A Fluorescence Resonance Energy Transfer Aptasensor for Aflatoxin B1 Based on Ligand-Induced ssDNA Displacement
Abstract
:1. Introduction
2. Results and Discussion
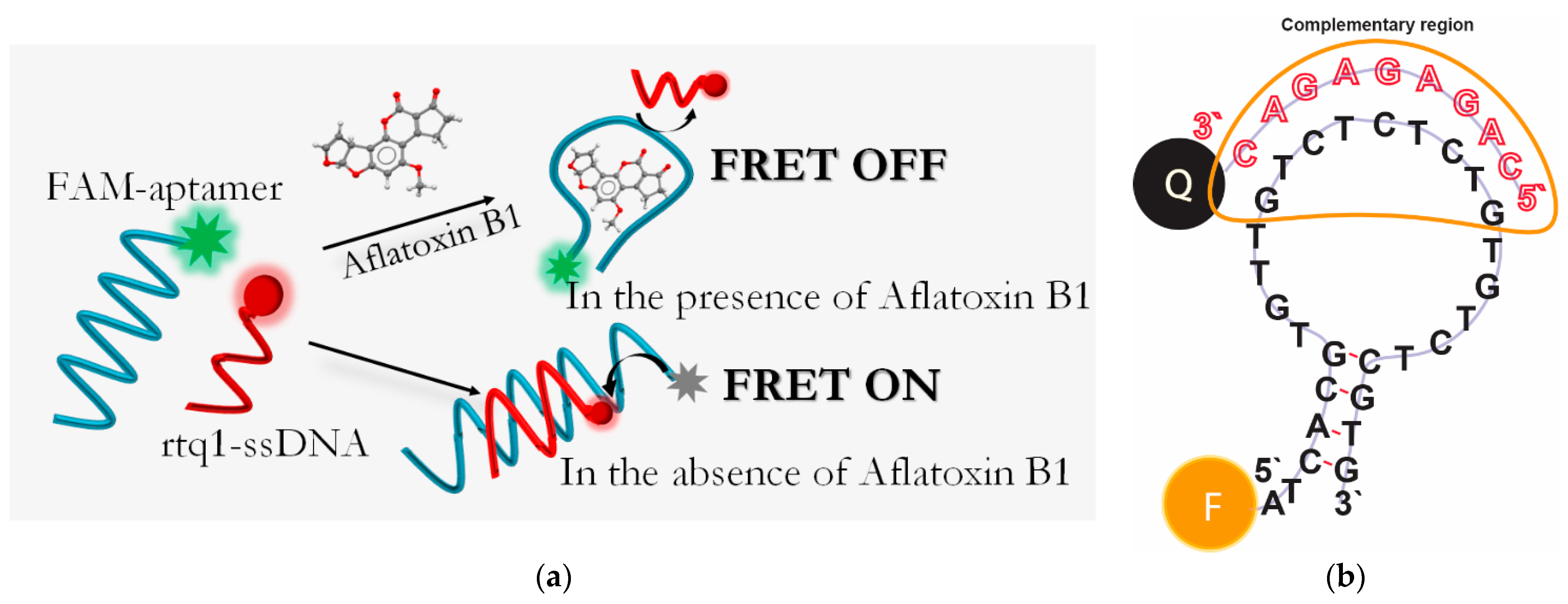
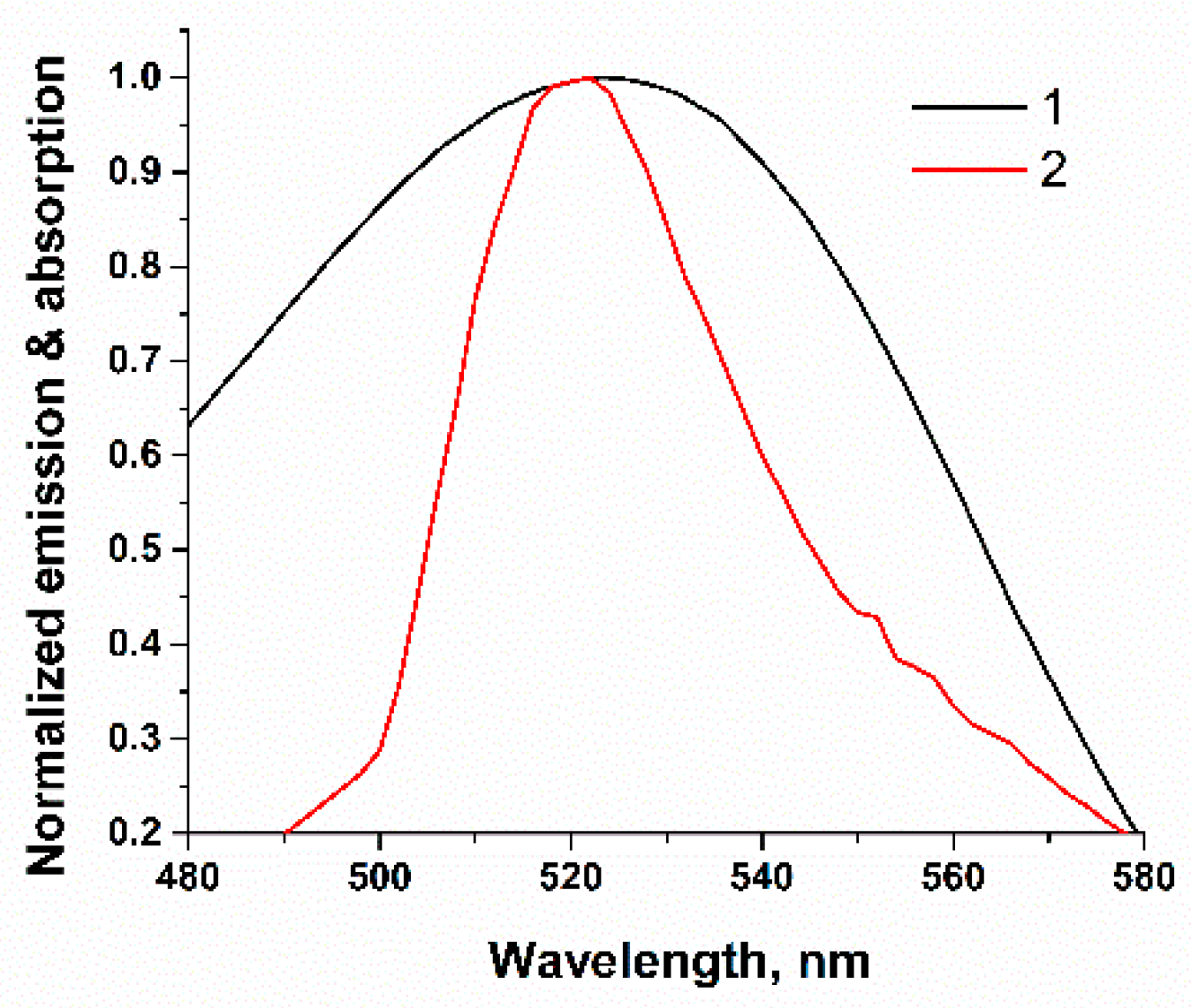
2.1. Principle of AFB1 Detection
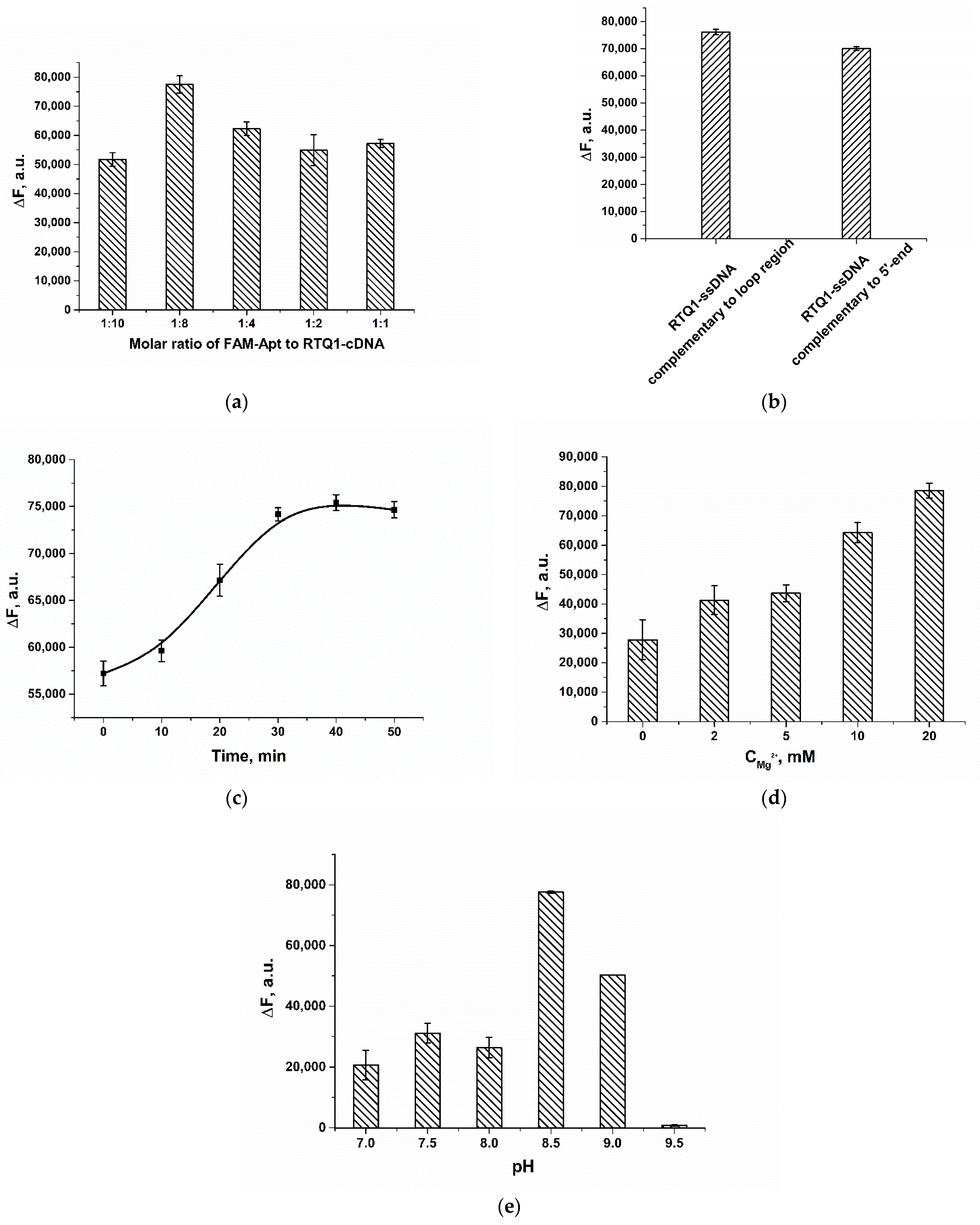
2.2. Optimization of Assay Conditions
2.3. Aflatoxin B1 Detection Performance
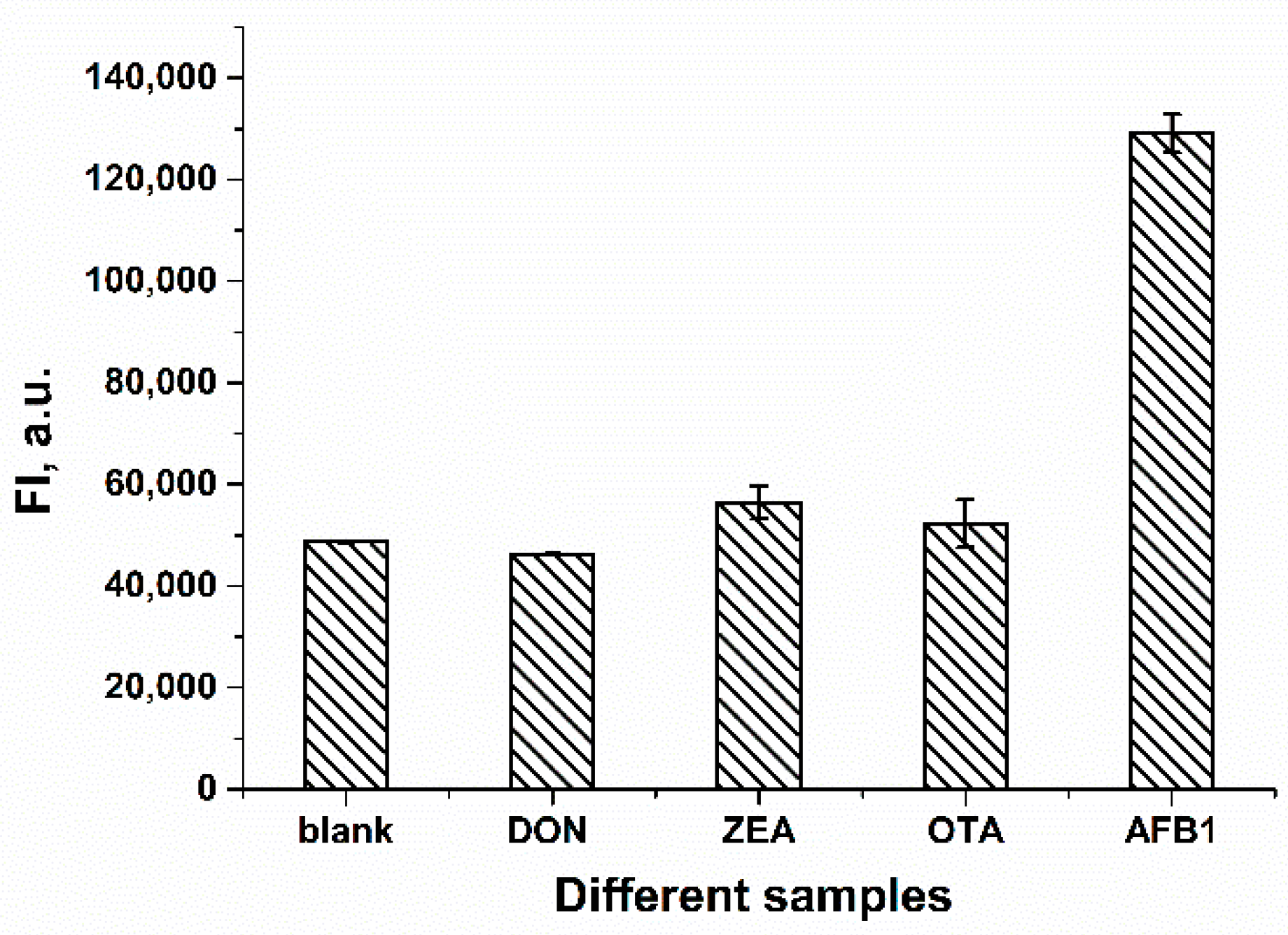
2.4. Selectivity of the FRET-Based Aptasensor in AFB1 Detection
2.5. Detection of AFB1 in Food Samples
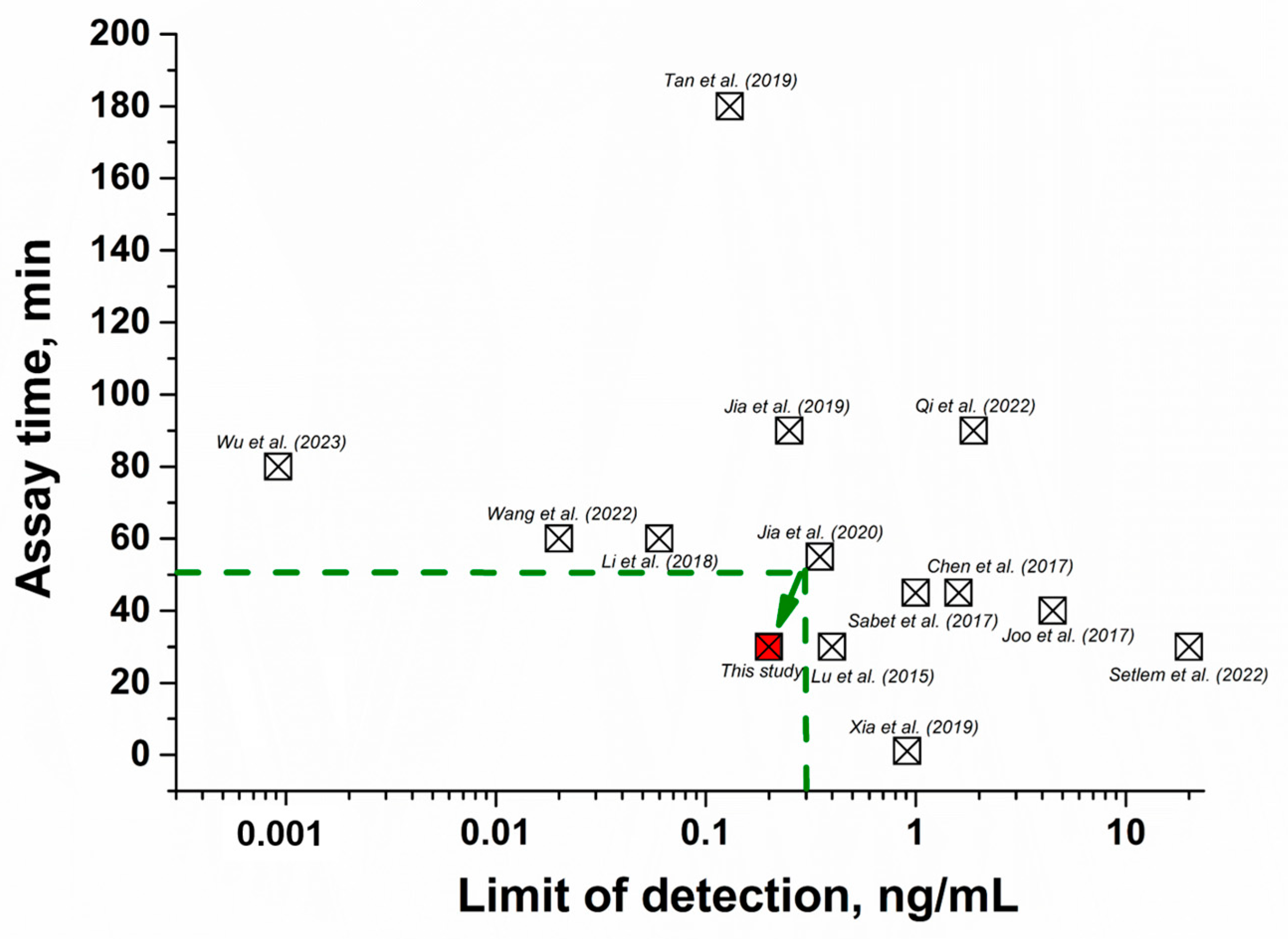
2.6. Comparison of the Developed FRET Aptasensor with Other Methods
3. Conclusions
4. Materials and Methods
4.1. Reagents and Materials
4.2. Apparatus
4.3. Optimization of Assay Conditions
4.4. Aflatoxin B1 Detection with FRET-Based Aptasensor
4.5. Selectivity Testing
4.6. Sample Preparation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sweeney, M.J.; Dobson, A.D.W. Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int. J. Food Microbiol. 1998, 43, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Okeke, C.A.; Ezekiel, C.N.; Sulyok, M.; Ogunremi, O.R.; Ezeamagu, C.O.; Šarkanj, B.; Warth, B.; Krska, R. Traditional processing impacts mycotoxin levels and nutritional value of ogi—A maize-based complementary food. Food Control 2018, 86, 224–233. [Google Scholar] [CrossRef]
- Mokhtarian, M.; Tavakolipour, H.; Bagheri, F.; Fernandes Oliveira, C.A.; Corassin, C.H.; Khaneghah, A.M. Aflatoxin B1 in the Iranian pistachio nut and decontamination methods: A systematic review. Qual. Assur. Saf. Crops Foods 2020, 12, 15–25. [Google Scholar] [CrossRef]
- Commission Regulation (EU). Commission Regulation (EU) 2023/915 of 25 April 2023 on Maximum Levels for Certain Contaminants in Food and Repealing Regulation (EC) No 1881/2006 (Text with EEA Relevance). Off. J. Eur. Union 2023. Available online: https://eur-lex.europa.eu/eli/reg/2023/915/oj (accessed on 25 October 2023).
- Wang, Y.; Zhao, G.; Li, X.; Liu, L.; Cao, W.; Wei, Q. Electrochemiluminescent competitive immunosensor based on polyethyleneimine capped SiO2 nanomaterials as labels to release Ru(bpy)32+ fixed in 3D Cu/Ni oxalate for the detection of aflatoxin B1. Biosens. Bioelectron. 2018, 101, 290–296. [Google Scholar] [CrossRef]
- Hamad, G.M.; Mehany, T.; Simal-Gandara, J.; Abou-Alella, S.; Esua, O.J.; Abdel-Wahhab, M.A.; Hafez, E.E. A review of recent innovative strategies for controlling mycotoxins in foods. Food Control 2023, 144, 109350. [Google Scholar] [CrossRef]
- Eaton, D.L.; Gallagher, E.P. Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 135–172. [Google Scholar] [CrossRef]
- Henry, S.H.; Bosch, F.X.; Troxell, T.C.; Bolger, P.M. Policy forum: Public health. Reducing liver cancer—Global control of aflatoxin. Science 1999, 286, 2453–2454. [Google Scholar] [CrossRef]
- Warth, B.; Petchkongkaew, A.; Sulyok, M.; Krska, R. Utilising an LC-MS/MS-based multi-biomarker approach to assess mycotoxin exposure in the Bangkok metropolitan area and surrounding provinces. Food Addit. Contam. Part A 2014, 31, 2040–2046. [Google Scholar] [CrossRef]
- Afsah-Hejri, L.; Rajaram, P.; O’Leary, J.; McGivern, J.; Baxter, R.; Mesbah, A.; Maboudian, R.; Ehsani, R. Identification of volatile organic compounds (VOCs) by SPME-GC-MS to detect Aspergillus flavus infection in pistachios. Food Control 2023, 154, 110033. [Google Scholar] [CrossRef]
- Xiong, Y.; Pei, K.; Wu, Y.; Duan, H.; Lai, W.; Xiong, Y. Plasmonic ELISA based on enzyme-assisted etching of Au nanorods for the highly sensitive detection of aflatoxin B1 in corn samples. Sens. Actuators B Chem. 2018, 267, 320–327. [Google Scholar] [CrossRef]
- Shao, Y.; Duan, H.; Guo, L.; Leng, Y.; Lai, W.; Xiong, Y. Quantum dot nanobead-based multiplexed immunochromatographic assay for simultaneous detection of aflatoxin B1 and zearalenone. Anal. Chim. Acta 2018, 1025, 163–171. [Google Scholar] [CrossRef]
- Duo, Y.; Xiang, Z.; Gao, G.; Luo, G.; Tang, B.Z. Biomedical application of aggregation-induced emission luminogen-based fluorescent sensors. TrAC Trends Anal. Chem. 2023, 167, 117252. [Google Scholar] [CrossRef]
- Wang, J.-X.; Yin, J.; Shekhah, O.; Bakr, O.M.; Eddaoudi, M.; Mohammed, O.F. Energy Transfer in Metal–Organic Frameworks for Fluorescence Sensing. ACS Appl. Mater. Interfaces 2022, 14, 9970–9986. [Google Scholar] [CrossRef] [PubMed]
- Amirjani, A.; Tsoulos, T.V.; Sajjadi, S.H.; Antonucci, A.; Wu, S.-J.; Tagliabue, G.; Haghshenas, D.F.; Boghossian, A.A. Plasmon-induced near-infrared fluorescence enhancement of single-walled carbon nanotubes. Carbon 2022, 194, 162–175. [Google Scholar] [CrossRef]
- Nazarzadeh Zare, E.; Khorsandi, D.; Zarepour, A.; Yilmaz, H.; Agarwal, T.; Hooshmand, S.; Mohammadinejad, R.; Ozdemir, F.; Sahin, O.; Adiguzel, S.; et al. Biomedical applications of engineered heparin-based materials. Bioact. Mater. 2024, 31, 87–118. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Lu, Q.; Duan, N.; Wang, Z. GO-amplified fluorescence polarization assay for high-sensitivity detection of aflatoxin B1 with low dosage aptamer probe. Anal. Bioanal. Chem. 2019, 411, 1107–1115. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Q. Aptamer structure switch fluorescence anisotropy assay for aflatoxin B1 using tetramethylrhodamine-guanine interaction to enhance signal change. Chin. Chem. Lett. 2020, 31, 1982–1985. [Google Scholar] [CrossRef]
- Li, Y.; Yu, H.; Zhao, Q. Aptamer fluorescence anisotropy assays for detection of aflatoxin B1 and adenosine triphosphate using antibody to amplify signal change. RSC Adv. 2022, 12, 7464–7468. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Huang, C.; Emery, B.P.; Sedgwick, A.C.; Bull, S.D.; He, X.-P.; Tian, H.; Yoon, J.; Sessler, J.L.; James, T.D. Förster resonance energy transfer (FRET)-based small-molecule sensors and imaging agents. Chem. Soc. Rev. 2020, 49, 5110–5139. [Google Scholar] [CrossRef]
- Cunningham, P.D.; Khachatrian, A.; Buckhout-White, S.; Deschamps, J.R.; Goldman, E.R.; Medintz, I.L.; Melinger, J.S. Resonance energy transfer in DNA duplexes labeled with localized dyes. J. Phys. Chem. B 2014, 118, 14555–14565. [Google Scholar] [CrossRef] [PubMed]
- Shaban, S.M.; Kim, D.H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. Engl. 2021, 60, 16800–16823. [Google Scholar] [CrossRef] [PubMed]
- Monserud, J.H.; Macri, K.M.; Schwartz, D.K. Toehold-Mediated Displacement of an Adenosine-Binding Aptamer from a DNA Duplex by its Ligand. Angew. Chem. Int. Ed. 2016, 55, 13710–13713. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, W.; Qian, J.; Wang, L.; Ren, Y.; Wang, Y.; Xu, M.; Huang, X. A FRET aptasensor for sensitive detection of aflatoxin B1 based on a novel donor–acceptor pair between ZnS quantum dots and Ag nanocubes. Anal. Methods 2021, 13, 462–468. [Google Scholar] [CrossRef]
- Aswani Kumar, Y.V.V.; Renuka, R.M.; Achuth, J.; Mudili, V.; Poda, S. Development of a FRET-based fluorescence aptasensor for the detection of aflatoxin B1 in contaminated food grain samples. RSC Adv. 2018, 8, 10465–10473. [Google Scholar] [CrossRef]
- Le, L.; Cruz-Aguado, J.A.; Penner, G.A. DNA Ligands for Aflatoxin and Zearalenone; Neoventures Biotechnology Inc.: London, ON, Canada, 2011. [Google Scholar]
- Sun, L.; Zhao, Q. Direct fluorescence anisotropy approach for aflatoxin B1 detection and affinity binding study by using single tetramethylrhodamine labeled aptamer. Talanta 2018, 189, 442–450. [Google Scholar] [CrossRef]
- Nasirian, V.; Chabok, A.; Barati, A.; Rafienia, M.; Arabi, M.S.; Shamsipur, M. Ultrasensitive aflatoxin B1 assay based on FRET from aptamer labelled fluorescent polymer dots to silver nanoparticles labeled with complementary DNA. Microchim. Acta 2017, 184, 4655–4662. [Google Scholar] [CrossRef]
- Lu, X.; Wang, C.; Qian, J.; Ren, C.; An, K.; Wang, K. Target-driven switch-on fluorescence aptasensor for trace aflatoxin B1 determination based on highly fluorescent ternary CdZnTe quantum dots. Anal. Chim. Acta 2019, 1047, 163–171. [Google Scholar] [CrossRef]
- Li, Y.; Sun, L.; Zhao, Q. Development of aptamer fluorescent switch assay for aflatoxin B1 by using fluorescein-labeled aptamer and black hole quencher 1-labeled complementary DNA. Anal. Bioanal. Chem. 2018, 410, 6269–6277. [Google Scholar] [CrossRef]
- Wang, L.; Yao, L.; Ma, Q.; Mao, Y.; Qu, H.; Zheng, L. Investigation on small molecule-aptamer dissociation equilibria based on antisense displacement probe. Food Sci. Hum. Wellness 2023, 12, 1257–1264. [Google Scholar] [CrossRef]
- Feng, B.; You, J.; Zhao, F.; Wei, M.; Liu, Y.; Yuan, K.; Suo, Z. A ratiometric fluorescent aptamer homogeneous biosensor based on hairpin structure aptamer for AFB1 detection. J. Fluoresc. 2022, 32, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, H.; Zhao, Q. A Simple Structure-Switch Aptasensor Using Label-Free Aptamer for Fluorescence Detection of Aflatoxin B1. Molecules 2022, 27, 4257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liang, X.; Liu, Y.; Wei, M.; Jin, H. A Novel Fluorescent Aptasensor Based on Dual-labeled DNA Nanostructure for Simultaneous Detection of Ochratoxin A and Aflatoxin B1. J. Fluoresc. 2023, 33, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, K.; Yu, J.; Shi, P. Complementary DNA Significantly Enhancing Signal Response and Sensitivity of a Molecular Beacon Probe to Aflatoxin B1. Biosensors 2023, 13, 195. [Google Scholar] [CrossRef]
- Murphy, M.C.; Rasnik, I.; Cheng, W.; Lohman, T.M.; Ha, T. Probing Single-Stranded DNA Conformational Flexibility Using Fluorescence Spectroscopy. Biophys. J. 2004, 86, 2530–2537. [Google Scholar] [CrossRef]
- Samokhvalov, A.V.; Safenkova, I.V.; Eremin, S.A.; Bonchuk, A.N.; Maksimenko, O.G.; Sluchanko, N.N.; Zherdev, A.V.; Dzantiev, B.B. Modulation of Aptamer–Ligand-Binding by Complementary Oligonucleotides: A G-Quadruplex Anti-Ochratoxin A Aptamer Case Study. Int. J. Mol. Sci. 2022, 23, 4876. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Zhao, Q. A signal-on electrochemical aptasensor for rapid detection of aflatoxin B1 based on competition with complementary DNA. Biosens. Bioelectron. 2019, 144, 111641. [Google Scholar] [CrossRef]
- Jia, Y.; Wu, F.; Liu, P.; Zhou, G.; Yu, B.; Lou, X.; Xia, F. A label-free fluorescent aptasensor for the detection of Aflatoxin B1 in food samples using AIEgens and graphene oxide. Talanta 2019, 198, 71–77. [Google Scholar] [CrossRef]
- Tan, H.; Ma, L.; Guo, T.; Zhou, H.; Chen, L.; Zhang, Y.; Dai, H.; Yu, Y. A novel fluorescence aptasensor based on mesoporous silica nanoparticles for selective and sensitive detection of aflatoxin B1. Anal. Chim. Acta 2019, 1068, 87–95. [Google Scholar] [CrossRef]
- Jia, Y.; Zhou, G.; Wang, X.; Zhang, Y.; Li, Z.; Liu, P.; Yu, B.; Zhang, J. A metal-organic framework/aptamer system as a fluorescent biosensor for determination of aflatoxin B1 in food samples. Talanta 2020, 219, 121342. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sun, D.-W.; Pu, H.; Wei, Q. A novel fluorescence biosensor based on CRISPR/Cas12a integrated MXenes for detecting Aflatoxin B1. Talanta 2023, 252, 123773. [Google Scholar] [CrossRef]
- Qi, X.; Lv, L.; Wei, D.; Lee, J.J.; Niu, M.; Cui, C.; Guo, Z.J.A.; Chemistry, B. Detection of aflatoxin B1 with a new label-free fluorescence aptasensor based on PVP-coated single-walled carbon nanohorns and SYBR Gold. Anal. Bioanal. Chem. 2022, 414, 3087–3094. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.; Baek, S.H.; Cheon, S.A.; Chun, H.S.; Choi, S.-W.; Park, T.J. Development of aflatoxin B1 aptasensor based on wide-range fluorescence detection using graphene oxide quencher. Colloids Surf. B Biointerfaces 2017, 154, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wen, F.; Li, M.; Guo, X.; Li, S.; Zheng, N.; Wang, J. A simple aptamer-based fluorescent assay for the detection of Aflatoxin B1 in infant rice cereal. Food Chem. 2017, 215, 377–382. [Google Scholar] [CrossRef]
- Lu, Z.; Chen, X.; Wang, Y.; Zheng, X.; Li, C.M. Aptamer based fluorescence recovery assay for aflatoxin B1 using a quencher system composed of quantum dots and graphene oxide. Microchim. Acta 2015, 182, 571–578. [Google Scholar] [CrossRef]
- Sabet, F.S.; Hosseini, M.; Khabbaz, H.; Dadmehr, M.; Ganjali, M.R. FRET-based aptamer biosensor for selective and sensitive detection of aflatoxin B1 in peanut and rice. Food Chem. 2017, 220, 527–532. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Yang, H.; Dong, Y.; Zhang, K.; Lu, Y.; Deng, R.; He, Q. Enzyme-free amplified and ultrafast detection of aflatoxin B1 using dual-terminal proximity aptamer probes. Food Chem. 2019, 283, 32–38. [Google Scholar] [CrossRef]
- Setlem, S.K.; Mondal, B.; Ramlal, S. A fluorescent aptasensor for the detection of Aflatoxin B1 by graphene oxide mediated quenching and release of fluorescence. J. Microbiol. Methods 2022, 193, 106414. [Google Scholar] [CrossRef]
- Terekhov, S.S.; Smirnov, I.V.; Stepanova, A.V.; Bobik, T.V.; Mokrushina, Y.A.; Ponomarenko, N.A.; Belogurov, A.A., Jr.; Rubtsova, M.P.; Kartseva, O.V.; Gomzikova, M.O.; et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc. Natl. Acad. Sci. USA 2017, 114, 2550–2555. [Google Scholar] [CrossRef]
- Masgutov, R.; Masgutova, G.; Mukhametova, L.; Garanina, E.; Arkhipova, S.S.; Zakirova, E.; Mukhamedshina, Y.O.; Margarita, Z.; Gilazieva, Z.; Syromiatnikova, V.; et al. Allogenic Adipose Derived Stem Cells Transplantation Improved Sciatic Nerve Regeneration in Rats: Autologous Nerve Graft Model. Front. Pharmacol. 2018, 9, 86. [Google Scholar] [CrossRef]
- Serebrennikova, K.V.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. A fluorescence immunosensor for ochratoxin A based on resonance energy transfer between fluorescein derivative and gold nanoparticles. J. Food Compos. Anal. 2022, 114, 104806. [Google Scholar] [CrossRef]
- Nan, M.; Bi, Y.; Xue, H.; Xue, S.; Long, H.; Pu, L.; Fu, G. Rapid Determination of Ochratoxin A in Grape and Its Commodities Based on a Label-Free Impedimetric Aptasensor Constructed by Layer-by-Layer Self-Assembly. Toxins 2019, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Bartosh, A.V.; Urusov, A.E.; Petrakova, A.V.; Kuang, H.; Zherdev, A.V.; Dzantiev, B.B. Highly sensitive lateral flow test with indirect labelling for zearalenone in baby food. Food Agric. Immunol. 2020, 31, 653–666. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, J.; Zhang, Z.; Zhang, Q.; Zhang, W.; Yu, L.; Jiang, J.; Chen, X.; Wang, X.; Li, P. Simultaneous Lateral Flow Immunoassay for Multi-Class Chemical Contaminants in Maize and Peanut with One-Stop Sample Preparation. Toxins 2019, 11, 56. [Google Scholar] [CrossRef]

| Samples | Added *, ng/g | Found, ng/g | Recovery, % |
|---|---|---|---|
| Wine | 3 | 2.3 ± 0.5 | 76.7 |
| 8 | 6.4 ± 1.5 | 80.0 | |
| 26 | 23.9 ± 4.6 | 91.9 | |
| Corn | 7.5 | 6.3 ± 2.5 | 84.0 |
| 20 | 17.3 ± 2.5 | 86.5 | |
| 65 | 55.3 ± 1.8 | 85.1 |
| Sensor Components | Linear Range | Limit of Detection | Assay Time | Sample/ Dilution | Minimum Detectable Concentration | Reference |
|---|---|---|---|---|---|---|
| FAM-Apt/RTQ1-cDNA | 2.5–208.3 ng/mL | 0.2 ng/mL | 30 min | Wine and corn flour/10 and 25 times | 7.5 ng/g for corn and 3 ng/g for wine | This study |
| A fluorescein amidite (FAM)-labeled AFB1-specific aptamer/graphene oxide | 4.5–300 ng/mL | 4.5 ng/mL | 40 min | Rice seeds/no dilution | 4.5 ng/mL | [45] |
| AFB1 aptamer modified with quaternized tetraphenylethene salt/graphene oxide | 0–3 ng/mL | 0.25 ng/mL | 90 min | milk, corn and rice/3 times | 1.92 ng/mL | [40] |
| Aptamers-modified mesoporous silica nanoparticles loaded with Rh6G | 0.5–50 ng/mL | 0.13 ng/mL | 3 h | Corn oil, corn/not mentioned | 1 ng/g | [41] |
| FAM-labeled AFB1 aptamer/cDNA modified with carboxytetramethylrhodamine | 5–100 ng/mL | 1.6 ng/mL | 45 min | Infant rice cereal samples/5 times | 5 ng/mL | [46] |
| Label-free aptamer/fluorescein-labeled complementary strand/quencher (BHQ1)-labeled complementary strand | 0.02–1000 ng/mL | 0.02 ng/mL | 60 min | Beer and corn flour/20 times | Not mentioned | [34] |
| TAMRA-labeled aptamer/metal-organic frameworks UiO-66-NH2 | 0–180 ng/mL | 0.35 ng/mL | 55 min | milk, corn and rice powder/3 times | 1.52 ng/mL | [42] |
| Aptamer modified with CdTe quantum dots/graphene oxide | 0.5 ng/mL to 50 µg/mL | 0.4 ng/mL | 30 min | Peanut oil/no dilution | 0.5 ng/mL | [47] |
| Aptamer-conjugated quantum dots adsorbed to Au nanoparticles | 3–125 ng/mL | 1 ng/mL | 45 min | Peanut and rice/not mentioned | ≈1.6 ng/mL | [48] |
| Two FAM-labeled aptamer/two black hole quencher-labeled anti-aptamer | 1–200 ng/mL | 0.91 ng/mL | 1 min | Peanut oil and broad bean paste/no dilution | 50 ng/mL | [49] |
| FAM-labeled aptamer/BHQ1-labeled cDNA | 0.06–156 ng/mL | 0.06 ng/mL | 1 h | Wine and maize flour/100 times | 0.2 ng/g for wine and 0.4 ng/g for maize | [31] |
| Alexa Fluor 488 labeled aptamer/graphene oxide sheets | 0.2–200 ng/mL | 20 ng/mL | 30 min | Groundnut/no dilution | 20 ng/mL | [50] |
| dual-AFB1 aptamers/Cas12a-crRNA/ssDNA-FAM/MXenes-Ti3C2Tx | 0.001–80 ng/mL | 0.92 pg/mL | 80 min | Peanut/no dilution | 1 ng/mL | [43] |
| SYBR Gold/aptamer/single-walled carbon nanohorns | 5–200 ng/mL | 1.89 ng/mL | 1.5 h | Soybean sauce/100 times | 10 ng/mL | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serebrennikova, K.V.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. A Fluorescence Resonance Energy Transfer Aptasensor for Aflatoxin B1 Based on Ligand-Induced ssDNA Displacement. Molecules 2023, 28, 7889. https://doi.org/10.3390/molecules28237889
Serebrennikova KV, Samokhvalov AV, Zherdev AV, Dzantiev BB. A Fluorescence Resonance Energy Transfer Aptasensor for Aflatoxin B1 Based on Ligand-Induced ssDNA Displacement. Molecules. 2023; 28(23):7889. https://doi.org/10.3390/molecules28237889
Chicago/Turabian StyleSerebrennikova, Kseniya V., Alexey V. Samokhvalov, Anatoly V. Zherdev, and Boris B. Dzantiev. 2023. "A Fluorescence Resonance Energy Transfer Aptasensor for Aflatoxin B1 Based on Ligand-Induced ssDNA Displacement" Molecules 28, no. 23: 7889. https://doi.org/10.3390/molecules28237889







