Antioxidant Activity, α-amylase Inhibition, Molecular Docking, and ADMET of Flavonoids from Fenugreek Seeds
Abstract
1. Introduction
2. Results and Discussion
2.1. HPLC-DAD-ESIMS Analysis
2.2. TPC, TFC, and Antioxidant Activity
2.3. Anti-α-amylase Activity
2.4. ADMET and Drug-Likeness Evaluation
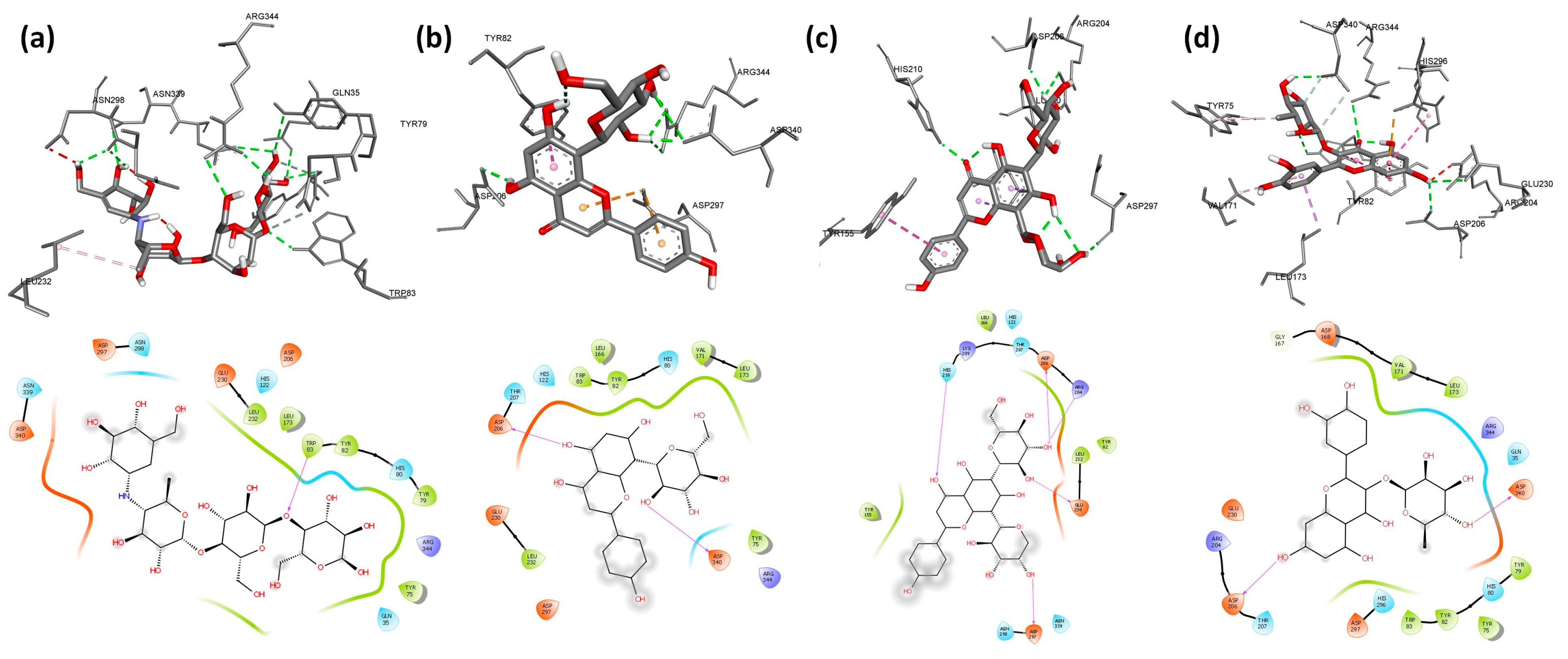
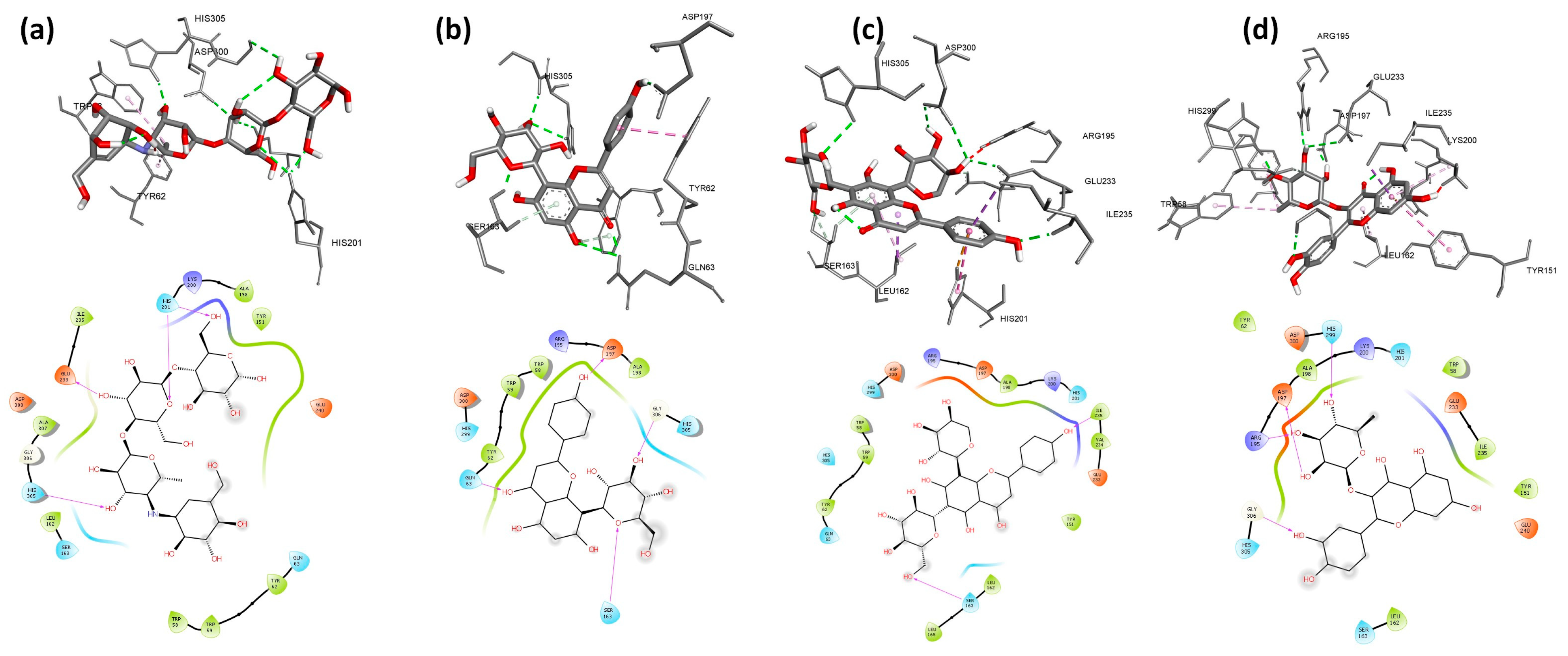
2.5. Molecular Docking
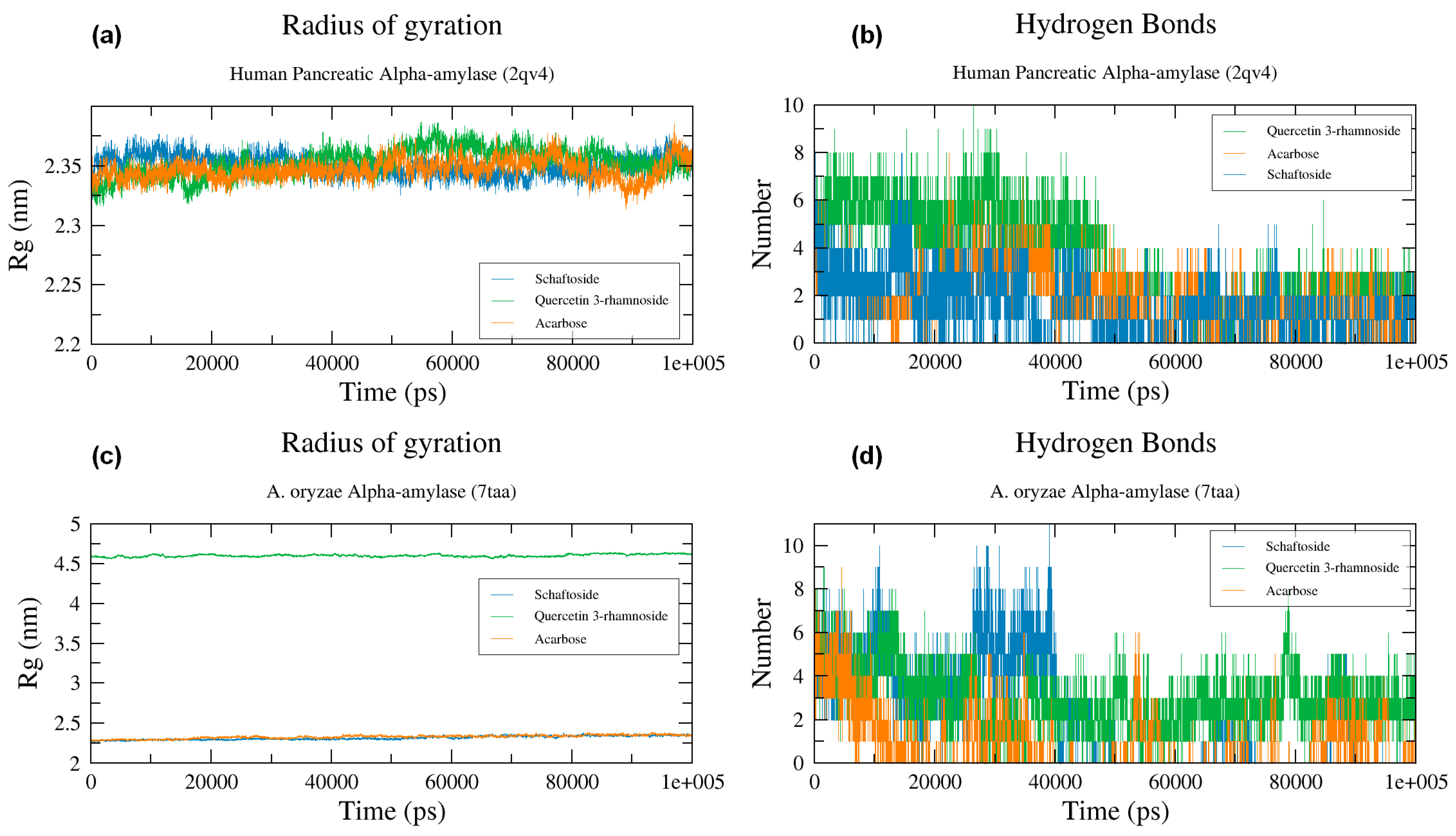
2.6. Molecular Dynamics Simulation
2.6.1. RMSD
2.6.2. RMSF
2.6.3. Radius of Gyration
2.6.4. Hydrogen Bonds
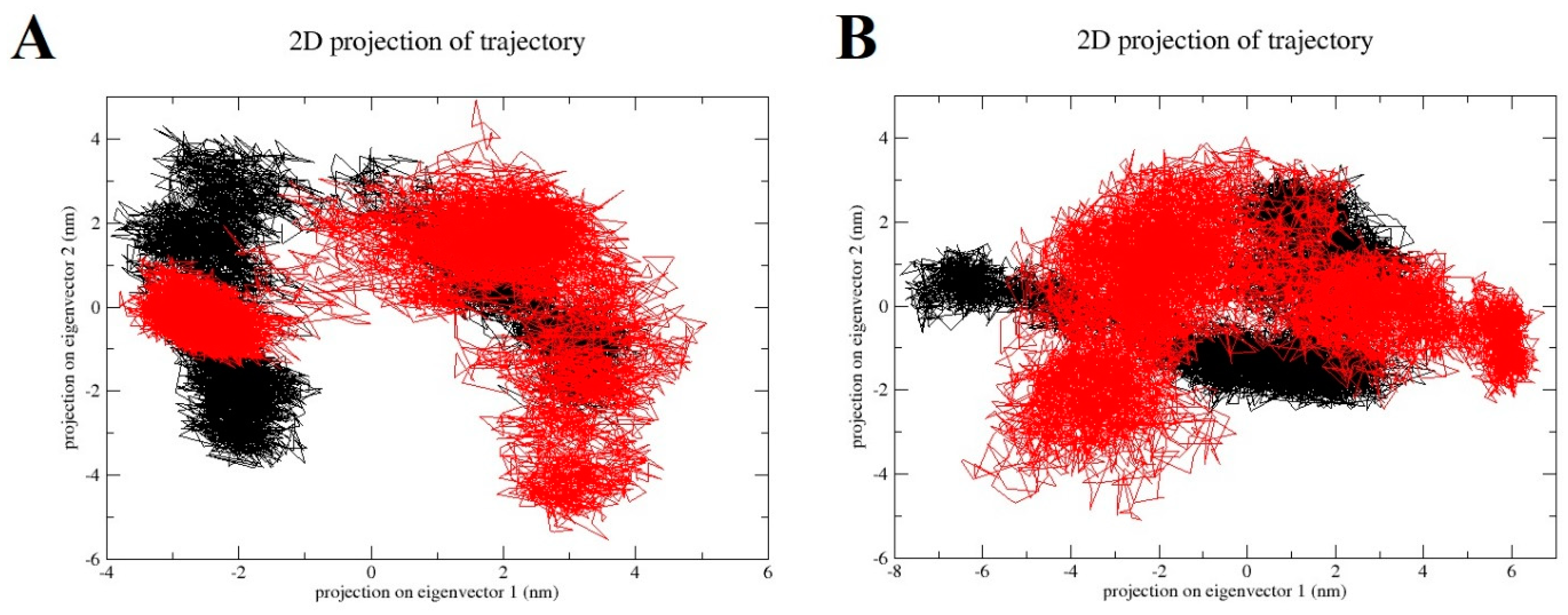
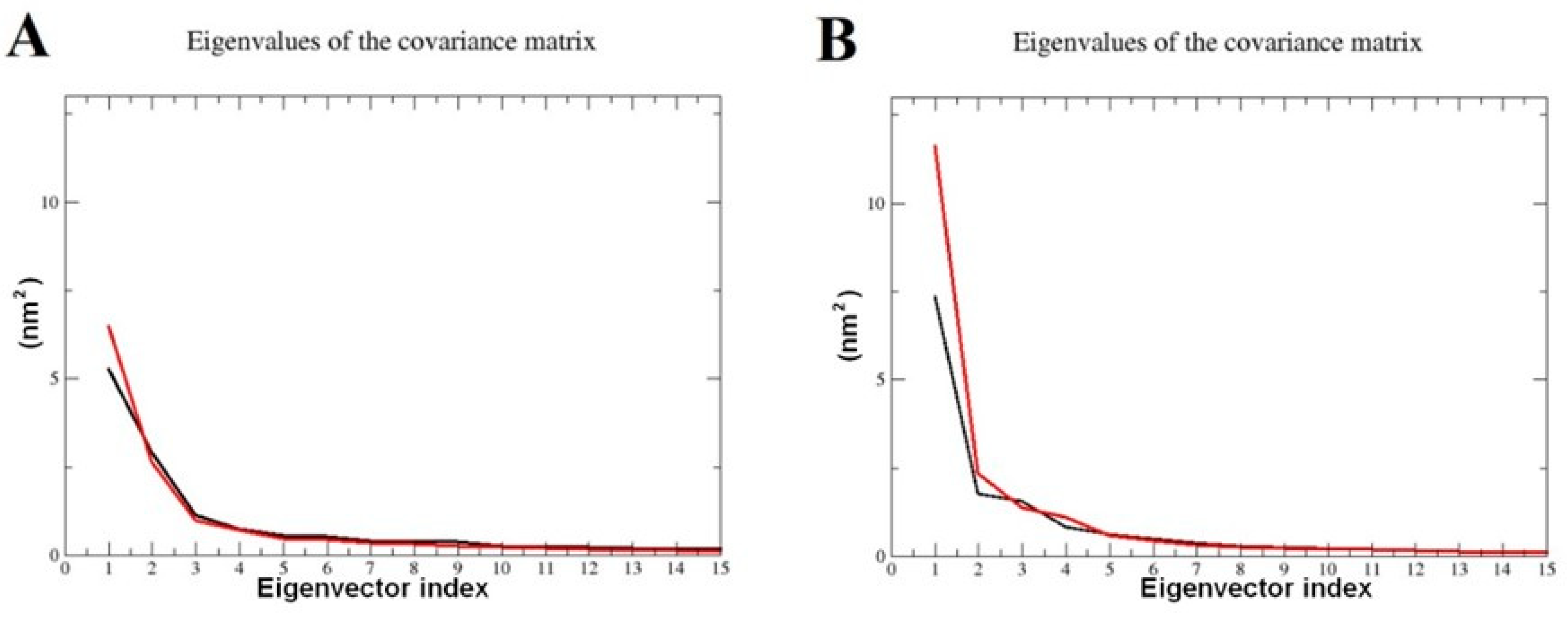
2.7. Principal Component Analysis (PCA)
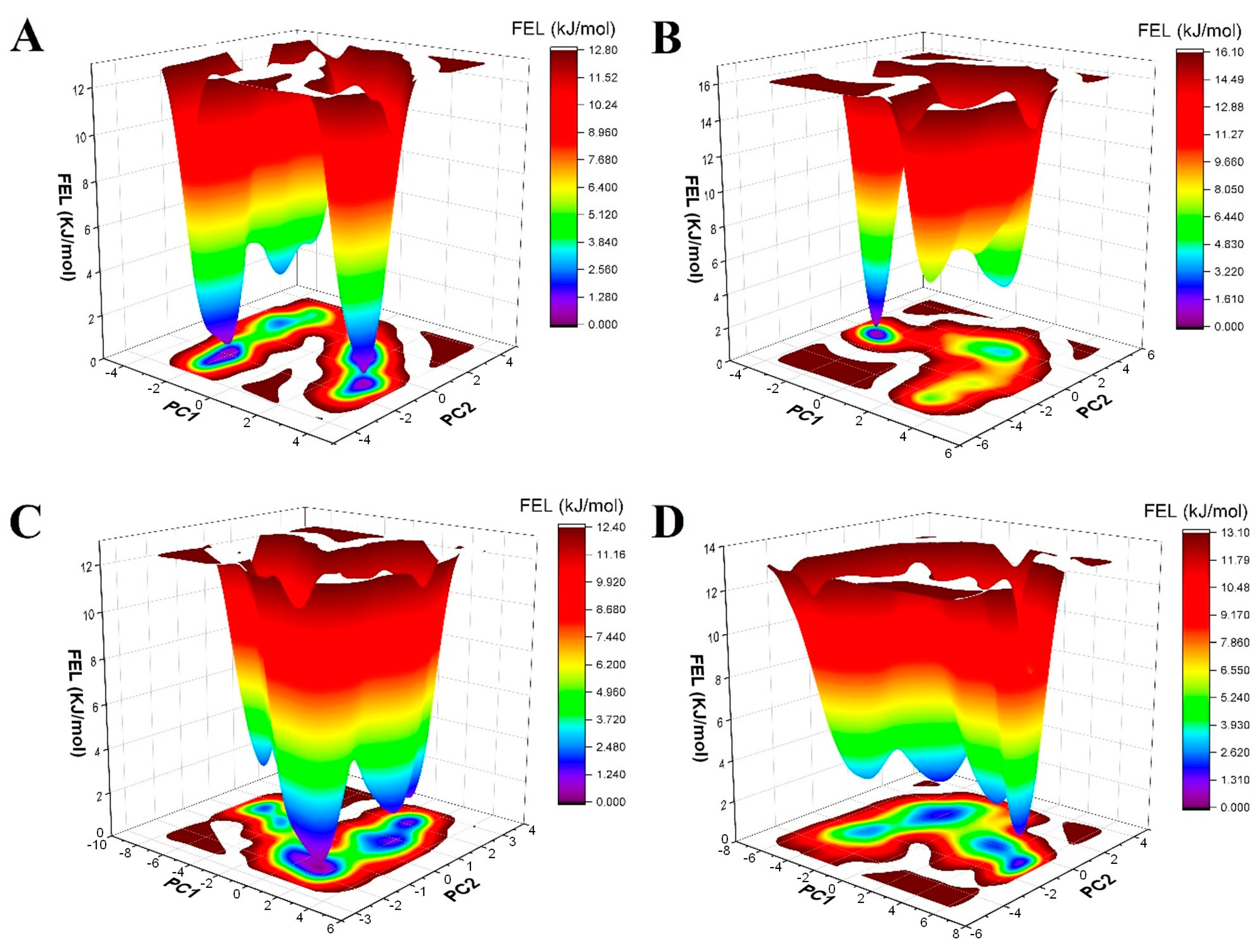
Free-Energy Landscape
3. Materials and Methods
3.1. Sample Preparation
3.1.1. Plant Material
3.1.2. Chemicals: Reagents and Standards
3.1.3. Plant Extract Preparation
3.2. Phytochemical Composition
3.2.1. Determination of Total Phenolic Contents (TPC)
3.2.2. Determination of Flavonoid Content (FC)
3.2.3. HPLC-DAD-ESIMS analysis
3.3. Anti-α-amylase Assay
3.4. Antioxidant Tests
3.4.1. DPPH Assay
3.4.2. Ferric-Reducing Antioxidant Power Assay
3.4.3. ABTS+ Scavenging Activity
3.4.4. Cupric Reducing Antioxidant Capacity (CUPRAC) Assay
3.5. ADMET and Drug-Likeness Evaluation
3.6. Molecular Docking
3.7. Molecular Dynamics Simulation
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rajhi, I.; Baccouri, B.; Rajhi, F.; Hammami, J.; Souibgui, M.; Mhadhbi, H.; Flamini, G. HS-SPME-GC–MS characterization of volatile chemicals released from microwaving and conventional processing methods of fenugreek seeds and flours. Ind. Crop. Prod. 2022, 182, 244–251. [Google Scholar] [CrossRef]
- Lohvina, H.; Sándor, M.; Wink, M. Effect of Ethanol Solvents on Total Phenolic Content and Antioxidant Properties of Seed Extracts of Fenugreek (Trigonella foenum-graecum L.) Varieties and Determination of Phenolic Composition by HPLC-ESI-MS. Diversity 2022, 14, 7. [Google Scholar] [CrossRef]
- Paramesha, M.; Priyanka, N.; Crassina, K.; Shetty, N.P. Evaluation of diosgenin content from eleven different Indian varieties of fenugreek and fenugreek leaf powder fortified bread. J. Food Sci. Technol. 2021, 58, 4746–4754. [Google Scholar] [CrossRef] [PubMed]
- Benayad, Z.; Gomez-Cordov, C.; Es-Safi, N.E. Identification and quantification of flavonoid glycosides from fenugreek (Trigonella foenum-graecum) germinated seeds by LC-DAD-ESI/MS analysis. J. Food Compos. Anal. 2014, 35, 21–29. [Google Scholar] [CrossRef]
- Beyzi, E.; Güneş, A. The Role of Agricultural Practises on Quality Characteristics of Fenugreek (Trigonella foenum-graecum L.) as Medicinal and Aromatic Plant. In Fenugreek, 1st ed.; Naeem, M.A., Aftab, T., Khan, M.M.A., Eds.; Springer: Singapore, 2021; pp. 63–81. [Google Scholar] [CrossRef]
- Shang, M.; Cai, S.; Han, J.; Li, J.; Zhao, Y.; Zheng, J.; Fan, W. Studies on flavonoids from Fenugreek (Trigonella foenum-graecum L.). Ch. J. MaT Med. 1998, 23, 614–626. [Google Scholar]
- Singh, U.; Chamoli, M.; Singh, K.P.; Ram, L.; Jangir, S.; Maheshwari, R.K. Amaing health benefit of fenugreek (Trigonella foenum-graecum L.). Int. J. Environ. Health 2022, 4, 19–27. [Google Scholar] [CrossRef]
- Naidu, M.M.; Shyamala, B.N.; Naik, J.P.; Sulochanamma, G.; Srinivas, P. Chemical composition and antioxidant activity of the husk and endosperm of fenugreek seeds. LWT 2011, 44, 451–456. [Google Scholar] [CrossRef]
- Ahmad, A.; Alghamdi, S.S.; Mahmood, K.; Afzal, M. Fenugreek a multipurpose crop: Potentialities and improvements. Saudi J. Biol. Sci. 2016, 23, 300–310. [Google Scholar] [CrossRef]
- Krol-Kogus, B.; Lamine, K.M.; Migas, P.; Boudjeniba, M.; Krauze-Baranowska, M. HPTLC determination of diosgenin in fenugreek seeds. Acta Pharm. 2018, 68, 97–107. [Google Scholar] [CrossRef]
- Sarwar, S.; Hanif, M.A.; Ayub, M.A.; Boakye, Y.D.; Agyare, C. Fenugreek. In Medicinal Plants of South Asia, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 257–271. [Google Scholar]
- Hilles, R.; Mahmood, A.S. A review on phytochemistry and pharmacological effects of Trigonella foenum-graecum. Adv. Herb. Med. 2016, 2, 61–67. [Google Scholar]
- Salam, S.G.A.; Rashed, M.M.; Ibrahim, N.A.; Emam, A.A.; Tahany, A.A.; Ammar, A.F. Phytochemical screening and in-vitro biological properties of unprocessed and household processed fenugreek (Trigonella foenum-graecum L.) seeds and leaves. Sci. Rep. 2023, 13, 7032. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.; Alkaabi, A.; Al-Falasi, S.; Daoud, S.A. Chemopreventive activities of Trigonella foenum-graecum (Fenugreek) against breast cancer. Cell Biol. Int. 2005, 29, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Yadav, S.S.; Kumar, S.; Narashiman, B. Ethnopharmacological, phytochemical and clinical studies on Fenugreek (Trigonella foenum-graecum L.). Food Biosci. 2022, 46, 101546. [Google Scholar] [CrossRef]
- Kaviarasan, S.; Naik, G.H.; Gangabhagirathi, R.; Anuradha, C.V.; Priyadarsini, K.I. In vitro studies on antiradical and antioxidant activities of fenugreek (Trigonella foenum-graecum) seeds. Food Chem. 2007, 103, 31–37. [Google Scholar] [CrossRef]
- Kahanovitz, L.; Sluss, P.M.; Russell, S.J. Type 1 diabetes–a clinical perspective. Point Care 2017, 16, 37–40. [Google Scholar] [CrossRef]
- Dilworth, L.; Facey, A.; Omoruyi, F. Diabetes Mellitus and Its Metabolic Complications: The Role of Adipose Tissues. Int. J. Mol. Sci. 2021, 22, 7644. [Google Scholar] [CrossRef]
- Jacobsen, I.B.; Henriksen, J.E.; Hother-Nielsen, O.; Vach, W.; Beck-Nielsen, H. Evidence-based insulin treatment in type 1 diabetes mellitus. Diabetes Res. Clin. Pract. 2009, 86, 1–10. [Google Scholar] [CrossRef]
- Kaur, N.; Kumar, V.; Nayak, S.K.; Wadhwa, P.; Kaur, P.; Sahu, S.K. Alpha- amylase as molecular target for treatment of diabetes mellitus: A comprehensive review. Chem. Biol. Drug Des. 2021, 98, 539–560. [Google Scholar] [CrossRef]
- Nia, S.; Khedidja, B.; Manel, L.; Israa, S.; Mohamed, Y. New Inhibition Detection Method to Evaluate the Human Salivary Alphaamylase Activity of Some Drugs, Molecular Docking, and SAR Studies. Antiinflamm. Antiallergy Agents Med. Chem. 2021, 20, 10–19. [Google Scholar] [CrossRef]
- Black, H.S. A Synopsis of the Associations of Oxidative Stress, ROS, and Antioxidants with Diabetes Mellitus. Antioxidants 2022, 11, 2003. [Google Scholar] [CrossRef]
- Rafiullah, M.; Benabdelkamel, H.; Masood, A.; Ekhzaimy, A.A.; Musambil, M.; Joy, S.S.; Alfadda, A.A. Urinary Proteome Differences in Patients with Type 2 Diabetes Pre and Post Liraglutide Treatment. Curr. Issues Mol. Biol. 2023, 45, 1407–1421. [Google Scholar] [CrossRef] [PubMed]
- Mwakalukwa, R.; Amen, Y.; Nagata, M.; Shimizu, K. Postprandial Hyperglycemia Lowering Effect of the Isolated Compounds from Olive Mill Wastes—An Inhibitory Activity and Kinetics Studies on α-Glucosidase and α-Amylase Enzymes. ACS Omega 2020, 5, 20070–20079. [Google Scholar] [CrossRef] [PubMed]
- Taslimi, P.; Aslan, H.E.; Demir, Y.; Oztaskin, N.; Maraş, A.; Gulçin, I.; Beydemir, S.; Goksu, S. Diarylmethanon, Bromophenol and Diarylmethane Compounds: Discovery of Potent Aldose Reductase, α-Amylase and α-Glycosidase Inhibitors as New Therapeutic Approach in Diabetes and Functional Hyperglycemia. Int. J. Biol. Macromol. 2018, 119, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Sidar, A.; Voshol, G.P.; Vijgenboom, E.; Punt, P.J. Novel Design of an α-Amylase with an N-Terminal CBM20 in Aspergillus niger Improves Binding and Processing of a Broad Range of Starches. Molecules 2023, 28, 5033. [Google Scholar] [CrossRef] [PubMed]
- Peyrot des Gachons, C.; Breslin, P.A. Salivary amylase: Digestion and metabolic syndrome. Curr. Diab. Rep. 2016, 16, 102. [Google Scholar] [CrossRef] [PubMed]
- Benguechoua, M.; Benarous, K.; Nia, S.; Yousfi, M. In Silico and In Vitro Studies of the Inhibitory Effect of Antihistamine Drug Cyproheptadine Hydrochloride on Human Salivary Alpha Amylase. Antiinflamm. Antiallergy Agents Med. Chem. 2021, 20, 233–238. [Google Scholar] [CrossRef]
- Lijun, S.; Yueyi, W.; Ming, M. Inhibition of α-amylase by polyphenolic compounds: Substrate digestion, binding interactions and nutritional intervention. Trends Food Sci. Technol. 2020, 104, 190–207. [Google Scholar] [CrossRef]
- Prasad, D.; Dandekar, A.S.; Kotmale, S.R.; Chavan, P.P.; Kadlag, S.V.; Sawant, D.D.; Ameeta, R. Insights into the Inhibition Mechanism of Human Pancreatic α-Amylase, a Type 2 Diabetes Target, by Dehydrodieugenol B Isolated from Ocimum tenuiflorum. ACS Omega 2021, 6, 1780–1786. [Google Scholar] [CrossRef]
- Etsassala, N.G.E.R.; Badmus, J.A.; Marnewick, J.L.; Egieyeh, S.; Iwuoha, E.I.; Nchu, F.; Hussein, A.A. Alpha-Glucosidase and Alpha-Amylase Inhibitory Activities, Molecular Docking, and Antioxidant Capacities of Plectranthus ecklonii Constituents. Antioxidants 2022, 11, 378. [Google Scholar] [CrossRef]
- Gazali, M.; Jolanda, O.; Husni, A.; Nurjanah; Majid, F.A.A.; Zuriat; Syafitri, R. In Vitro α-Amylase and α-Glucosidase Inhibitory Activity of Green Seaweed Halimeda tuna Extract from the Coast of Lhok Bubon, Aceh. Plants 2023, 12, 393. [Google Scholar] [CrossRef]
- Kim, H.-L.; Jung, Y.; Kim, H.I.; Sung, N.-Y.; Kim, M.-J.; Han, I.-J.; Kim, G.; Nho, E.Y.; Park, S.-Y.; Han, Y.; et al. Antidiabetic Effect of Fermented Mesembryanthemum crystallinum L. in db/db Mice Involves Regulation of PI3K-Akt Pathway. Curr. Issues Mol. Biol. 2023, 45, 6415–6431. [Google Scholar] [CrossRef] [PubMed]
- Kashtoh, H.; Baek, K.-H. New Insights into the Latest Advancement in α-Amylase Inhibitors of Plant Origin with Anti-Diabetic Effects. Plants 2023, 12, 2944. [Google Scholar] [CrossRef] [PubMed]
- Takahama, U.; Hirota, S. Interactions of flavonoids with α-amylase and starch slowing down its digestion. Food Funct. 2018, 9, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, R. Oxidative Stress and Antioxidants: Their Role in Human Disease, 1st ed.; Nova Biomedical Books: New York, NY, USA, 2009; p. 358. [Google Scholar]
- Munteanu, I.G.; Apetrei, C. Analytical methods used in determining antioxidant activity: A review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.P. Oxidative Stress in Health and Disease. Biomedicines 2023, 11, 2925. [Google Scholar] [CrossRef]
- Zujko, M.E.; Witkowska, A.M. Dietary Antioxidants and Chronic Diseases. Antioxidants 2023, 12, 362. [Google Scholar] [CrossRef]
- Juszczyk, G.; Mikulska, J.; Kasperek, K.; Pietrzak, D.; Mrozek, W.; Herbet, M. Chronic Stress and Oxidative Stress as Common Factors of the Pathogenesis of Depression and Alzheimer’s Disease: The Role of Antioxidants in Prevention and Treatment. Antioxidants 2021, 10, 1439. [Google Scholar] [CrossRef]
- Darenskaya, M.A.; Kolesnikova, L.I.; Kolesnikov, S.I. Oxidative Stress: Pathogenetic Role in Diabetes Mellitus and Its Complications and Therapeutic Approaches to Correction. Bull. Exp. Biol. Med. 2021, 171, 179–189. [Google Scholar] [CrossRef]
- Caturano, A.; D’Angelo, M.; Mormone, A.; Russo, V.; Mollica, M.P.; Salvatore, T.; Galiero, R.; Rinaldi, L.; Vetrano, E.; Marfella, R.; et al. Oxidative Stress in Type 2 Diabetes: Impacts from Pathogenesis to Lifestyle Modifications. Curr. Issues Mol. Biol. 2023, 45, 6651–6666. [Google Scholar] [CrossRef]
- Martemucci, G.; Costagliola, C.; Mariano, M.; D’andrea, L.; Napolitano, P.; D’Alessandro, A.G. Free Radical Properties, Source and Targets, Antioxidant Consumption and Health. Oxygen 2022, 2, 48–78. [Google Scholar] [CrossRef]
- Abeyrathne, E.D.N.S.; Nam, K.; Huang, X.; Ahn, D.U. Plant- and Animal-Based Antioxidants’ Structure, Efficacy, Mechanisms, and Applications: A Review. Antioxidants 2022, 11, 1025. [Google Scholar] [CrossRef] [PubMed]
- Benarous, K.; Djeridane, A.; Kameli, A.; Yousfi, M. Inhibition of Candida rugosa lipase by secondary metabolites extracts of three Algerian plants and their antioxydant activities. Curr. Enzyme Inhib. 2013, 9, 75–82. [Google Scholar] [CrossRef]
- Hoang, H.T.; Moon, J.-Y.; Lee, Y.-C. Natural Antioxidants from Plant Extracts in Skincare Cosmetics: Recent Applications, Challenges and Perspectives. Cosmetics 2021, 8, 106. [Google Scholar] [CrossRef]
- Chaudhary, P.; Janmeda, P.; Docea, A.O.; Yeskaliyeva, B.; Abdull Razis, A.F.; Modu, B.; Calina, D.; Sharifi-Rad, J. Oxidative stress, free radicals and antioxidants: Potential crosstalk in the pathophysiology of human diseases. Front. Chem. 2023, 11, 1158198. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, M.; Burzynska-Pedziwiatr, I.; Wozniak, L.A.; Bukowiecka-Matusiak, M. Impact of Polyphenols on Inflammatory and Oxidative Stress Factors in Diabetes Mellitus: Nutritional Antioxidants and Their Application in Improving Antidiabetic Therapy. Biomolecules 2023, 13, 1402. [Google Scholar] [CrossRef]
- Tuell, D.S.; Los, E.A.; Ford, G.A.; Stone, W.L. The Role of Natural Antioxidant Products That Optimize Redox Status in the Prevention and Management of Type 2 Diabetes. Antioxidants 2023, 12, 1139. [Google Scholar] [CrossRef]
- Król-Kogus, B.; Głód, D.; Krauze-Baranowska, M.; Matławska, I. Application of one- and two-dimensional highperformance liquid chromatography methodologies for the analysis of C-glycosylflavones from fenugreek seeds. J. Chromatogr. A 2014, 1367, 48–56. [Google Scholar] [CrossRef]
- Król-Kogus, B.; Głód, D.; Hałasa, R.; Krauze-Baranowska, M.; Pobłocka-Olech, L. 2D LC as a tool for standardization of Foenugraeci semen extracts containing compounds with anti-Helicobacter pylori Activity. Food Funct. 2021, 12, 2686–2692. [Google Scholar] [CrossRef]
- Omezzine, F.; Bouaziz, M.; Daami-Remadi, M.; Simmonds, M.S.J.; Haouala, R. Chemical composition and antifungal activity of Trigonella foenum-graecum L. varied with plant ploidy level and developmental stage. Arab. J. Chem. 2017, 10, S3622–S3631. [Google Scholar] [CrossRef]
- Khole, S.; Chatterjee, S.; Variyar, P.; Sharma, A.; Devssagayam, T.P.A.; Ghaskadbi, S. Bioactive constituents of germinated fenugreek seeds with strong antioxidant potential. J. Funct. Foods 2014, 6, 270–279. [Google Scholar] [CrossRef]
- Markham, U.R. Techniques of Flavonoid Identification; Academic Press: London, UK, 1982; p. 113. [Google Scholar]
- Bouhenni, H.; Doukani, K.; Hanganu, D.; Olah, N.K.; Sekeroglu, N.; Gezici, S.; Niculae, M. Comparative analysis on bioactive compounds and antioxidant activity of Algerian fenugreek (Trigonella foenum-graecum L.) and Syrian cumin (Cuminum cyminum L.) seeds. Herba Pol. 2021, 67, 18–34. [Google Scholar] [CrossRef]
- Bhanger, M.I.; Bukhari, S.B.; Memon, S. Antioxidative activity of extracts from a Fenugreek seeds (Trigonella foenum-graecum). Pak. J. Anal. Environ. 2008, 9, 6. [Google Scholar]
- Figueiredo, A.C.; Barroso, J.G.; Pedro, L.G.; Scheffer, J.J. Factors affecting secondary metabolite production in plants: Volatile components and essential oils. Flavour. Fragr. J. 2008, 23, 213–226. [Google Scholar] [CrossRef]
- Wijekoon, M.J.O.; Bhat, R.; Karim, A.A. Effect of extraction solvents on the phenolic compounds and antioxidant activities of bunga kantan (Etlingera elatior Jack.) inflorescence. J. Food Compos. Anal. 2011, 24, 615–619. [Google Scholar] [CrossRef]
- Kaki, Y.T.A.; Bennadja, S.; Chefrour, A. Revalorisation d’une essence endémique: Le sapin de Numidie (Abies numidica). Fl. Medit. 2013, 23, 123–129. [Google Scholar]
- Scarano, A.; Santino, A. The plant polyphenol metabolism as functional architecture and its nutritional exploitation. Int. J. Food Sci. Nutr. 2019, 8, 26–30. [Google Scholar] [CrossRef]
- Šamec, D.; Karalija, E.; Šola, I.; Vujčić Bok, V.; Salopek-Sondi, B. The Role of Polyphenols in Abiotic Stress Response: The Influence of Molecular Structure. Plants 2021, 10, 118. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, P.; Cheng, G.; Zhang, Y. A brief review of phenolic compounds identified from plants: Their extraction, analysis, and biological activity. Nat. Prod. Commun. 2022, 17, 1934578X211069721. [Google Scholar] [CrossRef]
- Al-Dabbagh, B.; Elhaty, I.A.; Al Hrout, A.; Al Sakkaf, R.; El-Awady, R.; Ashraf, S.S.; Amin, A. Antioxidant and anticancer activities of Trigonella foenum-graecum, Cassia acutifolia and Rhazya stricta. BMC Complement Altern. Med. 2018, 18, 240. [Google Scholar] [CrossRef]
- Iloki-Assanga, S.B.; Lewis-Luján, L.M.; Lara-Espinoza, C.L.; Gil-Salido, A.A.; Fernandez-Angulo, D.; Rubio-Pino, J.L.; Haines, D.D. Solvent effects on phytochemical constituent profiles and antioxidant activities, using four different extraction formulations for analysis of Bucida buceras L. and Phoradendron californicum. BMC Res. Notes 2015, 8, 396. [Google Scholar] [CrossRef] [PubMed]
- Zreen, Z.; Hameed, A.; Kiran, S.; Farooq, T.; Zaroog, M.S. A Comparative Study of Diospyros malabarica (Gaub) Extracts in Various Polarity-Dependent Solvents for Evaluation of Phytoconstituents and Biological Activities. Biomed. Res. Int. 2022, 2012, 4746223. [Google Scholar] [CrossRef]
- Özcan, M.M. A review on some properties of almond: Impact of processing, fatty acids, polyphenols, nutrients, bioactive properties, and health aspects. J. Food Sci. Technol. 2022, 60, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Engwa, G.A.; Nweke, F.N.; Nkeh-Chungag, B.N. Free Radicals, Oxidative Stress-Related Diseases and Antioxidant Supplementation. Altern. Ther. Health Med. 2022, 28, 114–128. [Google Scholar]
- Xu, X.; Liu, A.; Hu, S.; Ares, I.; Martínez-Larrañaga, M.R.; Wang, X.; Martínez, M.A. Synthetic phenolic antioxidants: Metabolism, hazards and mechanism of action. Food Chem. 2021, 353, 129488. [Google Scholar] [CrossRef]
- Deng, Y.; Yang, G.; Yue, J.; Qian, B.; Liu, Z.; Wang, D.; Zhao, Y. Influences of ripening stages and extracting solvents on the polyphenolic compounds, antimicrobial and antioxidant activities of blueberry leaf extracts. Food Control 2014, 38, 184–191. [Google Scholar] [CrossRef]
- Gulcin, İ. Antioxidants and antioxidant methods: An updated overview. Arch. Toxicol. 2020, 94, 651–715. [Google Scholar] [CrossRef]
- Rahmani, M.; Hamel, L.; Toumi-Benali, F.; Dif, M.M.; Moumen, F.; Rahmani, H. Determination of antioxidant activity, phenolic quantification of four varieties of fenugreek Trigonella foenum-graecum L. seed extract cultured in west Algeria. J. Mater. Environ. Sci. 2018, 9, 1656–1661. [Google Scholar]
- Akbari, S.; Abdurahman, N.H.; Yunus, R.M.; Alara, O.R.; Abayomi, O.O. Extraction, characterization and antioxidant activity of fenugreek (Trigonella Foenum-graecum) seed oil. Mater. Sci. Energy Technol. 2019, 2, 349–355. [Google Scholar] [CrossRef]
- Priya, V.; Jananie, R.K.; Vijayalakshmi, K. Studies on anti-oxidant activity of Trigonella foenum-graecum seed using in vitro models. Int. J. Pharm. Sci. Res. 2011, 2, 2704–2708. [Google Scholar] [CrossRef]
- Boeing, J.S.; Barizão, É.O.; Montanher, P.F.; de Cinque Almeida, V.; Visentainer, J.V. Evaluation of solvent effect on the extraction of phenolic compounds and antioxidant capacities from the berries: Application of principal component analysis. Chem. Cent. J. 2014, 8, 48. [Google Scholar] [CrossRef]
- Wissal, A.; Mohamed, A.; Fatouma, M.; Jalludin, O.; Manar, E.; Adnane, A.; Ayoub, A. Aantimicrobial and antioxidant activities of Trigonella Foenum-graecum essential oil from the of settat (Morocco). Pharmacologyonline 2021, 2, 434–442. [Google Scholar]
- Liu, Y.; Kakani, R.; Nair, M.G. Compounds in functional food fenugreek spice exhibit anti-inflammatory and antioxidant activities. Food Chem. 2012, 131, 1187–1192. [Google Scholar] [CrossRef]
- Ouissem, B.S.; Sabrina, B.; Lotfi, B.; Khellaf, R.; Chawki, B.; Ibrahim, D.; Fadila, B. HPLC Analysis and Antioxidant Properties of Algerian Lepidium draba Ethyl acetate Extract. J. Biol. Active Prod. Nature 2018, 8, 265–271. [Google Scholar] [CrossRef]
- Sharma, N.; Biswas, S.; Al-Dayan, N.; Alhegaili, A.S.; Sarwat, M. Antioxidant role of kaempferol in prevention of hepatocellular carcinoma. Antioxidants 2021, 10, 48. [Google Scholar] [CrossRef]
- Chen, J.; Yang, J.; Ma, L.; Li, J.; Shahzad, N.; Kim, C.K. Structure-antioxidant activity relationship of methoxy, phenolic hydroxyl, and carboxylic acid groups of phenolic acids. Sci. Rep. 2020, 10, 10. [Google Scholar] [CrossRef]
- Deka, H.; Choudhury, A.; Dey, B.K. An Overview on Plant Derived Phenolic Compounds and Their Role in Treatment and Management of Diabetes. J. Pharmacopunct. 2022, 25, 199–208. [Google Scholar] [CrossRef]
- Padhi, S.; Nayak, A.K.; Behera, A. Type II diabetes mellitus: A review on recent drug-based therapeutics. Biomed. Pharmacother. 2020, 131, 110708. [Google Scholar] [CrossRef]
- Jagadeesan, G.; Muniyandi, K.; Manoharan, A.L.; Nataraj, G.; Thangaraj, P. Understanding the bioaccessibility, α-amylase and α-glucosidase enzyme inhibition kinetics of Allmania nodiflora (L.) R. Br. ex Wight polyphenols during in vitro simulated digestion. Food Chem. 2022, 372, 131294. [Google Scholar] [CrossRef]
- Hichri, F.; Omri, A.; Hossan, A.S.M.; Ben Jannet, H. Alpha-glucosidase and amylase inhibitory effects of Eruca vesicaria subsp. longirostris essential oils: Synthesis of new 1, 2, 4-triazole-thiol derivatives and 1, 3, 4-thiadiazole with potential inhibitory activity. Pharm. Biol. 2019, 57, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Kazeem, M.I.; Adamson, J.O.; Ogunwande, I.A. Modes of inhibition of α -Amylase and α -Glucosidase by aqueous extract of Morinda lucida Benth Leaf. BioMed Res. Int. 2013, 2013, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, S.; Suresh, S.; Kadeppagari, R.K. Amylase inhibitors and their biomedical applications. Starch-Stärke 2013, 65, 535–542. [Google Scholar] [CrossRef]
- Etxeberria, U.; de la Garza, A.L.; Campión, J.; Martínez, J.A.; Milagro, F.I. Antidiabetic effects of natural plant extracts via inhibition of carbohydrate hydrolysis enzymes with emphasis on pancreatic alpha amylase. Expert Opin. Ther. Targets 2012, 16, 269–297. [Google Scholar] [CrossRef] [PubMed]
- Shawky, E.; Sobhy, A.A.; Ghareeb, D.A.; Eldin, S.M.S.; Selim, D.A. Comparative metabolomics analysis of bioactive constituents of the leaves of different Trigonella species: Correlation study to α-amylase and α-glycosidase inhibitory effects. Ind. Crop. Prod. 2022, 182, 114947. [Google Scholar] [CrossRef]
- Hemlata, B.; Pornima, G.; Tukaram, K.; Pankaj, B. In vitro anti-amylase activity of some Indian dietary spices. J. Appl. Biol. Biotechnol 2019, 7, 4–7. [Google Scholar] [CrossRef]
- Kamtekar, S.; Keer, V.; Patil, V. Estimation of phenolic content, flavonoid content, antioxidant and alpha amylase inhibitory activity of marketed polyherbal formulation. J. Appl. Pharm. Sci 2014, 4, 061–065. [Google Scholar] [CrossRef]
- Bansode, T.S.; Gupta, A.; Chaphalkar, S.R.; Salalkar, B.K. Integrating in-silico and in-vitro approaches to screen the antidiabetic drug from Trigonella foenum-graecum Linn. Int. J. Biochem. Res. Rev. 2016, 14, 1–10. [Google Scholar] [CrossRef]
- Laila, O.; Murtaza, I.; Muzamil, S.; Ali, S.I.; Ali, S.A.; Paray, B.A.; Mansoor, S. Enhancement of nutraceutical and antidiabetic potential of fenugreek (Trigonella foenum-graecum L.) Sprouts with natural elicitors. Saudi Pharm. J. 2022, 31, 1–13. [Google Scholar] [CrossRef]
- Amin, R.; Abdul-Ghani, A.S.; Suleiman, M.S. Effect of Trigonella foenum-graecum on intestinal absorption. Diabetes 1987, 36, 211A. [Google Scholar]
- Kim, Y.; Keogh, J.B.; Clifton, P.M. Polyphenols and glycemic control. Nutrients 2016, 8, 17. [Google Scholar] [CrossRef]
- Keskes, H.; Belhadj, S.; Jlail, L.; El Feki, A.; Sayadi, S.; Allouche, N. LC–MS–MS and GC–MS analyses of biologically active extracts of Tunisian Fenugreek (Trigonella foenum-graecum L.) Seeds. J. Food Meas. Charact. 2018, 12, 209–220. [Google Scholar] [CrossRef]
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef] [PubMed]
- Shashikumar, J.N.; Champawat, P.S.; Mudgal, V.D.; Jain, S.K. Role of fenugreek (Trigonella foenum-graecum) on in management of diabetes disease. J. Pharmacogn. Phytochem 2019, 8, 184–187. [Google Scholar]
- Mooventhan, A.; Nivethitha, L. A narrative review on evidence-based antidiabetic effect of fenugreek (Trigonella Foenum-graecum L.). Int. J. Nutr. Pharmacol. Neurol. Dis. 2017, 7, 84–87. [Google Scholar] [CrossRef]
- Kim, J.S.; Kwon, C.S.; Son, K.H. Inhibition of alpha-glucosidase and amylase by luteolin, a flavonoid. Biosci. Biotechnol. Biochem. 2000, 64, 2458–2461. [Google Scholar] [CrossRef] [PubMed]
- Ganeshpurkar, A.; Diwedi, V.; Bhardwaj, Y. In vitro α-amylase and α-glucosidase inhibitory potential of Trigonella foenum-graecum leaves extract. Int. Quart. J. Resea. Ayurveda 2013, 34, 109–111. [Google Scholar] [CrossRef]
- Xiao, J.; Capanoglu, E.; Jassbi, A.R.; Miron, A. Advance on the flavonoid C-glycosides and health benefits. Crit. Rev. Food Sci. Nutr. 2016, 56, 29–45. [Google Scholar] [CrossRef]
- Niknam, R.; Kiani, H.; Mousavi, Z.E.; Mousavi, M. Extraction, Detection, and Characterization of Various Chemical Components of Trigonella foenum-graecum L. (Fenugreek) Known as a Valuable Seed in Agriculture. In Fenugreek, 1st ed.; Naeem, M., Aftab, T., Khan, M.M.A., Eds.; Springer: Singapore, 2021; pp. 189–217. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Sehrawat, A.; Mishra, J.; Sidhu, I.S.; Navik, U.; Khullar, N.; Reddy, P.H. Oxidative stress in the pathophysiology of type 2 diabetes and related complications: Current therapeutics strategies and future perspectives. Free Radic. Biol. Med. 2022, 184, 114–134. [Google Scholar] [CrossRef]
- Fatima, H.; Shahid, M.; Pruitt, C.; Pung, M.A.; Mills, P.J.; Riaz, M.; Ashraf, R. Chemical Fingerprinting, Antioxidant, and Anti-Inflammatory Potential of Hydroethanolic Extract of Trigonella foenum-graecum. Antioxidants 2022, 11, 364. [Google Scholar] [CrossRef]
- He, Y.; Ding, C.; Wang, X.; Wang, H.; Suo, Y. Using response surface methodology to optimize countercurrent chromatographic separation of polyphenol compounds from fenugreek (Trigonella foenum-graecum L.) seeds. J. Liq. Chromatogr. Relat. 2015, 38, 29–35. [Google Scholar] [CrossRef]
- Serseg, T.; Benarous, K.; Serseg, M.; Rehman, H.M.; El Bakri, Y.; Goumri-Said, S. Discovery of inhibitors against SARS-CoV-2 associated fungal coinfections via virtual screening, ADMET evaluation, PASS, molecular docking, dynamics and pharmacophore studies. Arab. J. Basic Appl. Sci. 2022, 29, 337–350. [Google Scholar] [CrossRef]
- Kontoyianni, M.; McClellan, L.M.; Sokol, G.S. Evaluation of docking performance: Comparative data on docking algorithms. J. Med. Chem. 2004, 47, 558–565. [Google Scholar] [CrossRef]
- Lee, G.R.; Shin, W.H.; Park, H.B.; Shin, S.M.; Seok, C.O. Conformational sampling of flexible ligand-binding protein loops. Bull Korean Chem. Soc. 2012, 33, 770–774. [Google Scholar] [CrossRef][Green Version]
- Teague, S.J. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2003, 2, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Khan, R.J.; Jha, R.K.; Amera, G.M.; Jain, M.; Singh, E.; Pathak, A.; Singh, A.K. Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J. Biomol. Struct. Dyn. 2021, 39, 2679–2692. [Google Scholar] [CrossRef] [PubMed]
- Mosquera-Yuqui, F.; Lopez-Guerra, N.; Moncayo-Palacio, E.A. Targeting the 3CLpro and RdRp of SARS-CoV-2 with phytochemicals from medicinal plants of the Andean Region: Molecular docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2022, 40, 2010–2023. [Google Scholar] [CrossRef]
- Menéndez, C.A.; Accordino, S.R.; Gerbino, D.C.; Appignanesi, G.A. Hydrogen bond dynamic propensity studies for protein binding and drug design. PLoS ONE 2016, 11, e0165767. [Google Scholar] [CrossRef]
- Narang, S.S.; Suniba, S.; Deepti, G.; Bhupesh, G. Assessing the effect of D59P mutation in the DE loop region in amyloid aggregation propensity of β2-microglobulin: A molecular dynamics simulation study. J. Cell. Biochem. 2018, 119, 782–792. [Google Scholar] [CrossRef]
- D’Auria, M.; Mecca, M.; Bruno, M.R.; Todaro, L. Extraction Methods and Their Influence on Yield When Extracting Thermo-Vacuum-Modified Chestnut Wood. Forests 2021, 12, 73. [Google Scholar] [CrossRef]
- Müller, L.; Gnoyke, S.; Popken, A.M.; Böhm, V.V. Antioxidant capacity and related parameters of different fruit formulations. Food Sci. Technol. 2010, 43, 992–999. [Google Scholar] [CrossRef]
- Topçu, G.; Ay, A.; Bilici, A.; Sarıkürkcü, C.; Öztürk, M.; Ulubelen, A. A new flavone from antioxidant extracts of Pistacia terebinthus. Food Chem. 2007, 103, 816–822. [Google Scholar] [CrossRef]
- Zengin, G.; Cengiz, S.; Abdurrahman, A.; Ramazan, C. Olcay. A comprehensive study on phytochemical characterization of Haplophyllum myrtifolium Boiss. endemic to Turkey and its inhibitory potential against key enzymes involved in Alzheimer, skin diseases and type II diabetes. Ind. Crop. Prod. 2014, 53, 244–251. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant determinations by the use of a stable Free Radical. Nature 1958, 4617, 1119–1200. [Google Scholar] [CrossRef]
- Oyaizu, M. Studies on products of browning reactions: Antioxidative activities of browning reaction prepared from glucosamine. Jpn. J. Nutr. 1986, 44, 307–315. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Apak, R.; Guclu¨, K.; Ozyurek, M.; Karademir, S.E. Novel total antioxidant capacity index for dietary polyphenols and vitamins C and E, using their cupric ion reducing capability in the presence of neocuproin: CUPRAC Method. J. Agric. Food Chem. 2004, 52, 7970–7981. [Google Scholar] [CrossRef]
- Li, A.P. Screening for human ADME/Tox drug properties in drug discovery. Drug Discov. Today 2001, 6, 357–366. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Tang, Y. Sdmet SAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Bolton, E.E. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, 1388–1395. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Karatas, H.; Li, Y.; Liu, L.; Ji, J.; Lee, S.; Chen, Y.; Wang, S. Discovery of a highly potent, cell-permeable macrocyclic peptidomimetic (MM-589) targeting the WD repeat domain 5 protein (WDR5)–mixed lineage leukemia (MLL) protein–protein interaction. J. Med. Chem. 2017, 60, 4818–4839. [Google Scholar] [CrossRef] [PubMed]
- Nebeg, H.; Benarous, K.; Serseg, T.; Lazreg, A.; Hassani, H.; Yousfi, M. Seeds, leaves and roots of Thapsia garganica as a source of new potent lipases inhibitors: In vitro and in silico studies. Endocr. Metab. Immune. Disord. Drug Targets 2019, 19, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Serseg, T.; Benarous, K.; Yousfi, M. The inhibitory effect of three essential oils on Candida rugosa lipase: In vitro and in silico studies. J. Nat. Prod. 2020, 10, 208–215. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Mackerell, A.D. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Lindahl, A.; Hess Spoel, V.D. GROMACS 2021.5 Manual; 2022. [Google Scholar]
- Lemkul, J.A. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living. J. Comput Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
- Turner, P.J. XMGRACE, Version 5.1.19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005. [Google Scholar]
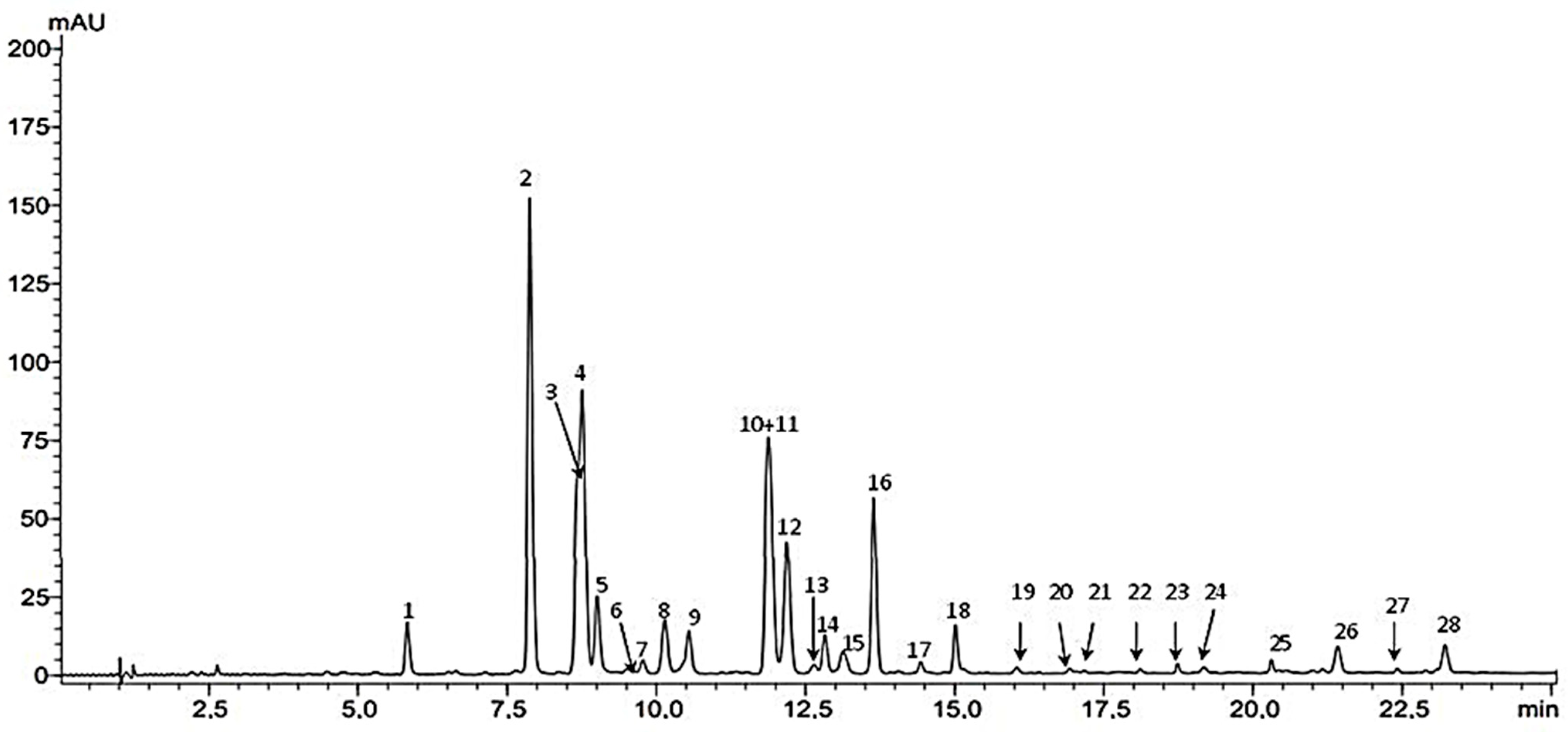



| Peak no. | tR min | UV [λmax nm] | ESI Scan (+) m/z 250–1200 | ESI Scan (−) m/z 250–1300 | Compound |
|---|---|---|---|---|---|
| 1 | 5.827 | 270, 334 | 595 | 593, 707 (TFA) | Vicenin-2 |
| 2 | 7.896 | 269, 333 | 565 | 563, 677 (TFA) | Vicenin-1 |
| 3 | 8.698 | 269, 333 | 565 | 563, 677 (TFA) | Schaftosid-2 |
| 4 | 8.763 | 270, 333 | 565 | 563, 677 (TFA) | Schaftoside-1 |
| 5 | 9.014 | 257, 268 (s), 347 | 449 | 447, 561 (TFA) | Isoorientin |
| 6 | 9.504 | 268, 324 | 949 | 947, 1061 (TFA) | KaempferolO-feruloyl-triglucoside/trigalactoside isomer |
| 7 | 9.779 | 271, 332 | 565 | 563, 677 (TFA) | Apigenin 6,8-C-pentoside-hexoside isomer |
| 8 | 10.138 | 255, 266 (s), 347 | 449 | 447, 561 (TFA) | Orientin |
| 9 | 10.554 | 270, 334 | 565 | 563, 677 (TFA) | Vicenin-3 |
| 10 | 11.890 | 270, 335 | 535 | 533, 647 (TFA) | Apigenin-C-di-(6/8)-pentoside isomer |
| 11 | 11.890 | 270, 335 | 433 | 431, 545 (TFA) | Isovitexin |
| 12 | 12.187 | 267, 335 | 433 | 431, 545 (TFA) | Vitexin |
| 13 | 12.594 | 253, 265, 315, 349 (s) | 935 | 933, 1047 (TFA) | Quercetin O-coumaroyl-tri-glucoside/trigalactoside isomer |
| 14 | 12.816 | 270, 335 | 535 | 533, 647 (TFA) | Apigenin-C-di-(6/8)-pentoside isomer |
| 15 | 13.130 | 270, 335 | 535 | 533, 647 (TFA) | Apigenin-C-di-(6/8)-pentoside isomer |
| 16 | 13.648 | 270, 334 | 535 | 533, 647 (TFA) | Apigenin-C-di-(6/8)-pentoside isomer |
| 17 | 14.434 | 270, 339 | 535 | 533, 647 (TFA) | Apigenin-C-di-(6/8)-pentoside isomer |
| 18 | 15.016 | 269, 331 | 535 | 533, 647 (TFA) | Apigenin-C-di-(6/8)-pentoside isomer |
| 19 | 16.024 | 269, 330 | 549 | 547, 661 (TFA) | Apigenin C-di-(6,8)-pentoside methyl ether |
| 20 | 16.976 | 267, 315 | - | - | Probably Unknown flavonoid ester |
| 21 | 17.181 | 250, 263, 327 (s), 361 | 449 | - | Quercetin 3-O-rhamnoside |
| 22 | 18.115 | 243, 314 | - | 647, 761 (TFA), 681, 795 (TFA) | Probably Unknown flavonoid ester |
| 23 | 18.730 | 274, 339 | - | - | Unknown flavone |
| 24 | 19.170 | 310 | - | - | Unknown |
| 25 | 20.303 | 274, 329 | - | 707, 875 | Unknown flavone |
| 26 | 21.425 | 270, 316 | 595 | 593, 707 (TFA) | p-coumaroyl-orientin/isoorientin isomer |
| 27 | 22.435 | 270, 315 | 579 | 577, 691 (TFA) | p-coumaroyl vitexin/isovitexin isomer |
| 28 | 23.211 | 270, 315 | 579 | 577, 691 (TFA) | p-coumaroyl vitexin/isovitexin isomer |
| Yield % | TPC (µgGAE/mg Extract) | TFC (µgQE/mg Extract) | α-Amylase Inhibition IC50 (µg/mL) | |
|---|---|---|---|---|
| Fenugreek seeds | 17.6 | 154.68 ± 1.50 | 37.69 ± 0.73 | 653.52 ± 3.24 |
| Acarbose | - | - | - | 3650.93 ± 10.70 |
| DPPH Inhibition IC50 (µg/mL) | FRAP A0.5 (µg/mL) | ABTS Inhibition IC50 (µg/mL) | CUPRAC A0.5 (µg/mL) | |
|---|---|---|---|---|
| Fenugreek seeds | 556.6 ±9.87 | >200 | 593.62 ± 9.35 | 451.90 ± 9.07 |
| BHA | 6.14 ± 0.41 | n.d | 1.81 ± 0.10 | - |
| BHT | 12.99 ± 0.41 | n.d | 1.29 ± 0.30 | - |
| Tannic acid | n.d | 5.39 ± 0.91 | n.d | n.d |
| Ascorbic acid | n.d | 6.77 ± 1.15 | n.d | n.d |
| Molecule | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug - likeness | Lipinski | - | + | - | - | - | - | - | - | + | - |
| Blood-Brain Barrier | - | - | - | - | - | - | - | - | - | - | |
| Absorption | Human Intestinal Absorption | Low | Low | Low | Low | Low | Low | Low | Low | Low | Low |
| Caco-2 | - | - | - | - | - | - | - | - | - | - | |
| Human oral bioavailability | - | - | - | - | - | - | - | - | - | - | |
| Skin Permeation | −9.14 cm/s | −8.79 cm/s | −9.14 cm/s | −8.42 cm/s | −11.30 cm/s | −11.30 cm/s | −11.53 cm/s | −11.30 cm/s | −8.79 cm/s | −16.29 cm/s | |
| Metabolism | P-glycoprotein Inhibitor | - | - | - | - | - | - | - | - | - | - |
| P-gp Substrate | - | - | - | - | + | + | - | + | - | + | |
| CYP450 1A2 Inhibitor | - | - | - | - | - | - | - | - | - | - | |
| CYP450 2C9 Inhibitor | - | - | - | - | - | - | - | - | - | - | |
| CYP450 2D6 Inhibitor | - | - | - | - | - | - | - | - | - | - | |
| CYP450 2C19 Inhibitor | - | - | - | - | - | - | - | - | - | - | |
| CYP450 3A4 Inhibitor | - | - | - | - | - | - | - | - | - | - | |
| Toxicity | AMES mutagenesis (probability) | +(0.610) | −(0.550) | +(0.710) | +(0.770) | +(0.510) | +(0.510) | +(0.600) | +(0.510) | +(0.540) | +(0.520) |
| Carcinogens (probability) | −(0.986) | −(0.986) | −(0.986) | −(0.986) | −(0.986) | −(0.986) | −(0.986) | −(0.986) | −(0.986) | −(1) | |
| Hepatotoxicity (probability) | +(0.600) | +(0.600) | +(0.600) | +(0.650) | +(0.675) | +(0.675) | +(0.650) | +(0.675) | +(0.675) | −(0.625) |
| A. oryzae | Human Pancreatic | Human Salivary | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | RR% | Affinity (Kcal/mol) | H-I | NHB | Hydrogen Bonds | RR% | Affinity | H-I | NHB | Hydrogen Bonds | RR% | Affinity | H-I | NHB | Hydrogen Bonds |
| C1: Isoorientin | 100 | −8.2 | 6 | 4 | Glu230 (1.90; 2.85 Å) Asp206 (2.64 Å) Asp233 (1.98 Å) | 58 | −8.2 | 4 | 3 | Arg195 (2.10 Å) Glu233 (2.93 Å) Asp300 (2.44 Å) | 100 | −8.7 | 7 | 4 | Ser163 (2.80; 2.01 Å) Lys200 (2.90 Å) Glu233 (2.79 Å) |
| C2: Isovitexin | 100 | −8.0 | 6 | 5 | Glu230 (2.04; 2.12; 2.92 Å) Asp206 (2.99 Å) Glu156 (2.86 Å) | 92 | −8.3 | 4 | 3 | Tyr62 (2.30 Å) Thr163 (2.21; 2.28 Å) | 54 | −8.6 | 6 | 2 | Ile235 (1.78 Å) His305 (2.62 Å) |
| C3: Orientin | 100 | −8.4 | 3 | 4 | Trp83 (2.47 Å) Arg344 (2.92 Å) Asp340 (2.91; 3.05 Å) | 100 | −8.5 | 5 | 3 | Tyr62 (2.62 Å) Asp197 (2.08 Å) Glu233 (2.16 Å) | 100 | −8.7 | 1 | 5 | Ser163 (2.40 Å) Arg195 (2.67 Å) His305 (2.22 Å) Glu233 (2.43; 2.30 Å) |
| C4: Quercetin 3-rhamnoside | 92 | −8.8 | 8 | 4 | His80 (2.58 Å) Asp340 (2.21 Å) Asp206 (2.46 Å) Glu230 (2.62 Å) | 100 | −8.6 | 6 | 4 | His305 (2.38 Å) Asp300 (2.23; 2.89 Å) Asp197 (2.13 Å) | 100 | −9.8 | 7 | 5 | Arg195 (2.58 Å) His299 (1.81 Å) Gly306 (2.13 Å) Asp197 (1.96 Å) Glu233 (2.93 Å) |
| C5: Schaftoside | 100 | −8.1 | 3 | 6 | Arg204 (1.95; 2.79 Å) His210 (2.12 Å) Asp206 (2.08 Å) Glu230 (2.05 Å) Asp297 (2.06 Å) | 72 | −8.7 | 6 | 5 | Thr163 (2.22 Å) Ile235 (2.42 Å) Trp59 (2.62 Å) His305 (2.51; 2.79 Å) | 100 | −9.9 | 5 | 6 | Ser163 (2.67 Å) Ile235 (2.05 Å) His305 (2.81 Å) Asp300 (2.40; 2.84 Å) Glu233 (2.18 Å) |
| C6: Vicenin1 | 100 | −7.8 | 3 | 1 | His210 (2.21 Å) | 72 | −8.4 | 6 | 5 | His101 (2.76 Å) Arg195 (2.51 Å) Lys200 (1.97 Å) Asp197 (2.12 Å) Glu233 (1.64 Å) | 98 | −8.7 | 2 | 6 | Gln63 (2.44; 2.71 Å) Ser163 (2.03 Å) His305 (1.78 Å) Asp197 (1.97 Å) Glu233 (2.92 Å) |
| C7: vicenin2 | 100 | −7.8 | 3 | 3 | His210 (2.23 Å) Asp206 (2.93 Å) Glu230 (1.95 Å) | 98 | −8.2 | 5 | 8 | Tyr151 (2.87 Å) Thr163 (2.22; 2.98 Å) Asp197 (1.74; 2.24 Å) Glu233 (2.57 Å) Asp300 (2.27; 2.71 Å) | 62 | −8.8 | 2 | 5 | Ser163 (2.29 Å) Arg195 (2.47 Å) His305 (2.51 Å) Gly306 (2.63 Å) Glu233 (2.20 Å) |
| C8: Vicenin3 | 100 | −8.0 | 3 | 2 | His210 (2.19 Å) Asp206 (2.63 Å) | 100 | −8.7 | 6 | 3 | Thr163 (2.23 Å) Ile235 (2.40 Å) Trp59 (2.03 Å) | 92 | −9.0 | 2 | 4 | Gln63 (2.46 Å) Ser163 (1.94 Å) Asp197 (1.98 Å) Glu233 (2.90 Å) |
| C9: Vitexin | 100 | −8.5 | 3 | 6 | Asp340 (1.97; 2.95 Å) Arg344 (1.95; 2.68; 2.89 Å) Asp206 (2.12 Å) | 98 | −8.2 | 5 | 4 | His101 (2.44 Å) Asp300 (1.89 Å) Asp197 (1.97; 2.61 Å) | 100 | −8.8 | 1 | 6 | Gln63 (1.97; 2.96 Å) Ser163 (1.92 Å) His305 (2.70 Å) Gly306 (2.11 Å) Asp197 (1.74 Å) |
| C10: Acarbose | 66 | −7.5 | 1 | 10 | Trp83 (2.19 Å) Asn339 (2.85 Å) Arg344 (2.34; 2.87 Å) Asp297 (2.24; 2.42 Å) Gln35 (2.92 Å) Tyr75 (2.47 Å) Tyr79 (2.92 Å) Asp340 (2.83 Å) | 100 | −8.3 | 3 | 6 | Ala106 (1.95 Å) His201 (2.52; 2.68 Å) Asp197 (2.50 Å) Asn105 (2.44 Å) Gln63 (2.09 Å) | 98 | −8.1 | 2 | 6 | His201 (2.77; 2.05 Å) His305 (2.06 Å) Glu233 (2.65 Å) Asp300 (2.71 Å) Gly306 (2.68 Å) |
| Enzyme | PDB ID | Resolution | Co-Crystalized Ligand | Grid Box Center | Grid Box |
|---|---|---|---|---|---|
| Human salivary α-amylase | 3DHP | 1.50 Å | Hydrolyzed substrate | 22*20*20 | 9.103*46.640*19.324 |
| Human pancreatic α-amylase | 2QV4 | 1.97 Å | Acarbose | 24*24*24 | 10.592*47.985*21.039 |
| Aspergillus oryzae α-amylase | 7TAA | 1.98 Å | Acarbose | 34*28*22 | 37.401*41.469*26.378 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khenifi, M.L.; Serseg, T.; Migas, P.; Krauze-Baranowska, M.; Özdemir, S.; Bensouici, C.; Alghonaim, M.I.; Al-Khafaji, K.; Alsalamah, S.A.; Boudjeniba, M.; et al. HPLC-DAD-MS Characterization, Antioxidant Activity, α-amylase Inhibition, Molecular Docking, and ADMET of Flavonoids from Fenugreek Seeds. Molecules 2023, 28, 7798. https://doi.org/10.3390/molecules28237798
Khenifi ML, Serseg T, Migas P, Krauze-Baranowska M, Özdemir S, Bensouici C, Alghonaim MI, Al-Khafaji K, Alsalamah SA, Boudjeniba M, et al. HPLC-DAD-MS Characterization, Antioxidant Activity, α-amylase Inhibition, Molecular Docking, and ADMET of Flavonoids from Fenugreek Seeds. Molecules. 2023; 28(23):7798. https://doi.org/10.3390/molecules28237798
Chicago/Turabian StyleKhenifi, Mohammed Lamine, Talia Serseg, Piotr Migas, Mirosława Krauze-Baranowska, Sadin Özdemir, Chawki Bensouici, Mohammed I. Alghonaim, Khattab Al-Khafaji, Sulaiman A. Alsalamah, Messaoud Boudjeniba, and et al. 2023. "HPLC-DAD-MS Characterization, Antioxidant Activity, α-amylase Inhibition, Molecular Docking, and ADMET of Flavonoids from Fenugreek Seeds" Molecules 28, no. 23: 7798. https://doi.org/10.3390/molecules28237798
APA StyleKhenifi, M. L., Serseg, T., Migas, P., Krauze-Baranowska, M., Özdemir, S., Bensouici, C., Alghonaim, M. I., Al-Khafaji, K., Alsalamah, S. A., Boudjeniba, M., Yousfi, M., Boufahja, F., Bendif, H., & Mahdid, M. (2023). HPLC-DAD-MS Characterization, Antioxidant Activity, α-amylase Inhibition, Molecular Docking, and ADMET of Flavonoids from Fenugreek Seeds. Molecules, 28(23), 7798. https://doi.org/10.3390/molecules28237798









