Carbon Nanostructured Immunosensing of Anti-SARS-CoV-2 S-Protein Antibodies
Abstract
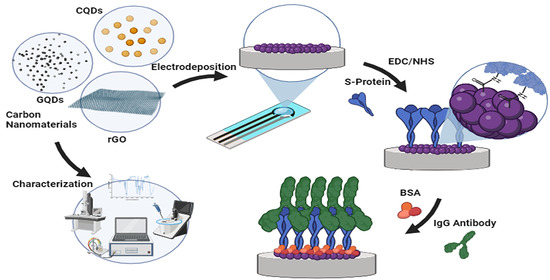
:1. Introduction
2. Results and Discussion
2.1. Selected Characterization of Graphene Quantum Dots (GQDs)
2.2. High-Resolution Transmission Electron Microscopy Characterization of Carbon Nanomaterials
2.3. Characterization of Electrochemical Carbon Nanomaterial SARS-CoV-2 Immunosensor
2.4. Assay Optimization
2.5. Carbon Nanomaterial-Modified SARS-CoV-2 Immunosensor Label-Free Detection of SARS-CoV-2 Antibodies
2.6. Reproducibility Study for the Detection of SARS-CoV-2-Specific IgG Antibody via GQD-Modified SARS-CoV-2 Immunosensor
2.7. SARS-CoV-2 Antibody Detection Immunosensor Using Other Carbon Nanomaterials
2.8. GQD-Modified SARS-CoV-2 Immunosensor Label-Free Detection of SARS-CoV-2 Antibodies in Real Sample
3. Materials and Instrumentation
3.1. Materials and Chemicals
3.2. Instrumentation
3.3. Synthesis of Graphene Quantum Dots
3.4. Synthesis of Carbon Quantum Dots
3.5. Fabrication of GQD-Functionalized Screen-Printed-Carbon Electrode Immunosensor
3.6. Electrochemical Detection of SARS-CoV-2 IgG and IgM Antibodies Using Carbon Nanomaterial-Modified SPCE’s
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Alimohamadi, Y.; Sepandi, M.; Taghdir, M.; Hosamirudsari, H. Determine the most common clinical symptoms in COVID-19 patients: A systematic review and meta-analysis. J. Prev. Med. Hyg. 2020, 61, E304–E312. [Google Scholar] [CrossRef] [PubMed]
- Giri, B.; Pandey, S.; Shrestha, R.; Pokharel, K.; Ligler, F.S.; Neupane, B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2020, 413, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, M. Recent progress in electrochemical immunosensors. Biosensors 2021, 11, 360. [Google Scholar] [CrossRef] [PubMed]
- Police Patil, A.V.; Chuang, Y.S.; Li, C.; Wu, C.C. Recent Advances in Electrochemical Immunosensors with Nanomaterial Assistance for Signal Amplification. Biosensors 2023, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Du Plooy, J.; Jahed, N.; Iwuoha, E.; Pokpas, K. Advances in paper-based electrochemical immunosensors: Review of fabrication strategies and biomedical applications. R. Soc. Open Sci. 2023, 10, 230940. [Google Scholar] [CrossRef]
- Wang, S.; Shu, J.; Lyu, A.; Huang, X.; Zeng, W.; Jin, T.; Cui, H. Label-Free Immunoassay for Sensitive and Rapid Detection of the SARS-CoV-2 Antigen Based on Functionalized Magnetic Nanobeads with Chemiluminescence and Immunoactivity. Anal. Chem. 2021, 93, 14238–14246. [Google Scholar] [CrossRef]
- Malla, P.; Liu, C.; Wu, W.; Kabinsing, P. Talanta Synthesis and Characterization of Au-Decorated Graphene Oxide Nanocomposite for Magneto-Electrochemical Detection of SARS-CoV-2 Nucleocapsid Gene. Talanta 2023, 262, 124701. [Google Scholar] [CrossRef]
- Farsaeivahid, N.; Grenier, C.; Nazarian, S.; Wang, M.L. A Rapid Label-Free Disposable Electrochemical Salivary Point-of-Care Sensor for SARS-CoV-2 Detection and Quantification. Sensors 2022, 23, 433. [Google Scholar] [CrossRef]
- Kumar, T.H.V.; Srinivasan, S.; Krishnan, V.; Vaidyanathan, R.; Anand, K.; Natarajan, S.; Veerapandian, M. Peptide-Based Direct Electrochemical Detection of Receptor Binding Domains of SARS-CoV-2 Spike Protein in Pristine Samples. Sens. Actuators B. Chem. 2023, 377, 133052. [Google Scholar] [CrossRef]
- Nascimento, E.D.; Fonseca, W.T.; Faça, V.M.; De Morais, B.P.; Silvestrini, V.C.; Pott-junior, H.; Teixeira, F.R.; Faria, R.C. COVID-19 Diagnosis by SARS-CoV-2 Spike Protein Detection in Saliva Using an Ultrasensitive Magneto-Assay Based on Disposable Electrochemical Sensor. Sens. Actuators B Chem. 2022, 353, 131128. [Google Scholar] [CrossRef] [PubMed]
- Amouzadeh, M.; Fern, J.P.; Mojena, D.; Acedo, P. An Ultrasensitive Molecularly Imprinted Polymer-Based Electrochemical Sensor for the Determination of SARS-CoV-2-RBD by Using Macroporous Gold Screen-Printed Electrode. Biosens. Bioelectron. 2022, 196, 113729. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Peng, Y.; Wang, L.; Wang, M.; Zhou, T.; Xiang, L.; Li, J.; Yang, J.; Li, G. Analytica Chimica Acta Target-Triggered Cascade Signal Amplification for Sensitive Electrochemical Detection of SARS-CoV-2 with Clinical Application. Anal. Chim. Acta 2022, 1208, 339846. [Google Scholar] [CrossRef] [PubMed]
- Ayankojo, A.G.; Boroznjak, R.; Reut, J.; Opik, A. Molecularly Imprinted Polymer Based Electrochemical Sensor for Quantitative Detection of SARS-CoV-2 Spike Protein. Sens. Actuators B Chem. 2022, 353, 131160. [Google Scholar] [CrossRef]
- Wu, P.; Liu, L.; Morgan, S.P.; Correia, R.; Korposh, S. (INVITED) Label-Free Detection of Antibodies Using Functionalised Long Period Grating Optical Fibre Sensors. Results Opt. 2021, 5, 100172. [Google Scholar] [CrossRef]
- Braz, B.A.; Hospinal-santiani, M.; Martins, G.; Gogola, J.L.; Thomaz-soccol, V.; Soccol, C.R. Gold-Binding Peptide as a Selective Layer for Electrochemical Detection of SARS-CoV-2 Antibodies. Talanta 2023, 257, 124348. [Google Scholar] [CrossRef]
- Timilsina, S.S.; Durr, N.; Jolly, P.; Ingber, D.E. Rapid Quantitation of SARS-CoV-2 Antibodies in Clinical Samples with an Electrochemical Sensor. Biosens. Bioelectron. 2023, 223, 115037. [Google Scholar] [CrossRef]
- Peng, R.; Pan, Y.; Li, Z.; Qin, Z.; Rini, J.M.; Liu, X. SPEEDS: A Portable Serological Testing Platform for Rapid Electrochemical Detection of SARS-CoV-2 Antibodies. Biosens. Bioelectron. 2022, 197, 113762. [Google Scholar] [CrossRef]
- Masterson, A.N.; Muhoberac, B.B.; Gopinadhan, A.; Wilde, D.J.; Deiss, F.T.; John, C.C.; Sardar, R. Multiplexed and High-Throughput Label-Free Detection of RNA/Spike Protein/IgG/IgM Biomarkers of SARS-CoV-2 Infection Utilizing Nanoplasmonic Biosensors. Anal. Chem. 2021, 93, 8754–8763. [Google Scholar] [CrossRef]
- Najjar, D.; Rainbow, J.; Timilsina, S.S.; Jolly, P.; De Puig, H.; Yafia, M.; Durr, N.; Sallum, H.; Alter, G.; Li, J.Z.; et al. A Lab-on-a-Chip for the Concurrent Electrochemical Detection of SARS-CoV-2 RNA and Anti-SARS-CoV-2 Antibodies in Saliva and Plasma. Nat. Biomed. Eng. 2022, 6, 968–978. [Google Scholar] [CrossRef]
- Rashed, M.Z.; Kopechek, J.A.; Priddy, M.C.; Hamorsky, K.T.; Palmer, K.E.; Mittal, N.; Valdez, J.; Flynn, J.; Williams, S.J. Rapid Detection of SARS-CoV-2 Antibodies Using Electrochemical Impedance-Based Detector. Biosens. Bioelectron. 2021, 171, 112709. [Google Scholar] [CrossRef] [PubMed]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef]
- Pokpas, K.; Jahed, N.; Bezuidenhout, P.; Smith, S.; Land, K.; Iwuoha, E. Nickel contamination analysis at cost-effective silver printed paper-based electrodes based on carbon black dimethylglyoxime ink as electrode modifier. J. Electrochem. Sci. Eng. 2022, 12, 153–164. [Google Scholar] [CrossRef]
- Ghaffari, N.; Pokpas, K.; Iwuoha, E.; Jahed, N. Sensitive Electrochemical Determination of Bisphenol a Using a Disposable, Electrodeposited Antimony-Graphene Nanocomposite Pencil Graphite Electrode (PGE) and Differential Pulse Voltammetry (DPV). Anal. Lett. 2023, 1–18. [Google Scholar] [CrossRef]
- Power, A.C.; Gorey, B.; Chandra, S.; Chapman, J. Carbon Nanomaterials and Their Application to Electrochemical Sensors: A Review. Nanotechnol. Rev. 2018, 7, 19–41. [Google Scholar] [CrossRef]
- Speranza, G. Carbon Nanomaterials: Synthesis, Functionalization and Sensing Applications. Nanomaterials 2021, 11, 967. [Google Scholar] [CrossRef] [PubMed]
- Tachi, S.; Morita, H.; Takahashi, M.; Okabayashi, Y.; Hosokai, T.; Sugai, T.; Kuwahara, S. Quantum Yield Enhancement in Graphene Quantum Dots via Esterification with Benzyl Alcohol. Sci. Rep. 2019, 9, 14115. [Google Scholar] [CrossRef]
- Hasan, M.T.; Gonzalez-Rodriguez, R.; Ryan, C.; Faerber, N.; Coffer, J.L.; Naumov, A.V. Photo-and Electroluminescence from Nitrogen-Doped and Nitrogen–Sulfur Codoped Graphene Quantum Dots. Adv. Funct. Mater. 2018, 28, 1804337. [Google Scholar] [CrossRef]
- Yuan, X.; Liu, Z.; Guo, Z.; Ji, Y.; Jin, M.; Wang, X. Cellular Distribution and Cytotoxicity of Graphene Quantum Dots with Different Functional Groups. Nanoscale Res. Lett. 2014, 9, 108. [Google Scholar] [CrossRef]
- Fan, T.; Zeng, W.; Tang, W.; Yuan, C.; Tong, S.; Cai, K.; Liu, Y.; Huang, W.; Min, Y.; Epstein, A.J. Controllable Size-Selective Method to Prepare Graphene Quantum Dots from Graphene Oxide. Nanoscale Res. Lett. 2015, 10, 55. [Google Scholar] [CrossRef]
- Choudhary, R.P.; Shukla, S.; Vaibhav, K.; Pawar, P.B.; Saxena, S. Optical Properties of Few Layered Graphene Quantum Dots. Mater. Res. Express 2015, 2, 095024. [Google Scholar] [CrossRef]
- Ramachandran, S.; Sathishkumar, M.; Kothurkar, N.K.; Senthilkumar, R. Synthesis and Characterization of Graphene Quantum Dots/Cobalt Ferrite Nanocomposite. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2018. [Google Scholar] [CrossRef]
- Ye, R.; Xiang, C.; Lin, J.; Peng, Z.; Huang, K.; Yan, Z.; Cook, N.P.; Samuel, E.L.G.; Hwang, C.C.; Ruan, G.; et al. Coal as an Abundant Source of Graphene Quantum Dots. Nat. Commun. 2013, 4, 2943. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, V.A.; Kaur, R.; Kumar, N.; Deep, A.; Rajesh, C.; Kim, K.H. Synthesis and Spectroscopic Studies of Functionalized Graphene Quantum Dots with Diverse Fluorescence Characteristics. RSC Adv. 2018, 8, 11446–11454. [Google Scholar] [CrossRef] [PubMed]
- Nezhad-Mokhtari, P.; Arsalani, N.; Ghorbani, M.; Hamishehkar, H. Development of Biocompatible Fluorescent Gelatin Nanocarriers for Cell Imaging and Anticancer Drug Targeting. J. Mater. Sci. 2018, 53, 10679–10691. [Google Scholar] [CrossRef]
- Kumar, S.; Ojha, A.K.; Ahmed, B.; Kumar, A.; Das, J.; Materny, A. Tunable (Violet to Green) Emission by High-Yield Graphene Quantum Dots and Exploiting Its Unique Properties towards Sun-Light-Driven Photocatalysis and Supercapacitor Electrode Materials. Mater. Today Commun. 2017, 11, 76–86. [Google Scholar] [CrossRef]
- Chung, P.P.; Wang, J.; Durandet, Y. Deposition Processes and Properties of Coatings on Steel Fasteners—A Review. Friction 2019, 7, 389–416. [Google Scholar] [CrossRef]
- Hou, J.; Bei, F.; Wang, M.; Ai, S. Electrochemical Determination of Malachite Green at Graphene Quantum Dots-Gold Nanoparticles Multilayers-Modified Glassy Carbon Electrode. J. Appl. Electrochem. 2013, 43, 689–696. [Google Scholar] [CrossRef]
- Raghav, R.; Srivastava, S. Immobilization Strategy for Enhancing Sensitivity of Immunosensors: L-Asparagine–AuNPs as a Promising Alternative of EDC–NHS Activated Citrate–AuNPs for Antibody Immobilization. Biosens. Bioelectron. 2016, 78, 396–403. [Google Scholar] [CrossRef]
- Wigman, L.; Remarchuk, T.; Gomez, S.; Kumar, A.; Dong, M.W.; Medley, C.D. Byproducts of commonly used coupling reagents: Origin, toxicological evaluation and methods for determination. Am. Pharm. Rev. 2014, 17, 1–16. [Google Scholar]
- Qin, D.; Jiang, X.; Mo, G.; Feng, J.; Yu, C.; Deng, B. A Novel Carbon Quantum Dots Signal Amplification Strategy Coupled with Sandwich Electrochemiluminescence Immunosensor for the Detection of Ca15-3 in Human Serum. ACS Sens. 2019, 4, 504–512. [Google Scholar] [CrossRef]










| Replication | Spiked (ng/mL) | Detected (ng/mL) | RSD% |
|---|---|---|---|
| 1 | 5 | 5.22 | 104.4 |
| 2 | 5 | 5.16 | 103.2 |
| 3 | 5 | 4.51 | 90.2 |
| Platform | Target Antibody | Slope | R2 | LOD (ng/mL) | Reference |
|---|---|---|---|---|---|
| Pept/AuNP-SPCE | Anti-S | 1.059 | 0.984 | 75 | [16] |
| prGOx-Au-coated multiplex sensor chip | Anti-SARS-CoV-2 | N/A | N/A | N/A | [17] |
| Streptavidin/biotin SPCE | ALP-anti human IgG and IgM | N/A | N/A | 1.64 | [18] |
| Au-TNP wells | anti human IgG and IgM | N/A | N/A | 89 aM | [19] |
| Multiplexed sensor chip | anti human IgG | N/A | N/A | 2.3 copies/µL | [20] |
| Interdigitated Au | Anti-SARS-CoV-2 monoclonal | N/A | 0.9 | 1000 | [21] |
| GO-paper-based carbone electrode | S-protein IgG and IgM | 6.1137 | 0.995 | 0.14 | [22] |
| SPCE | S-protein IgG | 0.1046 | 0.9129 | 41.140 | This work |
| GQD-SPCE | S-protein IgG | 2.1216 | 0.9885 | 2.028 | This work |
| CQD-SPCE | S-protein IgG | 0.9677 | 0.9429 | 4.447 | This work |
| rGO-SPCE | S-protein IgG | 0.5807 | 0.9740 | 7.410 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
du Plooy, J.; Kock, B.; Jahed, N.; Iwuoha, E.; Pokpas, K. Carbon Nanostructured Immunosensing of Anti-SARS-CoV-2 S-Protein Antibodies. Molecules 2023, 28, 8022. https://doi.org/10.3390/molecules28248022
du Plooy J, Kock B, Jahed N, Iwuoha E, Pokpas K. Carbon Nanostructured Immunosensing of Anti-SARS-CoV-2 S-Protein Antibodies. Molecules. 2023; 28(24):8022. https://doi.org/10.3390/molecules28248022
Chicago/Turabian Styledu Plooy, Jarid, Branham Kock, Nazeem Jahed, Emmanuel Iwuoha, and Keagan Pokpas. 2023. "Carbon Nanostructured Immunosensing of Anti-SARS-CoV-2 S-Protein Antibodies" Molecules 28, no. 24: 8022. https://doi.org/10.3390/molecules28248022