The Integration of the Metabolome and Transcriptome for Dendrobium nobile Lindl. in Response to Methyl Jasmonate
Abstract
:1. Introduction
2. Results
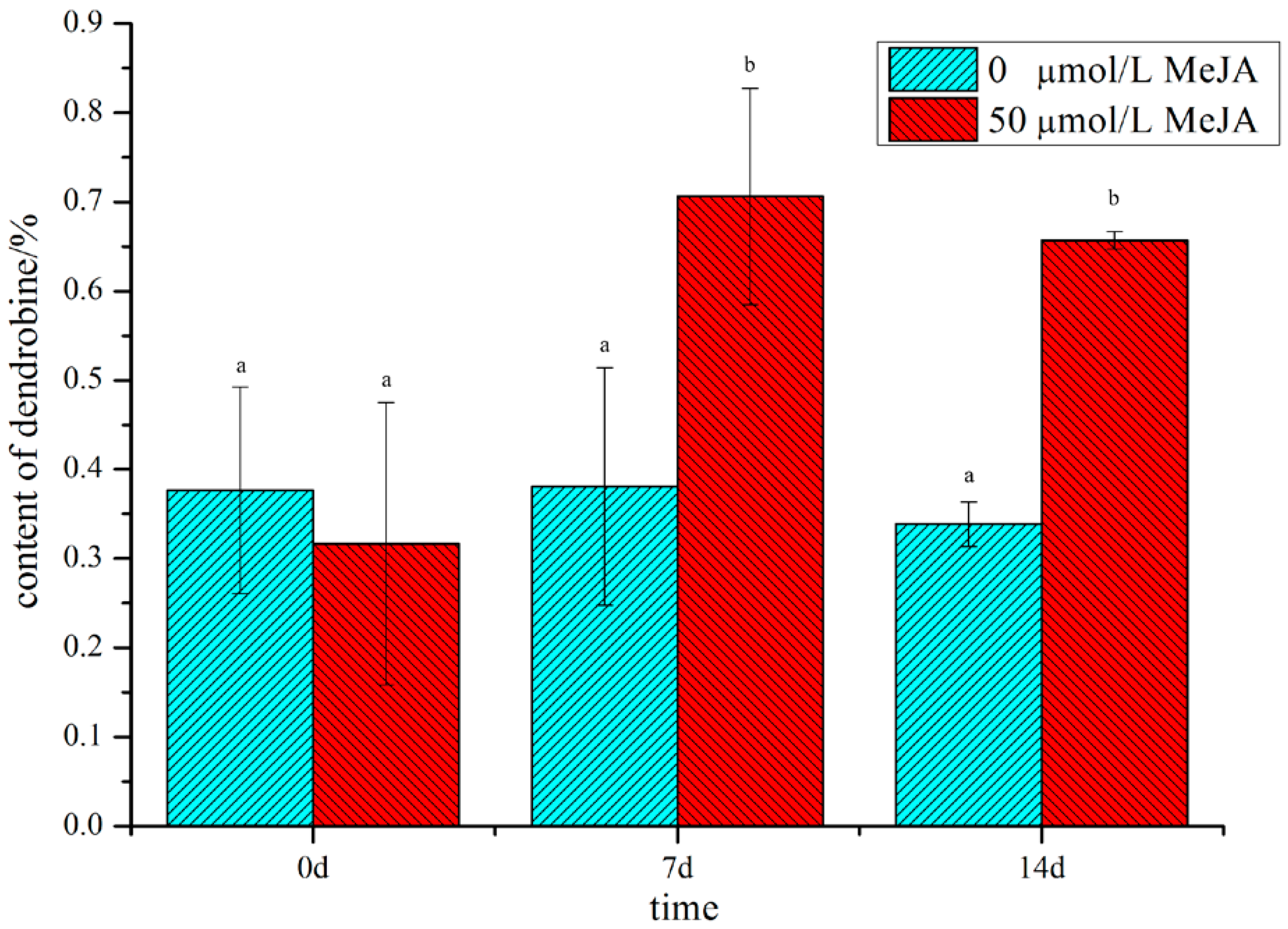
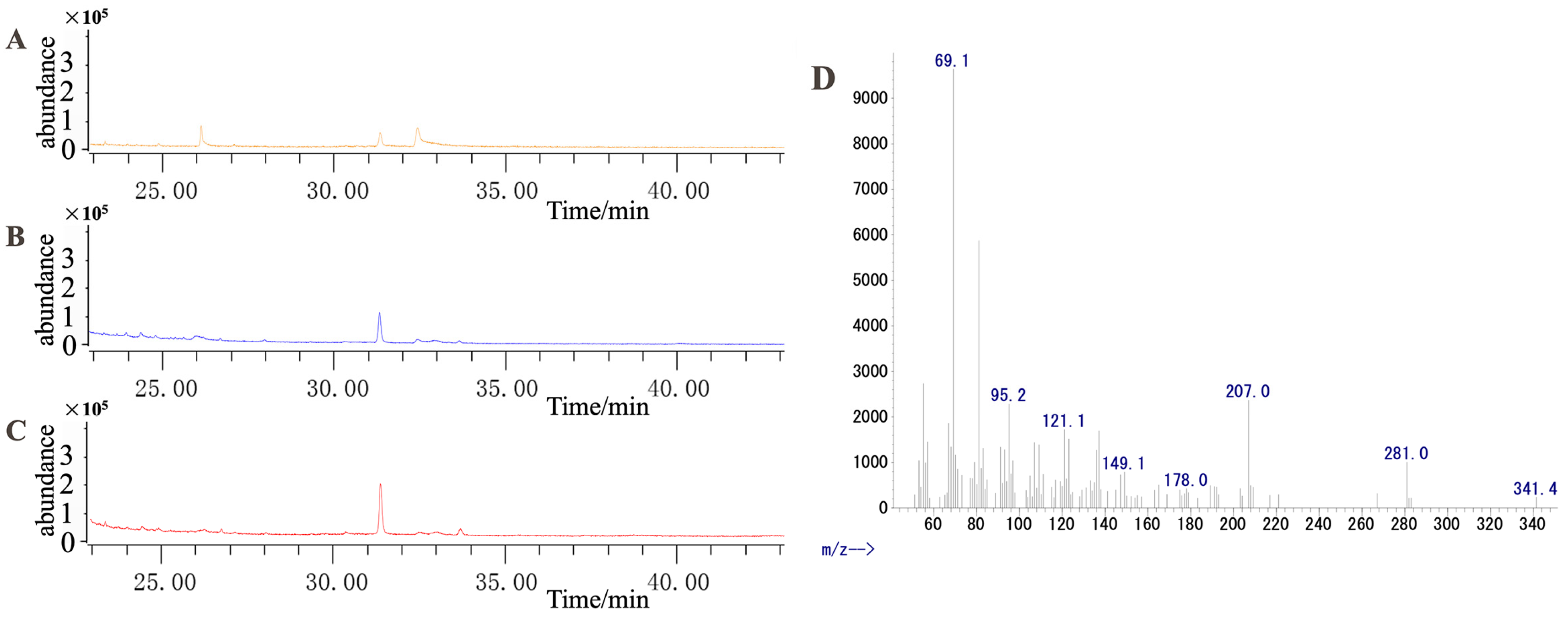
2.1. Analysis of the Dendrobine Content Following MeJA Treatment
2.2. Metabonomic Profiling
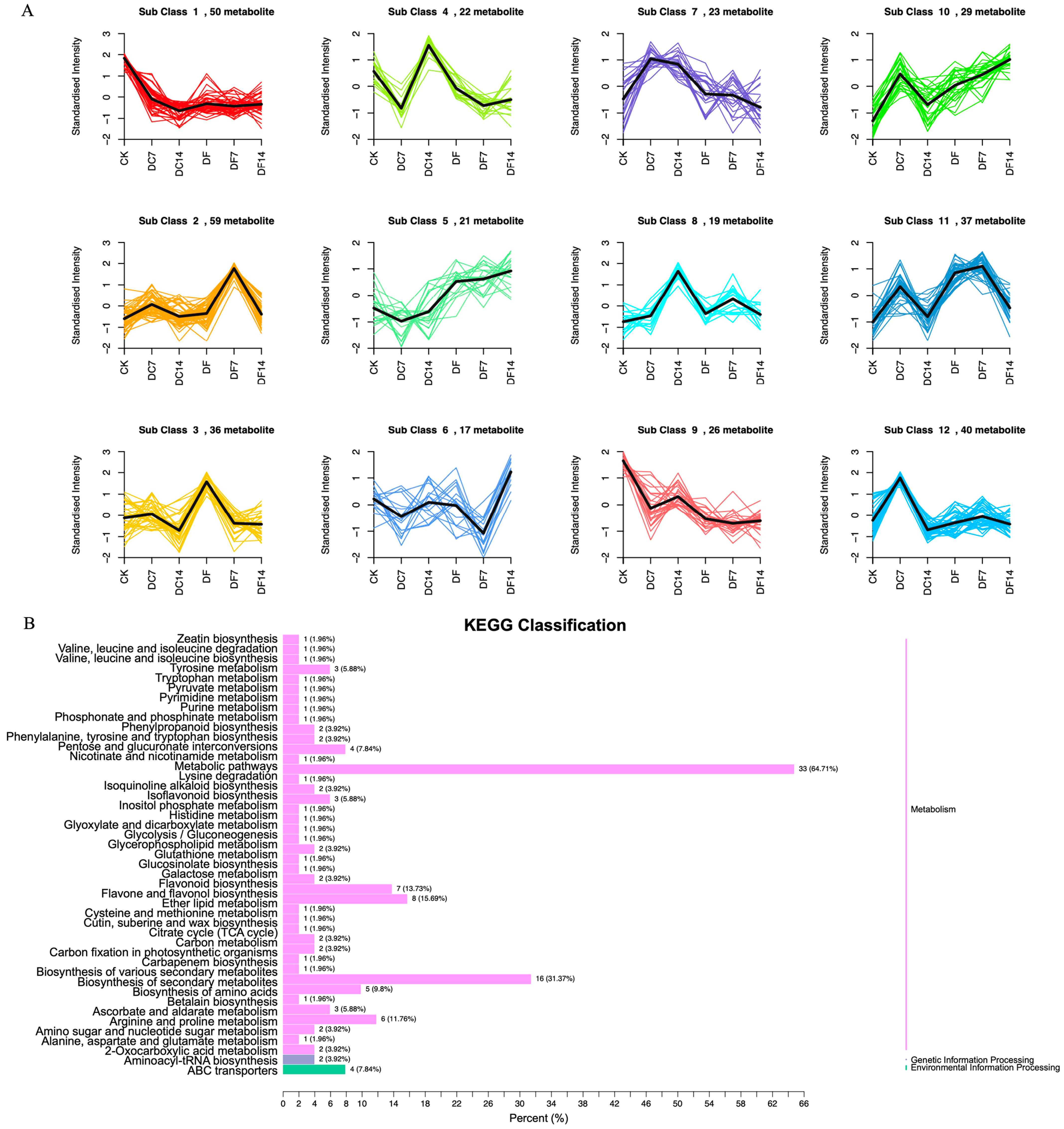
2.3. Identification of Differentially Accumulated Metabolites (DAMs)
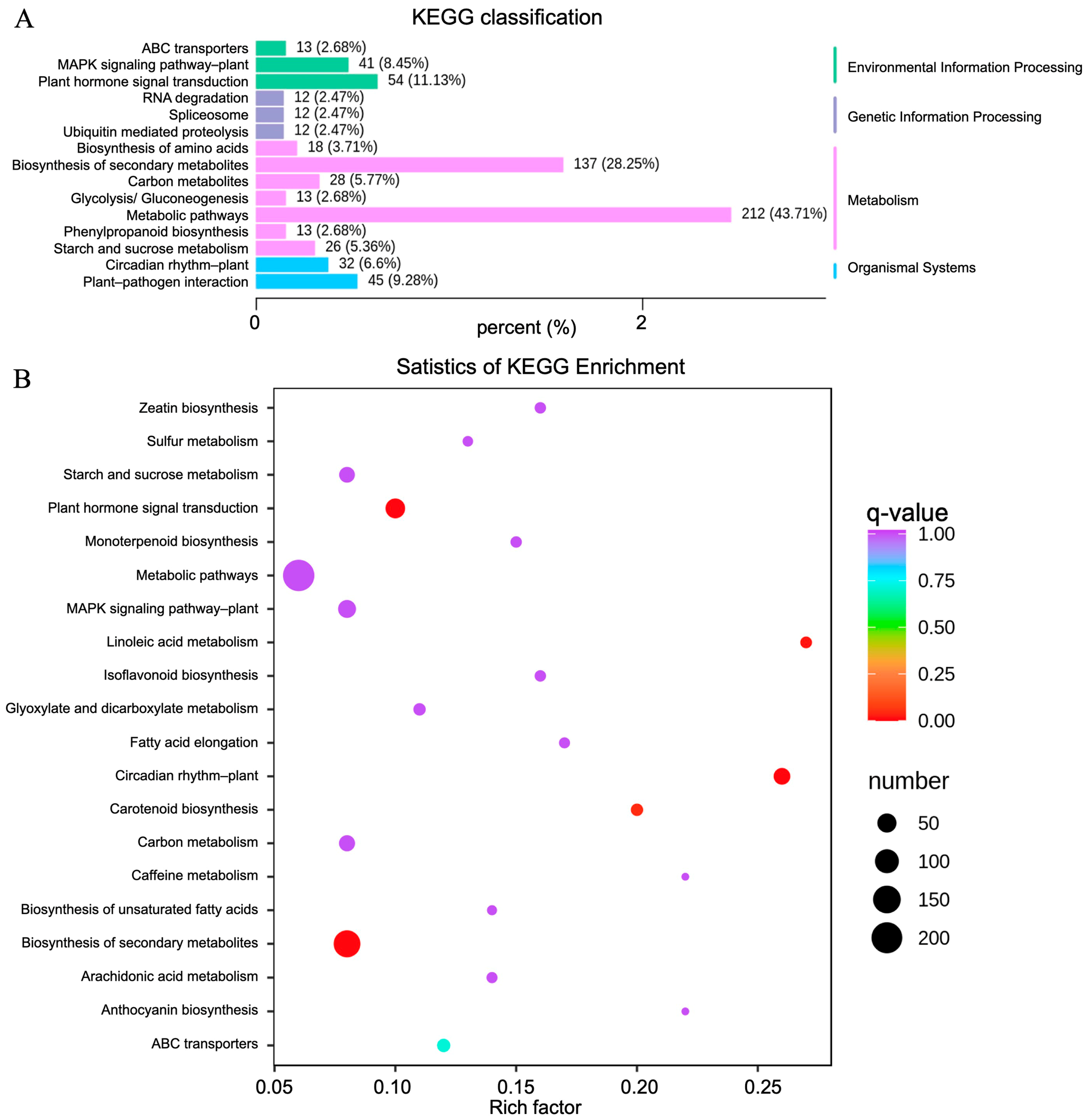
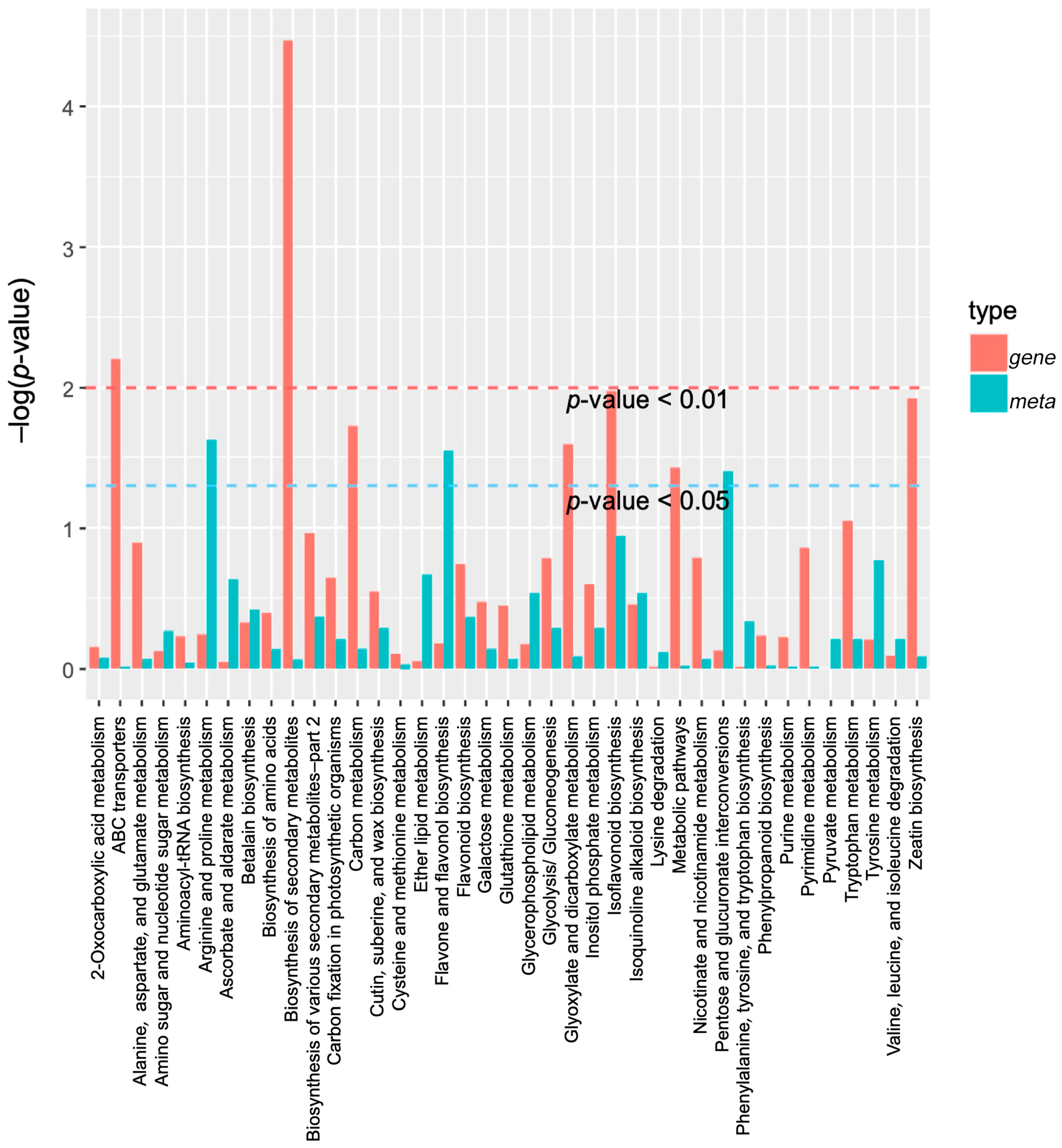
2.4. DAMs Identification and Enrichment Analyses
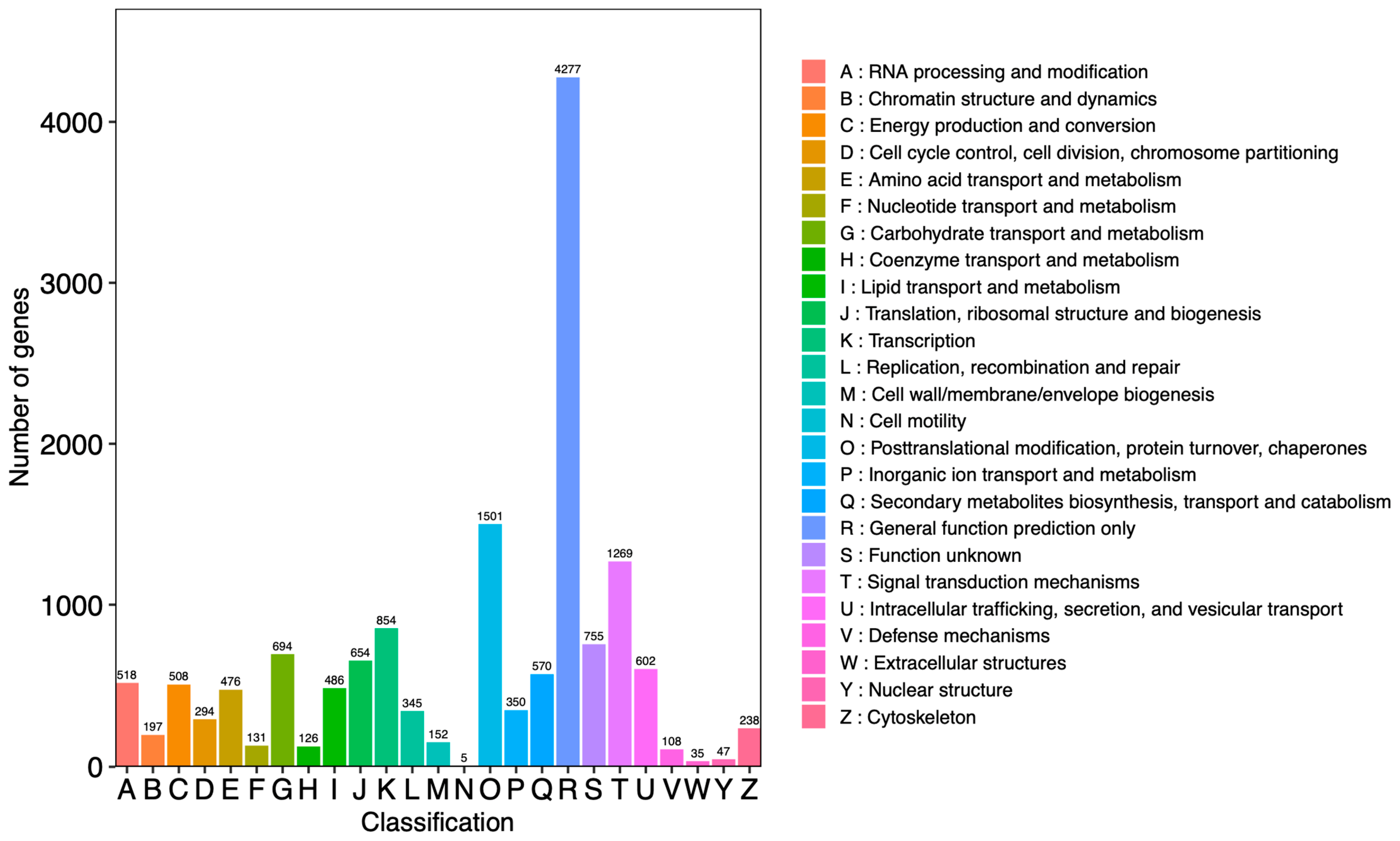
2.5. RNA Sequencing, Assembly, and Functional Annotation
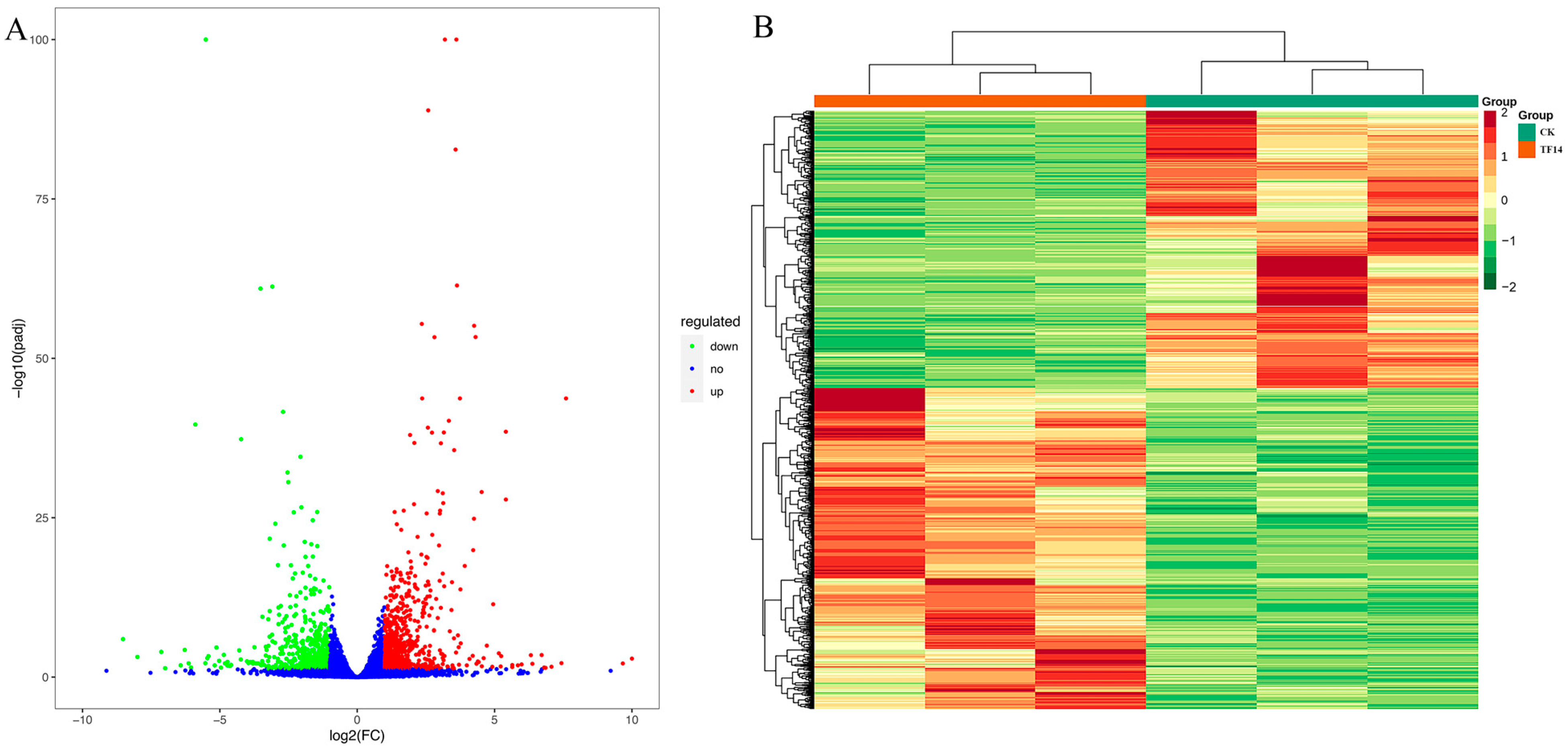
2.6. Identification and Functional Enrichment Analyses of DEGs
2.7. Identification of Unigenes Related to the Biosynthetic Pathways of Terpenoids and Polysaccharides
2.8. qRT-PCR Validation of Differentially Expressed Genes
2.9. Correlation Analysis of the Metabolome and Transcriptome Data
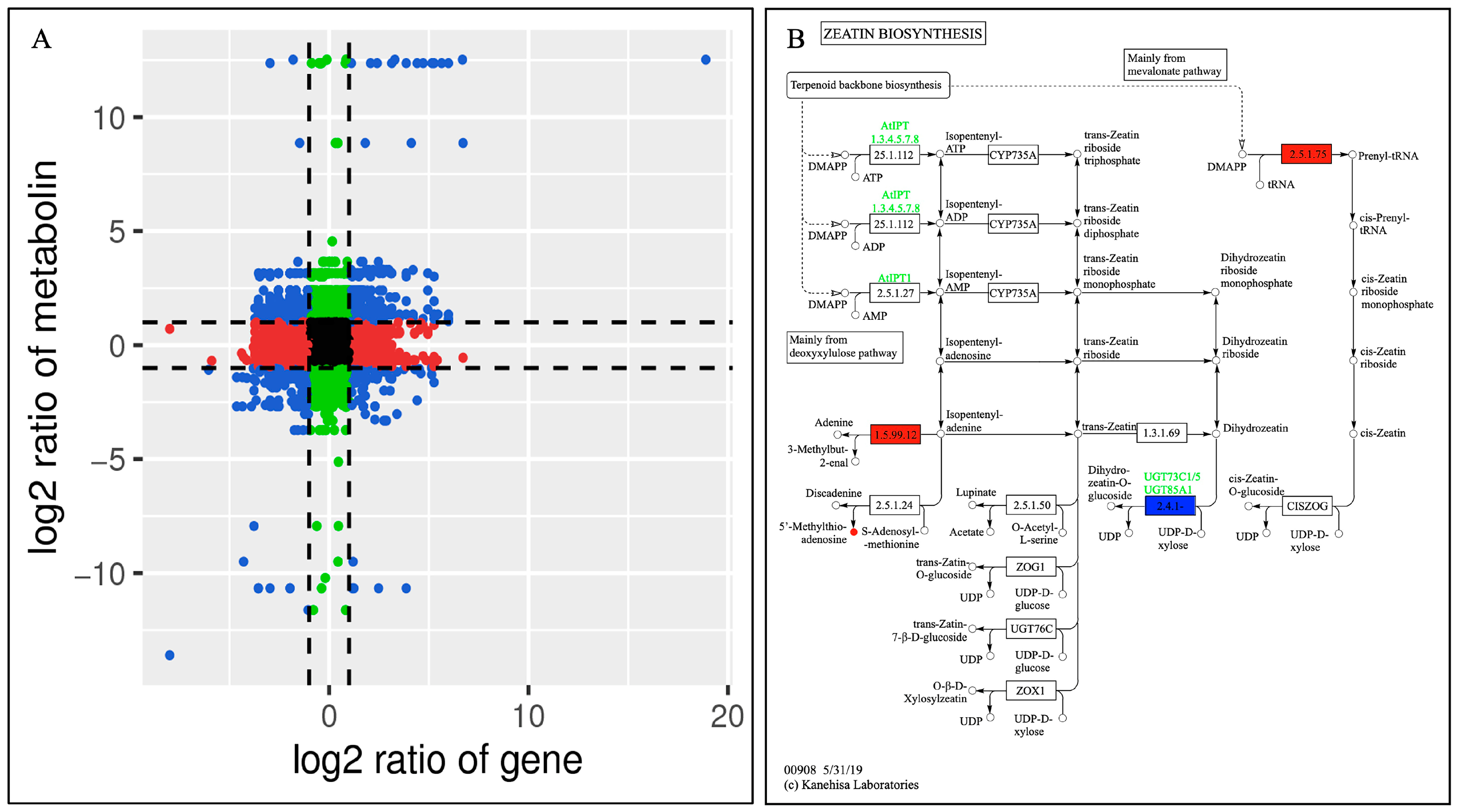
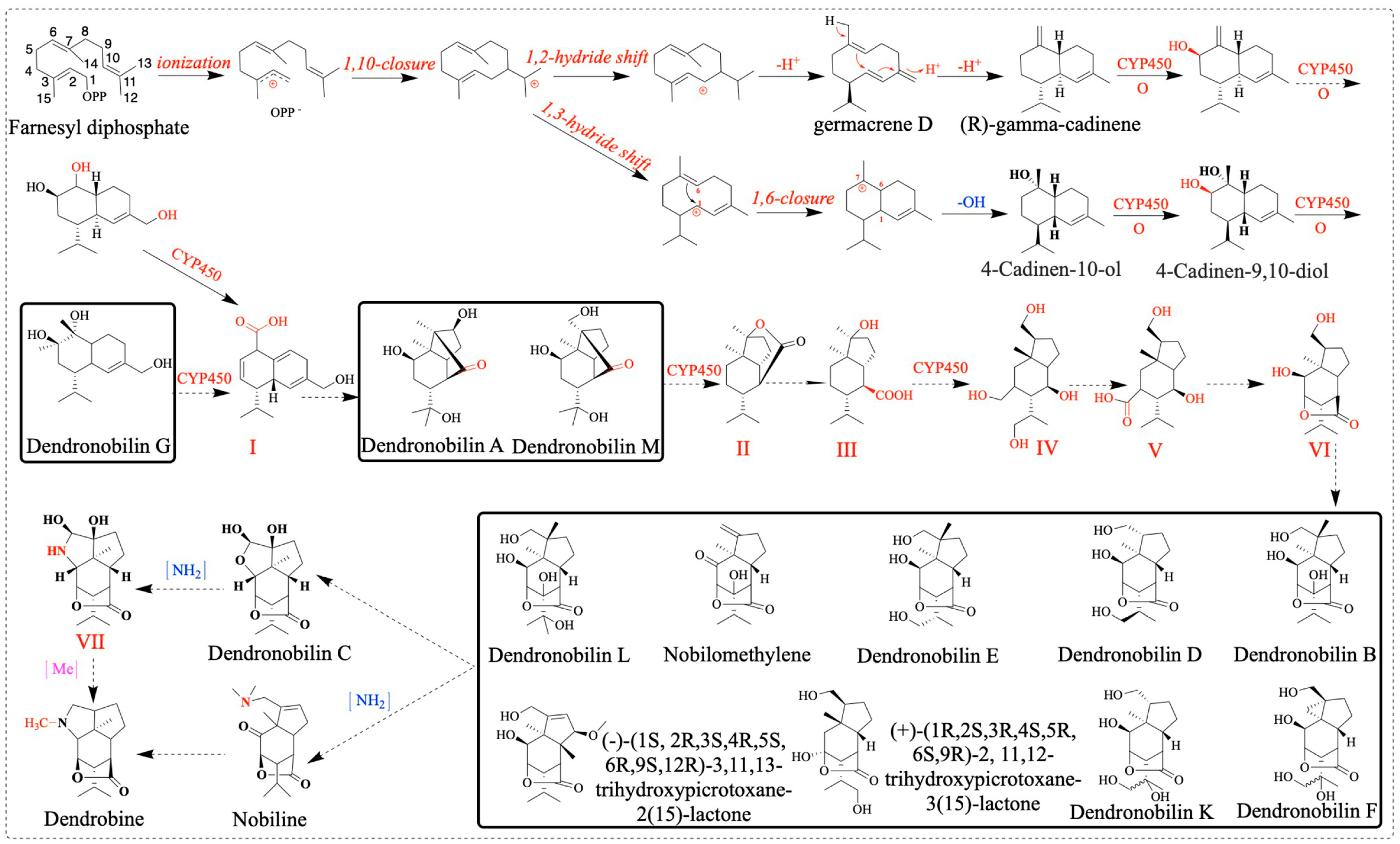
2.10. Dendrobine Biosynthetic Pathway Activation in Response to MeJA
2.11. Functional Validation of DnSQS-2
3. Discussion
3.1. Metabolome Analysis of D. nobile
3.2. Transcriptome Analysis of D. nobile
3.3. Metabolome and Transcriptome Analysis of the Metabolite Biosynthesis Pathway
4. Materials and Methods
4.1. Plant Materials and Culture Conditions
4.2. Sample Extraction and Wide-Target Metabolomics Analysis
4.2.1. Sample Preparation for Widely Targeted Metabolic Profiling
4.2.2. Determination of the Dendrobine Content in Samples
4.2.3. UPLC–MS/MS Conditions
4.2.4. Characterization and Quantification of Metabolites
4.2.5. KEGG Annotation and KEGG Enrichment Analysis of Metabolites
4.3. RNA Extraction and Transcriptome Analysis
4.3.1. RNA Extraction and Illumina Sequencing
4.3.2. Gene Functional Annotation and Expression Levels Analysis
4.3.3. Sequence Analysis, Gene Expression in Yeast, and Product Analysis by GC–MS
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bhattacharyya, P.; Kumaria, S. Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplified polymorphic DNA. Plant Syst. Evol. 2015, 301, 201–210. [Google Scholar] [CrossRef]
- Li, Q.; Liu, C.; Huang, C.; Wang, M.; Long, T.; Liu, J.; Shi, J.; Shi, J.; Li, L.; He, Y.; et al. Transcriptome and Metabonomics Analysis Revealed the Molecular Mechanism of Differential Metabolite Production of Dendrobium nobile Under Different Epiphytic Patterns. Front. Plant Sci. 2022, 13, 868472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, M.; Wu, Q.; Shi, J.S. Dendrobium nobile Lindl. Alkaloids Ameliorate Aβ25-35-Induced Synaptic Deficits by Targeting Wnt/β-Catenin Pathway in Alzheimer’s Disease Models. J. Alzheimers Dis. 2022, 86, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tu, H.; Zhu, J.; Liang, A.; Huo, P.; Shan, K.; He, J.; Zhao, M.; Chen, X.; Lei, X. Dendrobium nobile Lindl. polysaccharides improve follicular development in PCOS rats. Int. J. Biol. Macromol. 2020, 149, 826–834. [Google Scholar] [CrossRef]
- Wang, P.; Chen, X.; Wang, H.; Huang, S.; Cai, C.; Yuan, J.; Zhu, G.; Xu, X.; Mei, W.; Dai, H. Four New Picrotoxane-Type Sesquiterpenes from Dendrobium nobile Lindl. Front. Chem. 2019, 7, 812. [Google Scholar] [CrossRef]
- Wang, Y.; Liao, X.; Zhou, C.; Hu, L.; Wei, G.; Huang, Y.; Lei, Z.; Ren, Z.; Liu, Z.; Liu, Z. Identification of C-glycosyl flavones and quality assessment in Dendrobium nobile. Rapid Commun. Mass Spectrom. 2021, 35, e9012. [Google Scholar] [CrossRef]
- Xu, J.; Han, Q.B.; Li, S.L.; Chen, X.J.; Wang, X. Chemistry, bioactivity and quality control of Dendrobium, a commonly used tonic herb in traditional Chinese medicine. Phytochem. Rev. 2013, 12, 341–367. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, Q.; Lu, Y.L.; Gong, Q.H.; Zhang, F.; Shi, J.S. Protective effects of Dendrobium nobile Lindl. alkaloids on amyloid beta (25–35)-induced neuronal injury. Neural Regen. Res. 2017, 12, 1131–1136. [Google Scholar] [CrossRef]
- Li, Z.; Xiang, J.; Hu, D.; Song, B. Naturally potential antiviral agent polysaccharide from Dendrobium nobile Lindl. Pestic. Biochem. Physiol. 2020, 167, 104598. [Google Scholar] [CrossRef]
- Luo, A.; He, X.; Zhou, S.; Fan, Y.; He, T.; Chun, Z. In vitro antioxidant activities of a water-soluble polysaccharide derived from Dendrobium nobile Lindl. extracts. Int. J. Biol. Macromol. 2009, 45, 359–363. [Google Scholar] [CrossRef]
- Long, Y.; Wang, W.; Zhang, Y.; Zhang, S.; Li, Z.; Deng, J.; Li, J. Dendrobium nobile Lindl Polysaccharides Attenuate UVB-induced Photodamage by Regulating Oxidative Stress, Inflammation and MMPs Expression in Mice Model. Photochem. Photobiol. 2023, 99, 1269–1281. [Google Scholar] [CrossRef]
- Wang, J.H.; Luo, J.P.; Zha, X.Q. Structural features of a pectic polysaccharide from the stems of Dendrobium nobile Lindl. Carbohydr. Polym. 2010, 81, 1–7. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Mei, N.; Ma, C.; Lou, Z.; Lv, W.; He, G. Protective effects of polysaccharide from Dendrobium nobile against ethanol-induced gastric damage in rats. Int. J. Biol. Macromol. 2018, 107, 230–235. [Google Scholar] [CrossRef]
- Nie, X.; Chen, Y.; Li, W.; Lu, Y. Anti-aging properties of Dendrobium nobile Lindl.: From molecular mechanisms to potential treatments. J. Ethnopharmacol. 2020, 257, 112839. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Suzuki, M.; Hayakawa, Y.; Aoki, K.; Nakamura, H.; Nagase, H.; Hirata, Y. Total synthesis of dl-dendrobine. J. Am. Chem. Soc. 1972, 94, 8278–8280. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.Y.; Chen, X.Y.; Guo, S.X.; Wang, B.C.; Li, B. Recent advances and new insights in biosynthesis of dendrobine and sesquiterpenes. Appl. Microbiol. Biotechnol. 2021, 105, 6597–6606. [Google Scholar] [CrossRef]
- Li, Q.; Ding, G.; Li, B.; Guo, S.X. Transcriptome Analysis of Genes Involved in Dendrobine Biosynthesis in Dendrobium nobile Lindl. Infected with Mycorrhizal Fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 316. [Google Scholar] [CrossRef]
- Pan, F.; Wu, M.; Hu, W.; Liu, R.; Yan, H.; Xiang, Y. Genome-Wide Identification and Expression Analyses of the bZIP Transcription Factor Genes in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 20, 2203. [Google Scholar] [CrossRef]
- Bhambhani, S.; Kondhare, K.R.; Giri, A.P. Diversity in Chemical Structures and Biological Properties of Plant Alkaloids. Molecules 2021, 26, 3374. [Google Scholar] [CrossRef]
- Nokihara, K.; Okada, Y.; Ohata, S.; Monden, Y. Transcriptome Analysis Reveals Key Genes Involved in Weevil Resistance in the Hexaploid Sweetpotato. Plants 2021, 10, 1535. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, Y.; Wang, G.; Wang, T.; Cao, F. Integrated analysis of the transcriptome and metabolome in young and mature leaves of Ginkgo biloba L. Ind. Crop. Prod. 2020, 143, 111906. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, B.; Bu, J.; Guo, J.; Chen, T.; Ma, Y.; Tang, J.; Cui, G.; Huang, L. Transcriptomic Insight into Terpenoid Biosynthesis and Functional Characterization of Three Diterpene Synthases in Scutellaria barbata. Molecules 2018, 23, 2952. [Google Scholar] [CrossRef]
- Liu, Y.; Patra, B.; Singh, S.K.; Paul, P.; Zhou, Y.; Li, Y.; Wang, Y.; Pattanaik, S.; Yuan, L. Terpenoid indole alkaloid biosynthesis in Catharanthus roseus: Effects and prospects of environmental factors in metabolic engineering. Biotechnol. Lett. 2021, 43, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, W.; Liu, Y.; Meng, X.; Su, X.; Cao, M.; Wu, L.; Yu, N.; Xing, S.; Peng, D. Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between Protocorm-like bodies and leaves. BMC Genom. 2021, 22, 579. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tong, S.; Ma, T.; Xi, Z.; Liu, J. Chromosome-level genome assembly of Sichuan pepper provides insights into apomixis, drought tolerance, and alkaloid biosynthesis. Mol. Ecol. Resour. 2021, 21, 2533–2545. [Google Scholar] [CrossRef]
- Mao, X.; Ge, M.; Wang, X.; Yu, J.; Li, X.; Liu, B.; Chen, F. Transcriptomics and Metabolomics Analyses Provide Novel Insights into Glucose-Induced Trophic Transition of the Marine Diatom Nitzschia laevis. Mar. Drugs 2021, 19, 426. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xu, J.; Zheng, Y.; Zhang, Y.; Cai, Y. Transcriptomic and Metabolomic Analyses Reveals That Exogenous Methyl Jasmonate Regulates Galanthamine Biosynthesis in Lycoris longituba Seedlings. Front. Plant Sci. 2021, 12, 713795. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Han, C.; Liu, Z.; Ni, M.; Wu, Q.; Yu, F. Transcriptomic and Metabolomic Analysis Unravels the Molecular Regulatory Mechanism of Fatty Acid Biosynthesis in Styrax tonkinensis Seeds under Methyl Jasmonate Treatment. Int. J. Mol. Sci. 2022, 23, 6190. [Google Scholar] [CrossRef]
- Zhan, X.; Luo, X.; He, J.; Zhang, C.; Liao, X.; Xu, X.; Feng, S.; Yu, C.; Jiang, Z.; Meng, Y.; et al. Bioactive compounds induced in Physalis angulata L. by methyl-jasmonate: An investigation of compound accumulation patterns and biosynthesis-related candidate genes. Plant Mol. Biol. 2020, 103, 341–354. [Google Scholar] [CrossRef]
- Armstrong, R.A. Should Pearson’s correlation coefficient be avoided? Ophthalmic Physiol. Opt. 2019, 39, 316–327. [Google Scholar] [CrossRef]
- Picaud, S.; Olsson, M.E.; Brodelius, M.; Brodelius, P.E. Cloning, expression, purification and characterization of recombinant (+)-germacrene D synthase from Zingiber officinale. Arch. Biochem. Biophys. 2006, 452, 17–28. [Google Scholar] [CrossRef]
- Yamazaki, M.; Matsuo, M.; Arai, K. Biosynthesis of dendrobine. Chem. Pharm. Bull. 1966, 14, 1058–1059. [Google Scholar] [CrossRef]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, H.; Geng, P.; Lin, Y.; Liu, Z.; Zhang, L.; Ma, J.; Zhou, Y.; Wang, X.; Wen, C. Pharmacokinetic study of dendrobine in rat plasma by ultra-performance liquid chromatography tandem mass spectrometry. Biomed. Chromatogr. 2016, 30, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Shi, F.G.; Fang, C.; Shi, J.S. Metabolic characterization of a potent natural neuroprotective agent dendrobine in vitro and in rats. Acta Pharmacol. Sin. 2021, 43, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Chen, X.; Lee, C.; Sze, S.; Feng, Y.; Yang, Z.; Chen, H.; Li, S.; Zhang, L.; Wei, G. Dendrobine targeting JNK stress signaling to sensitize chemotoxicity of cisplatin against non-small cell lung cancer cells in vitro and in vivo. Phytomedicine 2019, 53, 18–27. [Google Scholar] [CrossRef]
- Li, L.S.; Lu, Y.L.; Nie, J.; Xu, Y.Y.; Zhang, W.; Yang, W.J.; Gong, Q.H.; Lu, Y.F.; Lu, Y.; Shi, J.S. Dendrobium nobile Lindl alkaloid, a novel autophagy inducer, protects against axonal degeneration induced by Aβ25-35 in hippocampus neurons in vitro. CNS Neurosci. Ther. 2017, 23, 329–340. [Google Scholar] [CrossRef]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined Analysis of the Fruit Metabolome and Transcriptome Reveals Candidate Genes Involved in Flavonoid Biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef]
- Chakka, V.P.; Zhou, T. Carboxymethylation of polysaccharides: Synthesis and bioactivities. Int. J. Biol. Macromol. 2020, 165, 2425–2431. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Huang, G.; Yang, Z.; Hou, Y. Antioxidant activity of Momordica charantia polysaccharide and its derivatives. Int. J. Biol. Macromol. 2019, 138, 673–680. [Google Scholar] [CrossRef]
- Joseph, A.A.; Pardo-Vargas, A.; Seeberger, P.H. Total Synthesis of Polysaccharides by Automated Glycan Assembly. J. Am. Chem. Soc. 2020, 142, 8561–8564. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Huang, L.; Chen, M.; Zeng, W.; Feng, Z.; Huang, S.; Liu, T. Integrated Analysis of the Transcriptome and Metabolome Reveals Genes Involved in Terpenoid and Flavonoid Biosynthesis in the Loblolly Pine (Pinus taeda L.). Front. Plant Sci. 2021, 12, 729161. [Google Scholar] [CrossRef] [PubMed]
- Chacon, D.S.; Torres, T.M.; da Silva, I.B.; de Araújo, T.F.; Roque, A.A.; Pinheiro, F.; Selegato, D.; Pilon, A.; Reginaldo, F.P.S.; da Costa, C.T.; et al. Erythrina velutina Willd. alkaloids: Piecing biosynthesis together from transcriptome analysis and metabolite profiling of seeds and leaves. J. Adv. Res. 2021, 34, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Li, K.; Yao, F.; Sun, L.; Liu, Y. Comparative transcriptome analyses on terpenoids metabolism in field- and mountain-cultivated ginseng roots. BMC Plant Biol. 2019, 19, 82. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Ma, S.; Tay, T.; Geiger-Schuller, K.R.; Kirchgatterer, P.C.; Verdine, V.K.; Guo, B.; Arias-Garcia, M.A.; Allen, W.E.; Singh, A.; et al. A transcription factor atlas of directed differentiation. Cell 2023, 186, 209–229.e226. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, L.; Chang, H.; Cai, B.; Gao, H.; Chen, G.; Hou, W.; Jappar, Z.; Yan, Y. Research progress on extraction, purification, structure and biological activity of Dendrobium officinale polysaccharides. Front. Nutr. 2022, 9, 965073. [Google Scholar] [CrossRef] [PubMed]
- Boeckx, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol oxidase in leaves: Is there any significance to the chloroplastic localization? J. Exp. Bot. 2015, 66, 3571–3579. [Google Scholar] [CrossRef]
- Khan, B.R.; Adham, A.R.; Zolman, B.K. Peroxisomal Acyl-CoA oxidase 4 activity differs between Arabidopsis accessions. Plant Mol. Biol. 2012, 78, 45–58. [Google Scholar] [CrossRef]
- Dai, H.; Zhu, Z.; Wang, Z.; Zhang, Z.; Kong, W.; Miao, M. Galactinol synthase 1 improves cucumber performance under cold stress by enhancing assimilate translocation. Hortic. Res. 2022, 9, uhab063. [Google Scholar] [CrossRef]
- Zhao, D.; Luan, Y.; Shi, W.; Zhang, X.; Meng, J.; Tao, J. A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging. Plant Sci. 2021, 303, 110765. [Google Scholar] [CrossRef]
- Jia, Q.; Brown, R.; Köllner, T.G.; Fu, J.; Chen, X.; Wong, G.K.; Gershenzon, J.; Peters, R.J.; Chen, F. Origin and early evolution of the plant terpene synthase family. Proc. Natl. Acad. Sci. USA 2022, 119, e2100361119. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, G.; Yamamura, Y.; Chen, F.; Fu, J. Divergent Evolution of the Diterpene Biosynthesis Pathway in Tea Plants (Camellia sinensis) Caused by Single Amino Acid Variation of ent-Kaurene Synthase. J. Agric. Food Chem. 2020, 68, 9930–9939. [Google Scholar] [CrossRef] [PubMed]
- Lackus, N.D.; Morawetz, J.; Xu, H.; Gershenzon, J.; Dickschat, J.S.; Köllner, T.G. The Sesquiterpene Synthase PtTPS5 Produces (1S,5S,7R,10R)-Guaia-4(15)-en-11-ol and (1S,7R,10R)-Guaia-4-en-11-ol in Oomycete-Infected Poplar Roots. Molecules 2021, 26, 555. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, L.; Liu, J. A Memetic Algorithm Based on Probability Learning for Solving the Multidimensional Knapsack Problem. IEEE Trans. Cybern. 2022, 52, 2284–2299. [Google Scholar] [CrossRef] [PubMed]
- Aisporna, A.; Benton, H.P.; Chen, A.; Derks, R.J.E.; Galano, J.M.; Giera, M.; Siuzdak, G. Neutral Loss Mass Spectral Data Enhances Molecular Similarity Analysis in METLIN. J. Am. Soc. Mass Spectrom. 2022, 33, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Vicini, A.; Salzer, L.; Stanstrup, J.; Badia, J.M.; Neumann, S.; Stravs, M.A.; Verri Hernandes, V.; Gatto, L.; Gibb, S.; et al. A Modular and Expandable Ecosystem for Metabolomics Data Annotation in R. Metabolites 2022, 12, 173. [Google Scholar] [CrossRef]
- Ara, T.; Sakurai, N.; Takahashi, S.; Waki, N.; Suganuma, H.; Aizawa, K.; Matsumura, Y.; Kawada, T.; Shibata, D. TOMATOMET: A metabolome database consists of 7118 accurate mass values detected in mature fruits of 25 tomato cultivars. Plant Direct 2021, 5, e00318. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Shi, G.; Shen, X.; Ren, H.; Rao, Y.; Weng, S.; Tang, X. Kernel principal component analysis and differential non-linear feature extraction of pesticide residues on fruit surface based on surface-enhanced Raman spectroscopy. Front. Plant Sci. 2022, 13, 956778. [Google Scholar] [CrossRef]
- Su, D.; Yang, Y.; Zeng, Q.; Liao, L.; Chen, C.; Yang, M.; Zhu, G.; Zhang, R.W.; Ai, Z.; Li, Y.; et al. Two Species Origins Comparison of Herba Patriniae Based on Their Ingredients Profile by UPLC-QTOF/MS/MS and Orthogonal Partial Least Squares Discriminant Analysis. Chem. Biodivers. 2022, 19, e202100961. [Google Scholar] [CrossRef]
- Carpinetti, P.A.; Fioresi, V.S.; Ignez da Cruz, T.; de Almeida, F.A.N.; Canal, D.; Ferreira, A.; Ferreira, M. Efficient method for isolation of high-quality RNA from Psidium guajava L. tissues. PLoS ONE 2021, 16, e0255245. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.S.; Luu, P.L.; Qu, W.; Maddugoda, M.; Huschtscha, L.; Reddel, R.; Chenevix-Trench, G.; Toso, M.; Kench, J.G.; Horvath, L.G.; et al. Guidelines for whole genome bisulphite sequencing of intact and FFPET DNA on the Illumina HiSeq X Ten. Epigenet. Chromatin 2018, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The Genome of Dendrobium officinale Illuminates the Biology of the Important Traditional Chinese Orchid Herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Gill, N.; Dhillon, B. RNA-seq Data Analysis for Differential Expression. Methods Mol. Biol. 2022, 2391, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three Differential Expression Analysis Methods for RNA Sequencing: Limma, EdgeR, DESeq2. J. Vis. Exp. 2021, 175, e62528. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ali, F.; Khan, A.; Muhammad, S.A.; Hassan, S.S.U. Quantitative Real-Time Analysis of Differentially Expressed Genes in Peripheral Blood Samples of Hypertension Patients. Genes 2022, 13, 187. [Google Scholar] [CrossRef] [PubMed]



| Classification | Quantity of Metabolites | Classification | Quantity of Metabolites |
|---|---|---|---|
| Flavonoids | 178 | Others | 24 |
| Phenolic acids | 81 | Lignans | 17 |
| Lipids | 77 | Quinones | 11 |
| Amino acids and derivatives | 57 | Terpenoids | 9 |
| Alkaloids | 41 | Vitamin | 9 |
| Nucleotides and derivatives | 37 | Stilbene | 6 |
| Saccharides and alcohols | 33 | Tannins | 2 |
| Organic acids | 32 | Xanthone | 1 |
| Group | Down a | Up a | Total a | Group | Down b | Up b | Total b |
|---|---|---|---|---|---|---|---|
| CK_vs_DC7 | 60 | 81 | 141 | CK_vs_TC7 | 233 | 164 | 397 |
| CK_vs_DC14 | 68 | 49 | 117 | CK_vs_TC14 | 313 | 540 | 853 |
| CK_vs_DF | 64 | 77 | 141 | CK_vs_TF | 288 | 416 | 704 |
| CK_vs_DF7 | 68 | 97 | 165 | CK_vs_TF7 | 76 | 116 | 192 |
| CK_vs_DF14 | 82 | 52 | 134 | CK_vs_TF14 | 624 | 722 | 1346 |
| DC7_vs_DC14 | 82 | 28 | 110 | TC7_vs_TC14 | 508 | 854 | 1362 |
| DC7_vs_DF7 | 38 | 52 | 90 | TC7_vs_TF7 | 33 | 96 | 129 |
| DC14_vs_DF14 | 49 | 51 | 100 | TC14_vs_TF14 | 272 | 79 | 351 |
| DF_vs_DF7 | 27 | 62 | 89 | TF_vs_TF7 | 411 | 366 | 777 |
| DF_vs_DF14 | 48 | 21 | 69 | TF_vs_TF14 | 212 | 130 | 342 |
| DF7_vs_DF14 | 78 | 25 | 103 | TF7_vs_TF14 | 405 | 484 | 889 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, D.; Li, B.; Wu, B.; Fu, D.; Li, Z.; Wei, H.; Guo, S.; Ding, G.; Wang, B. The Integration of the Metabolome and Transcriptome for Dendrobium nobile Lindl. in Response to Methyl Jasmonate. Molecules 2023, 28, 7892. https://doi.org/10.3390/molecules28237892
Gong D, Li B, Wu B, Fu D, Li Z, Wei H, Guo S, Ding G, Wang B. The Integration of the Metabolome and Transcriptome for Dendrobium nobile Lindl. in Response to Methyl Jasmonate. Molecules. 2023; 28(23):7892. https://doi.org/10.3390/molecules28237892
Chicago/Turabian StyleGong, Daoyong, Biao Li, Bin Wu, Deru Fu, Zesheng Li, Haobo Wei, Shunxing Guo, Gang Ding, and Bochu Wang. 2023. "The Integration of the Metabolome and Transcriptome for Dendrobium nobile Lindl. in Response to Methyl Jasmonate" Molecules 28, no. 23: 7892. https://doi.org/10.3390/molecules28237892
APA StyleGong, D., Li, B., Wu, B., Fu, D., Li, Z., Wei, H., Guo, S., Ding, G., & Wang, B. (2023). The Integration of the Metabolome and Transcriptome for Dendrobium nobile Lindl. in Response to Methyl Jasmonate. Molecules, 28(23), 7892. https://doi.org/10.3390/molecules28237892







