NMR and Docking Calculations Reveal Novel Atomistic Selectivity of a Synthetic High-Affinity Free Fatty Acid vs. Free Fatty Acids in Sudlow’s Drug Binding Sites in Human Serum Albumin
Abstract
:1. Introduction
2. Results and Discussion
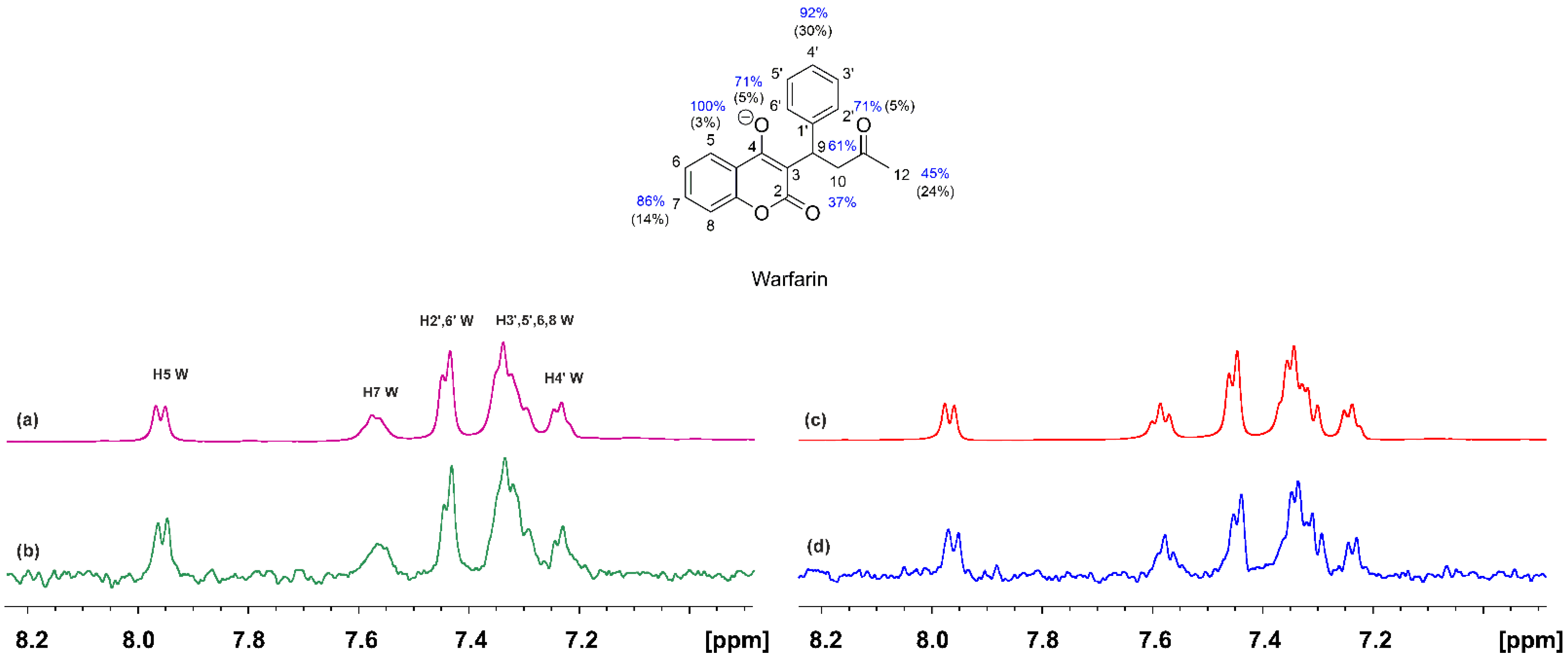
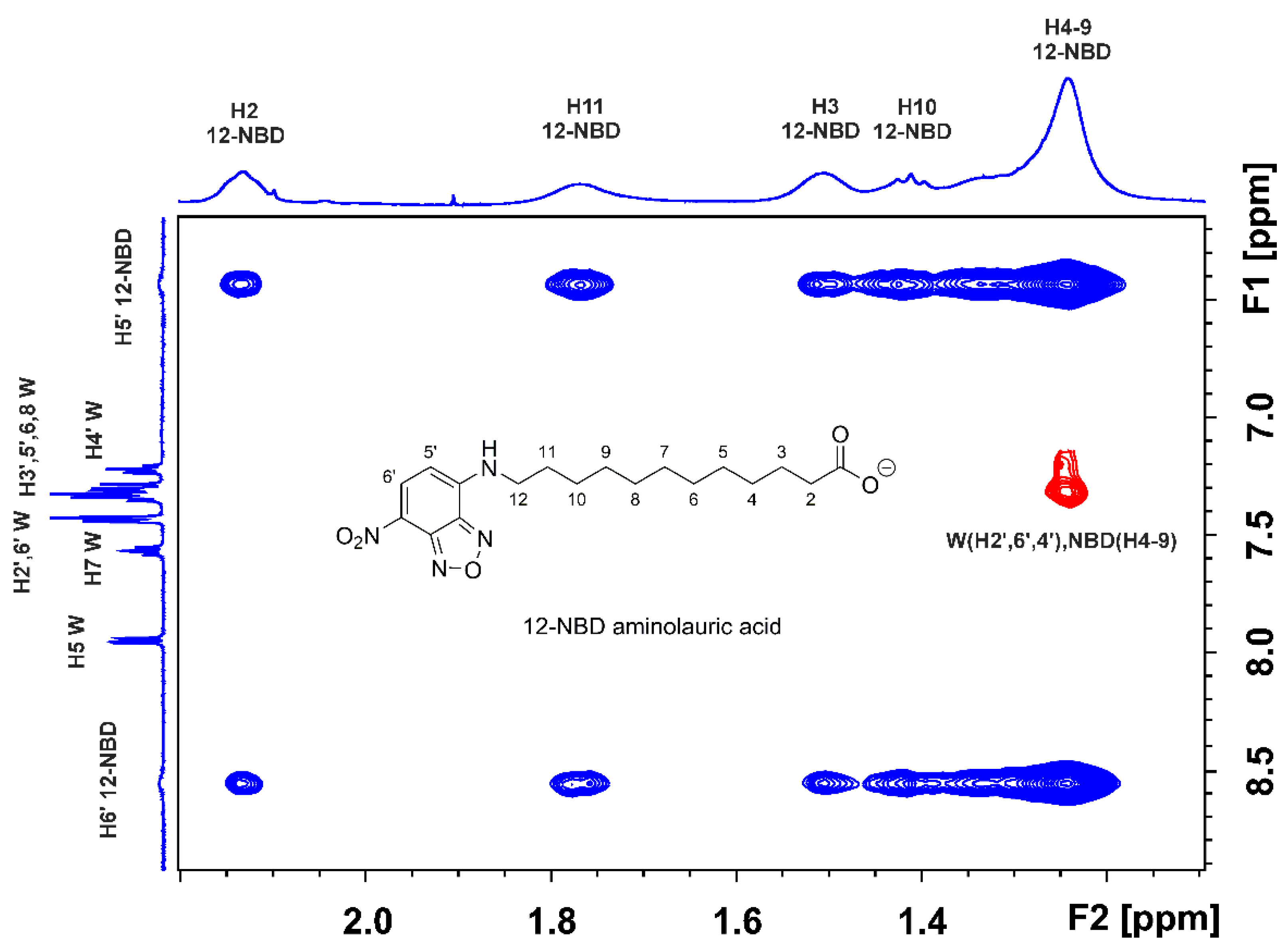
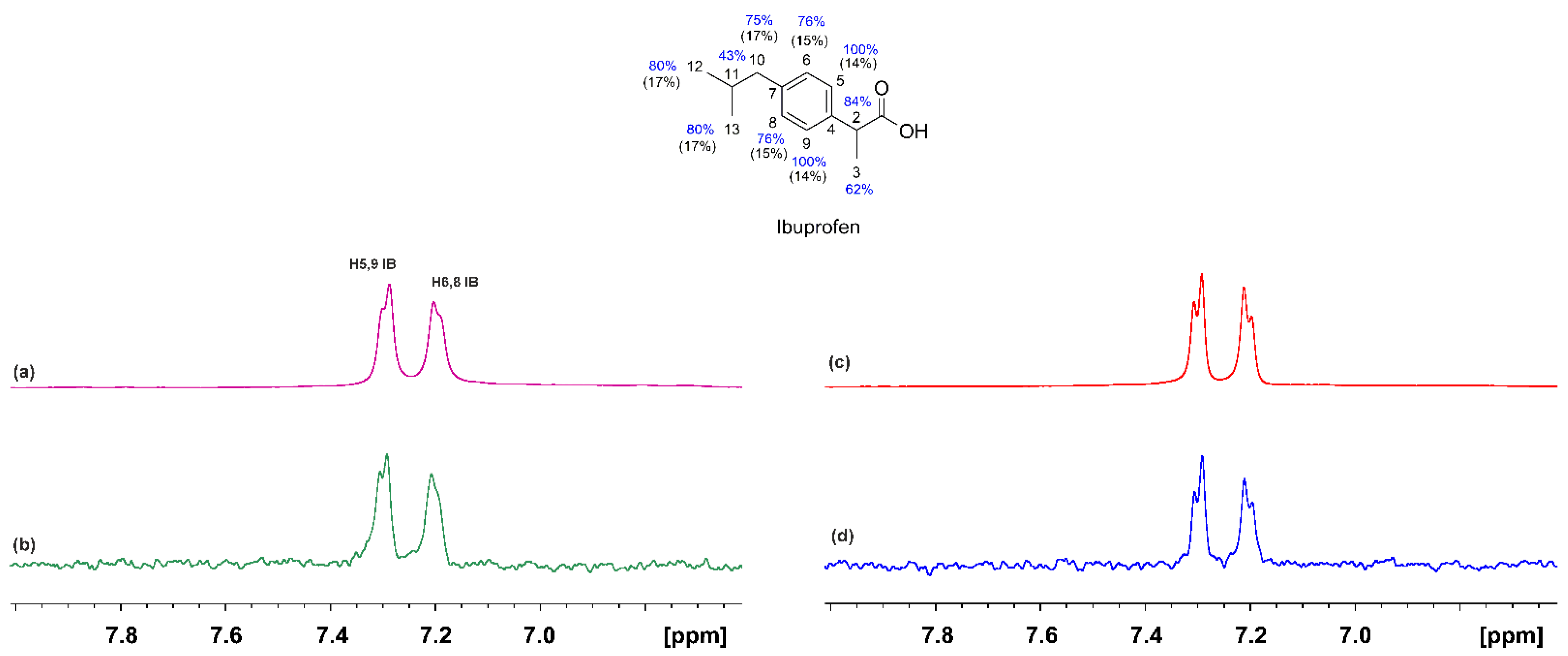
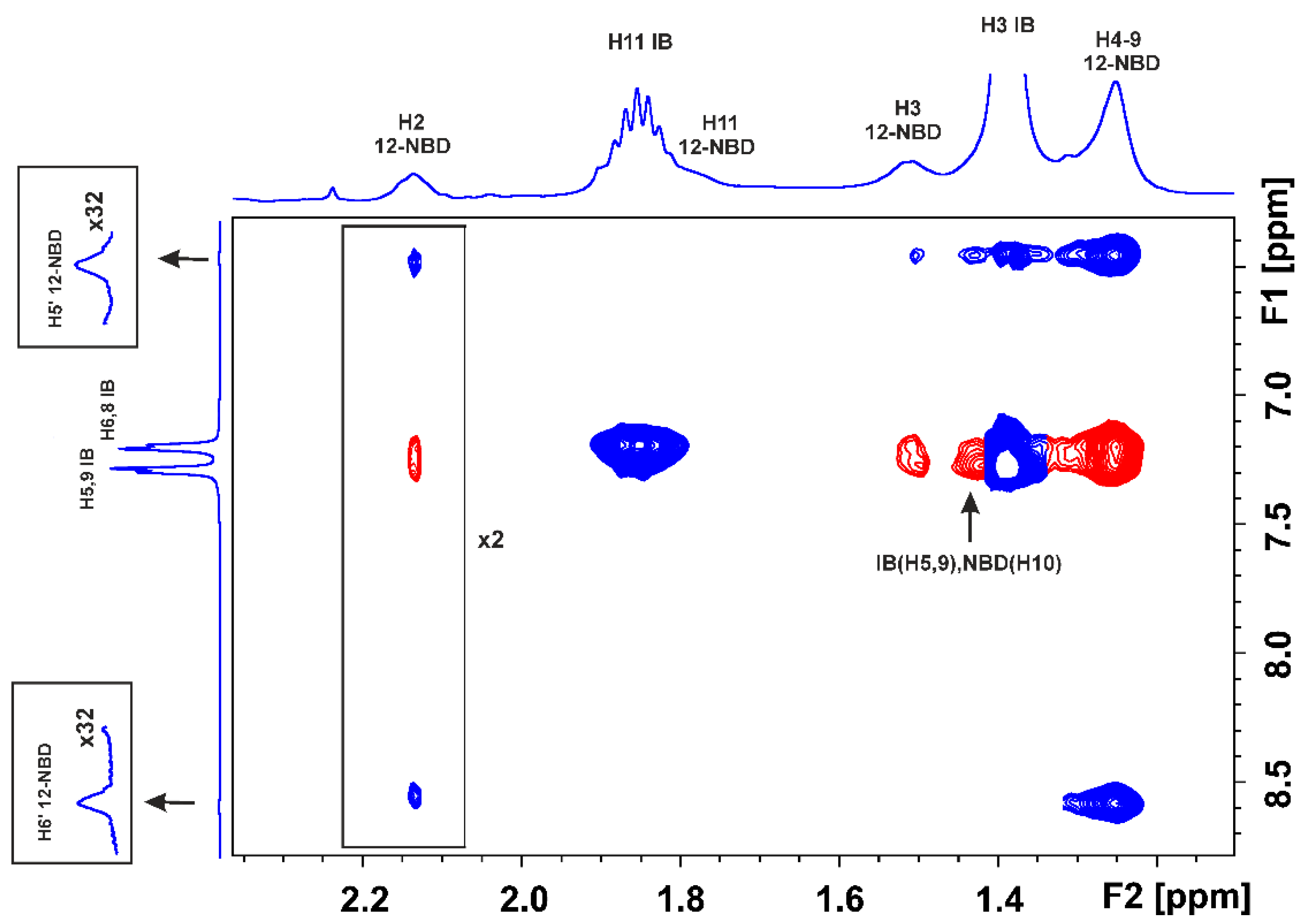
2.1. STD and INPHARMA NMR Competition Experiments of NBD-C12 FA with Warfarin and Ibuprofen
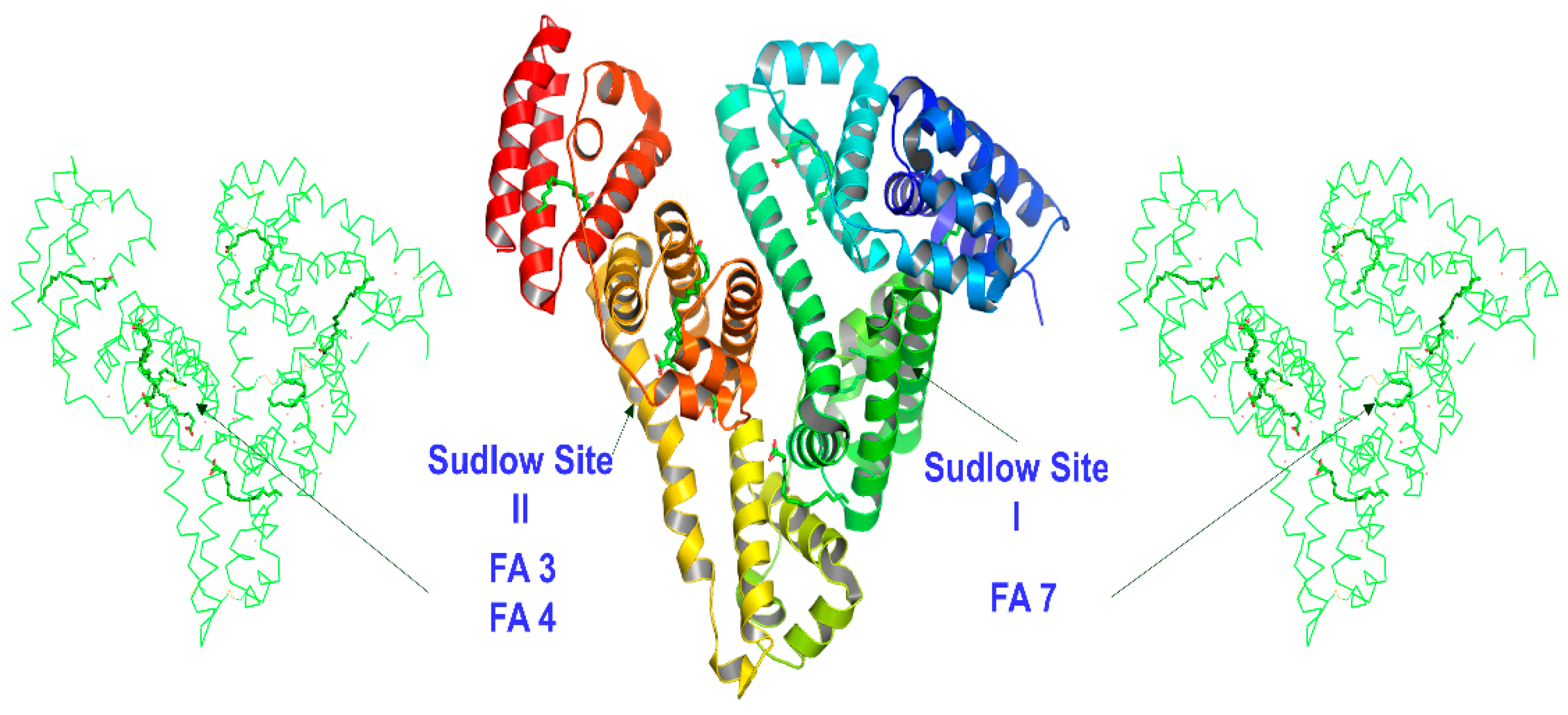
2.1.1. The FA7 Binding Site
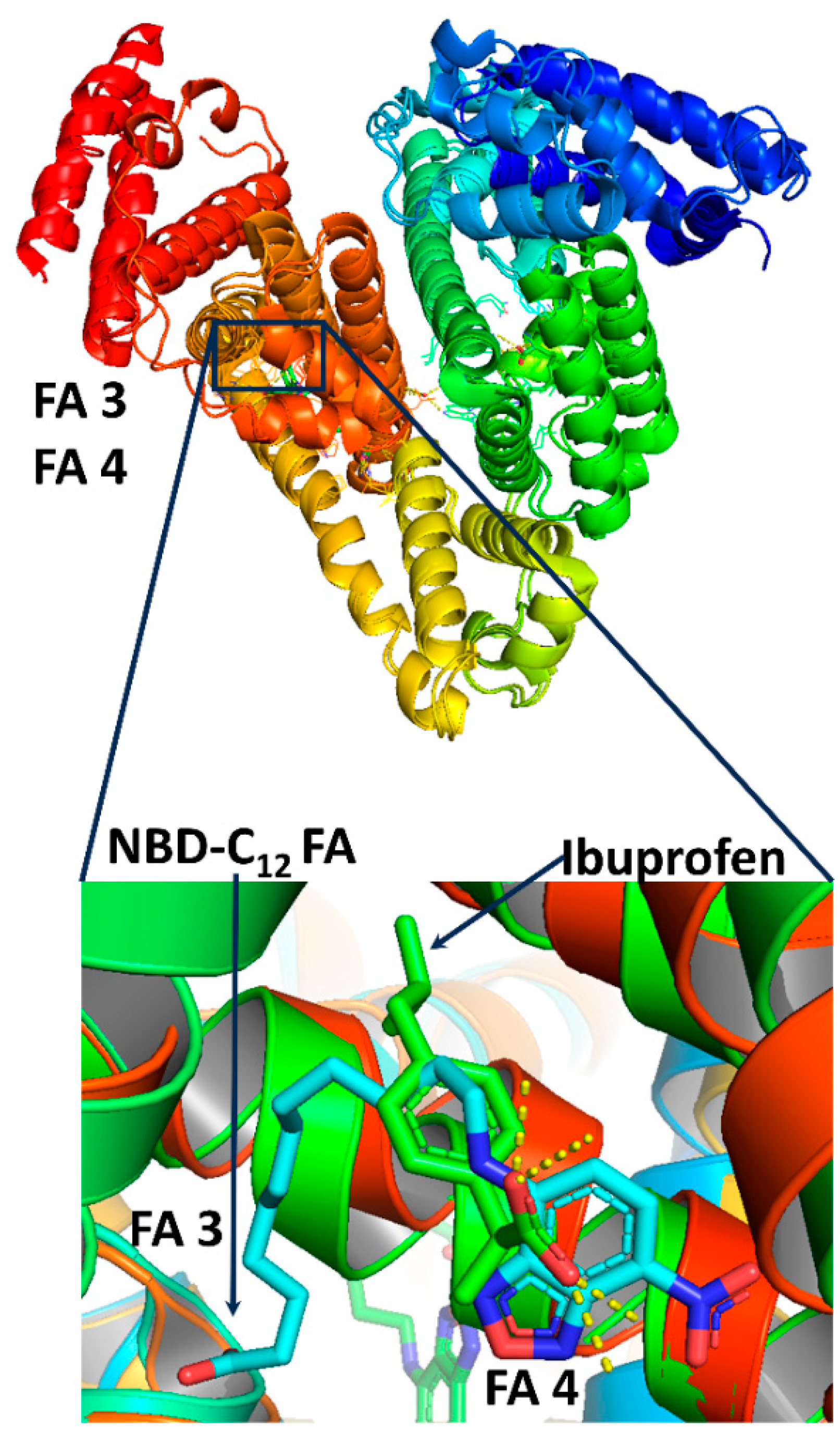
2.1.2. The FA3 and FA4 Binding Sites
2.2. Docking Calculations
- The (7-nitrobenz-2-oxa-1,3-diazol-4-yl)-C12 fatty acid [17] and the drug warfarin interact weakly through the FA’s methylene groups and the drug’s phenyl butyl moiety in the binding site FA7.
- The NBD-C12 fatty acid interacts with the drug ibuprofen in the binding sites FA3 and FA4.
2.2.1. The FA7 Binding Site
2.2.2. The FA3 and FA4 Binding Sites
2.3. A Unified Atomic Level for the Selectivity of NBD-C12 FA and Short, Medium, and Long Mono- and Polyunsaturated Free Fatty Acids
3. Material and Methods
3.1. Chemicals and Reagents
3.2. NMR Experiments
3.3. Computational Methods
4. Conclusions
- The limited number of negative interligand NOEs between H4–9 protons of NBD-C12 FA and protons of the phenyl ring of warfarin and the absence of common inter-NOEs between the aromatic rings of the two ligands were interpreted in terms of a short-range negative allosteric competitive binding of NBD-C12 FA with the amino acids Ser-202, Lys-199, Trp-214, and warfarin with Arg-218 and Tyr-411 in the wide binding site FA7.
- The extensive number of interligand NOEs between H2, H3, and H4–9 of NBD-C12 FA and the aromatic protons H5,9 and H6,8 of ibuprofen was interpreted in terms of a competitive binding mode with Ser-342, Arg-348, Arg-485, Arg-410, and Tyr-411 in the binding sites FA3 and FA4.
- The self-docking protocol of the ligands NBD-C12 FA, warfarin, and ibuprofen on the X-ray HSA–ligand structure allowed us to define the search space as precisely as possible and, thus, accurately define electrostatic and hydrogen bond interactions between ligands and HSA.
- Compared to short-, medium-, and long-chain mono- and polyunsaturated FFAs, the NBD-C12 FA has the unique structural characteristics of interacting with amino acids of both the internal and external clusters in Sudlow’s binding site I. In Sudlow’s binding site II, the NBD-C12 FA interacts with amino acids in both FA3 and FA4.
- X-ray and NMR-based docking calculations with site-specific docking has been proven to constitute a very successful method to elucidate and describe the generated electrostatic and H-bonded interactions between the ligands and the HSA protein at an atomic level.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hodgson, J. All about Albumin: Biochemistry, Genetics and Medical Applications; Academic Press: San Diego, CA, USA, 2001. [Google Scholar]
- Wenskowsky, L.; Wagner, M.; Reusch, J.; Schreuder, H.; Matter, H.; Opatz, T.; Petry, M.S. Resolving binding events on the multifunctional human serum albumin. Chem. Med. Chem. 2020, 15, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Heath, R.J. Structural and biochemical features of human serum albumin essential for eukaryotic cell culture. Int. J. Mol. Sci. 2021, 22, 8411. [Google Scholar] [CrossRef] [PubMed]
- Di Massi, A. Human serum albumin: From molecular aspects to biotechnological applications. Int. J. Mol. Sci. 2023, 24, 4081. [Google Scholar] [CrossRef] [PubMed]
- Curry, S.; Brick, P.; Franks, N.P. Fatty acid binding to human serum albumin: New insights from crystallographic studies. Biochim. Biophys. Acta 1999, 1441, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.A.; Grűne, T.; Curry, S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef]
- Petitpas, I.; Grűne, T.; Bhattacharya, A.A.; Curry, S. Crystal structures of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids. J. Mol. Biol. 2001, 314, 955–960. [Google Scholar] [CrossRef]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal structure analysis of warfarin binding to human serum albumin anatomy of drug site I. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar]
- Krenzel, E.S.; Chen, Z.; Hamilton, J.A. Correspondence of fatty acid and drug binding sites on human serum albumin: A two-dimensional nuclear magnetic resonance study. Biochemistry 2013, 52, 1559–1567. [Google Scholar] [CrossRef]
- Itoh, T.; Saura, Y.; Tsuda, Y.; Yamada, H. Stereoselectivity and enantiomer-enantiomer interactions in the binding of ibuprofen to human serum albumin. Chirality 1997, 9, 643–649. [Google Scholar] [CrossRef]
- Murray, C.W.; Hartshorn, M.J. New applications for structure-based drug design. In Comprehensive Medicinal Chemistry II; Elsevier: Amsterdam, The Netherlands, 2007; Volume 4, Chapter 4.31; pp. 775–806. [Google Scholar]
- Alexandri, Ε.; Primikyri, A.; Papamokos, G.; Venianakis, T.; Gkalpinos, V.G.; Tzakos, A.G.; Karydis-Messinis, A.; Moschovas, D.; Avgeropoulos, A.; Gerothanassis, I.P. NMR and computational studies reveal novel aspects in molecular recognition of unsaturated fatty acids with non-labeled serum albumin. FEBS J. 2022, 289, 5617–5636. [Google Scholar] [CrossRef] [PubMed]
- Hernychova, L.; Alexandri, E.; Tzakos, A.G.; Zatloukalova, M.; Primikyri, A.; Gerothanassis, I.P.; Uhrik, L.; Šebela, M.; Kopečný, D.; Jedinák, L. Serum albumin as a primary non-covalent binding protein for nitro-oleic acid. Int. J. Biol. Macromol. 2022, 203, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Alexandri, E.; Venianakis, T.; Primikyri, A.; Papamokos, G.; Gerothanassis, I.P. Molecular basis for the selectivity of DHA and EPA in Sudlow’s drug binding sites in human serum albumin with the combined use of NMR and docking calculations. Molecules 2023, 28, 3724. [Google Scholar] [CrossRef] [PubMed]
- Gerothanassis, I.P. Ligand-observed in-tube NMR in natural products research: A review on enzymatic biotransformations, and in-cell NMR spectroscopy. Arab. J. Chem. 2023, 16, 104536. [Google Scholar] [CrossRef]
- Wenskowsky, L.; Schreuder, H.; Derdau, V.; Matter, H.; Volkmar, J.; Nazaré, M.; Opatz, T.; Petry, S. Identification and characterization of a single high-affinity fatty acid binding site in human serum albumin. Angew. Chem. Int. Ed. 2018, 57, 1044–1048. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Claasen, B.; Axmann, M.; Meinecke, R.; Meyer, B. Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin alpha(IIb)beta3 in native platelets than in liposomes. J. Am. Chem. Soc. 2005, 127, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Monaco, S.; Tailford, L.E.; Juge, N.; Angulo, J. Differential epitope mapping by STD NMR spectroscopy to reveal the nature of protein-ligand contacts. Angew. Chem. Int. Ed. 2017, 56, 15289–15293. [Google Scholar] [CrossRef]
- Monaco, S.; Ramírez-Cárdenas, J.; Carmona, A.T.; Robina, I.; Angulo, J. Inter-ligand STD NMR: An efficient 1D NMR approach to probe relative orientation of ligands in a multi-subsite protein binding pocket. Pharmaceuticals 2022, 15, 1030. [Google Scholar] [CrossRef]
- Potenza, D.; Vasile, F.; Belvisi, L.; Civera, M.; Araldi, E.M. STD and tr-NOESY NMR study of receptor-ligand interactions in living cancer cells. Chem. Bio. Chem. 2011, 12, 695–699. [Google Scholar] [CrossRef]
- Mari, S.; Invernizzi, C.; Spitaleri, A.; Alberici, L.; Ghitti, M.; Bordignon, C.; Traversari, C.; Rizzardi, G.-P.; Musco, G. 2D Tr-NOESY experiments interrogate and rank ligand-receptor interactions in living human cancer cells. Angew. Chem. Int. Ed. 2010, 49, 1071–1074. [Google Scholar] [CrossRef]
- Primikyri, A.; Sayyad, N.; Quilici, G.; Vrettos, E.I.; Lim, K.; Chi, S.-W.; Musco, G.; Gerothanassis, I.P.; Tzakos, A.G. Probing the interaction of a quercetin bioconjugate with Bcl-2 in living human cancer cells with in-cell NMR spectroscopy. FEBS Lett. 2018, 592, 3367–3379. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pedregal, V.M.; Reese, M.; Meiler, J.; Bloomers, M.J.J.; Griesinger, C.; Carlomagno, T. The INPHARMA method: Protein-mediated interligand NOEs for pharmacophore mapping. Angew. Chem. Int. Ed. 2005, 44, 4172–4175. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, T. NMR in natural products: Understanding conformation, configuration and receptor interactions. Nat. Prod. Rep. 2012, 29, 536–554. [Google Scholar] [CrossRef]
- Primikyri, A.; Papamokos, G.; Venianakis, T.; Sakka, M.; Kontogianni, V.; Gerothanassis, I.P. Structural basis of artemisinin binding sites in serum albumin with the combined use of NMR and docking calculations. Molecules 2022, 27, 5912. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Fehske, K.J.; Schläfer, U.; Wollert, U.; Müller, W.E. Characterization of an important drug binding area on human serum albumin including the high-affinity binding sites of warfarin and azapropazone. Mol. Pharmacol. 1982, 21, 387–393. [Google Scholar]
- Kragh-Hansen, U. Evidence for a large and flexible region of human serum albumin possessing high affinity binding sites for salicylate, warfarin, and other ligands. Mol. Pharnacol. 1988, 34, 160–171. [Google Scholar]
- Rafols, C.; Amezqueta, S.; Fuguet, E.; Bosch, E. Molecular interactions between warfarin and human (HSA) or bovine (BSA) serum albumin evaluated by isothermal titration carolimetry (ITC), fluorescence spectrometry (FS) and frontal analysis capillary electrophoresis (FA/CE). J. Pharm. Biomed. Anal. 2018, 150, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, B.; Bartucci, R.; Pey, A.L.; Guzzi, R. Warfarin increases thermal resistance of albumin through stabilization of the protein lobe that includes its binding site. Arch. Biochem. Biophys. 2019, 676, 10813. [Google Scholar] [CrossRef] [PubMed]
- Cox, P.B.; Gupta, R. Contemporary computational applications and tools in drug discovery. ACS Med. Chem. Lett. 2022, 13, 1016–1029. [Google Scholar] [CrossRef] [PubMed]
- Feher, M.; Williams, C.I. Effect of input differences on the results of docking calculations. J. Chem. Inf. Model. 2009, 49, 1704–1714. [Google Scholar] [CrossRef] [PubMed]
- Moitessier, N.; Englebienne, P.; Lee, D.; Lawandi, J.; Corbeil, C.R. Towards the development of universal, fast and highly accurate docking/scoring methods: A long way to go. Brit. J. Pharmacol. 2008, 153, S7–S26. [Google Scholar] [CrossRef]
- Schaduangrat, N.; Lampa, S.; Simeon, S.; Gleeson, M.P.; Spjuth, O.; Nantasenamat, C. Towards reproducible computational drug discovery. J. Cheminform. 2020, 12, 9. [Google Scholar] [CrossRef]
- Koehler Leman, J.; Lyskov, S.; Lewis, S.M.; Adolf-Bryfogle, J.; Alford, R.F.; Barlow, K.; Ben-Aharon, Z.; Farrell, D.; Fell, J.; Hansen, W.A. Ensuring scientific reproducibility in bio-macromolecular modeling via extensive, automated benchmarks. Nat. Commun. 2021, 12, 6947. [Google Scholar] [CrossRef]
- Gentile, F.; Agrawal, V.; Hsing, M.; Ton, A.-T.; Ban, F.; Norinder, U.; Gleave, M.E.; Cherkasov, A. Deep Docking: A deep learning platform for augmentation of structure-based drug discovery. ACS Centr. Sci. 2020, 6, 939–949. [Google Scholar] [CrossRef]
- Papamokos, G.V. The nature of the biological material and the irreproducibility problem in biomedical research. EMBO J. 2019, 38, e101011. [Google Scholar] [CrossRef]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Fanali, G.; Narciso, P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Vaid, T.M.; Wu, F.-J.; Chalmers, D.K.; Scott, D.J.; Gooley, P.R. INPHARMA-based determination of ligand binding modes at α1-adrenergic receptors explains the molecular basis of subtype selectivity. Chem. Eur. J. 2020, 26, 11796–11805. [Google Scholar] [CrossRef] [PubMed]
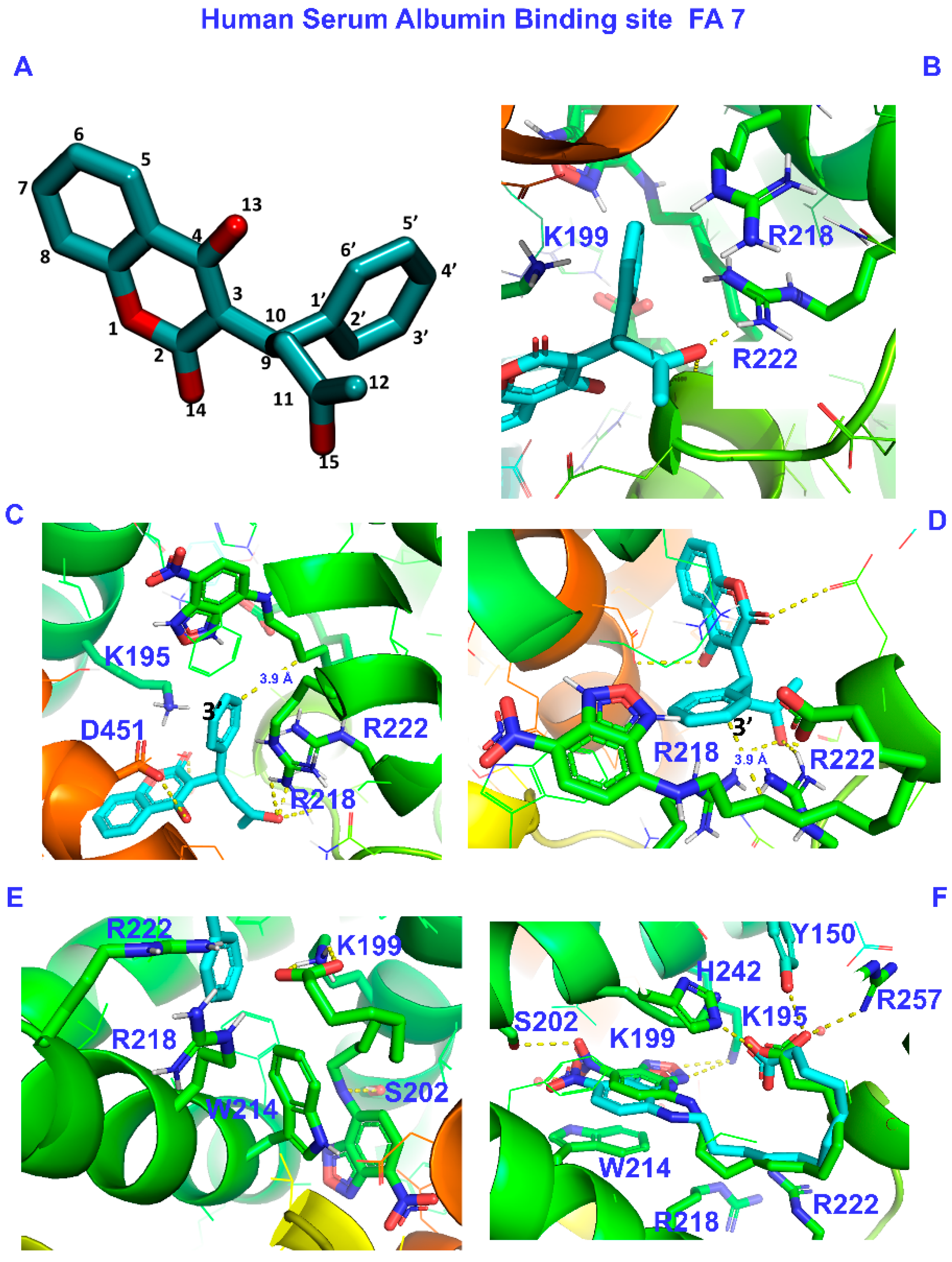
| HSA/Amino Acid Group | Dist. (Å) | Pose /Predicted Affinity (kcal/mol) | |
|---|---|---|---|
| |||
| - | K-199 | 6/−5.5 | |
| O15 | R-218/NH2 η 1 | 3.0 | |
| O15 | R-222/NH2 η 2 | 3.1 | |
| O13 | D-451/OD2 | 3.3 | |
| C4′ | K-195/N ζ | 3.6 | |
| C3′ | NBD/C10 | 3.9 | |
| |||
| COO− | K-199 | 2.1 | 6/−6.5 |
| COO− | R-222 | 3.4 | |
| COO− | R-218 | 4.0 | |
| Aromatic ring-C2′ | W-214 | 3.5 | |
| NH | S-202 | 1.9 |
| FA7 | ||
|---|---|---|
| Ligand | Inner Cluster | External Cluster |
| Tyr-150, His-242, Arg-257 | Lys-195, Lys-199, Arg-218, Arg-222 | |
| DHA (docking) a | His-242, Arg-257 (−7.0 kcal/mol) | Lys-199, Arg-218, Arg-222 (−7.0 kcal/mol) |
| EPA (docking) a | His-242, Arg-257 (−6.7 kcal/mol) | Lys-199, Arg-218, Arg-222 (−6.8 kcal/mol) |
| Warfarin (docking) b | His-242, Arg-257 (−7.0 kcal/mol) | Arg-218, Arg-222 (−7.7 kcal/mol) |
| NBD-C12 (X-ray) | His-242, Ser-202 NO2 | Lys-199, Trp-214 COO− |
| NBD-C12 (docking) | His-242, Ser-202 NO2 | Lys-199, Trp-214 COO− |
| (−7.3 kcal/mol) | ||
| Warfarin in the presence of NBD-C12 (docking) | - | Lys-195, Arg-218, Arg-222 |
| (−5.5 kcal/mol) | ||
| NBD-C12 in the presence of warfarin (docking) | Ser-202, Trp-214 | Lys-199, Arg-218, Arg-222 |
| NO2 | COO− | |
| (−6.5 kcal/mol) | ||
| FA3 | FA4 | ||
|---|---|---|---|
| Cluster-1 | Cluster-2 | ||
| DHA (docking) a | Ser-342, Arg-348, Arg-485 (−8.3 kcal/mol) | Arg-410, Tyr-411 (−7.5 kcal/mol) | Ser-419, Thr-422 (−7.8 kcal/mol) |
| EPA (docking) a | Ser-342, Arg-348, Arg-485 (−7.9 kcal/mol) | Arg-410, Tyr-411 (−7.0 kcal/mol) | Ser-419, Thr-422 (−7.8 kcal/mol) |
| Ibuprofen (docking) b | Ser-342, Arg-348, Arg-485 (−7.2 kcal/mol) | Arg-410, Tyr-411 (−7.3 kcal/mol) | |
| Ibuprofen (X-ray) | Arg-410, Tyr-411 | ||
| NBD-C12 (X-ray) | Ser-342, Arg-348, Arg-485 COO− | Arg-410, Tyr-411 NO2 | |
| NBD-C12 (docking) | Ser-342, Arg-348, Arg-485 COO− | Arg-410, Tyr-411 NO2 | |
| (−8.3 kcal/mol) | |||
| Ibuprofen in the presence of NBD-C12 (docking) | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venianakis, T.; Primikyri, A.; Opatz, T.; Petry, S.; Papamokos, G.; Gerothanassis, I.P. NMR and Docking Calculations Reveal Novel Atomistic Selectivity of a Synthetic High-Affinity Free Fatty Acid vs. Free Fatty Acids in Sudlow’s Drug Binding Sites in Human Serum Albumin. Molecules 2023, 28, 7991. https://doi.org/10.3390/molecules28247991
Venianakis T, Primikyri A, Opatz T, Petry S, Papamokos G, Gerothanassis IP. NMR and Docking Calculations Reveal Novel Atomistic Selectivity of a Synthetic High-Affinity Free Fatty Acid vs. Free Fatty Acids in Sudlow’s Drug Binding Sites in Human Serum Albumin. Molecules. 2023; 28(24):7991. https://doi.org/10.3390/molecules28247991
Chicago/Turabian StyleVenianakis, Themistoklis, Alexandra Primikyri, Till Opatz, Stefan Petry, Georgios Papamokos, and Ioannis P. Gerothanassis. 2023. "NMR and Docking Calculations Reveal Novel Atomistic Selectivity of a Synthetic High-Affinity Free Fatty Acid vs. Free Fatty Acids in Sudlow’s Drug Binding Sites in Human Serum Albumin" Molecules 28, no. 24: 7991. https://doi.org/10.3390/molecules28247991





