Future Antimicrobials: Natural and Functionalized Phenolics
Abstract
:1. Introduction
1.1. Classification of Major Classes of Antibacterial Phenolics
1.2. Biosynthesis, Natural and Synthetic Sources
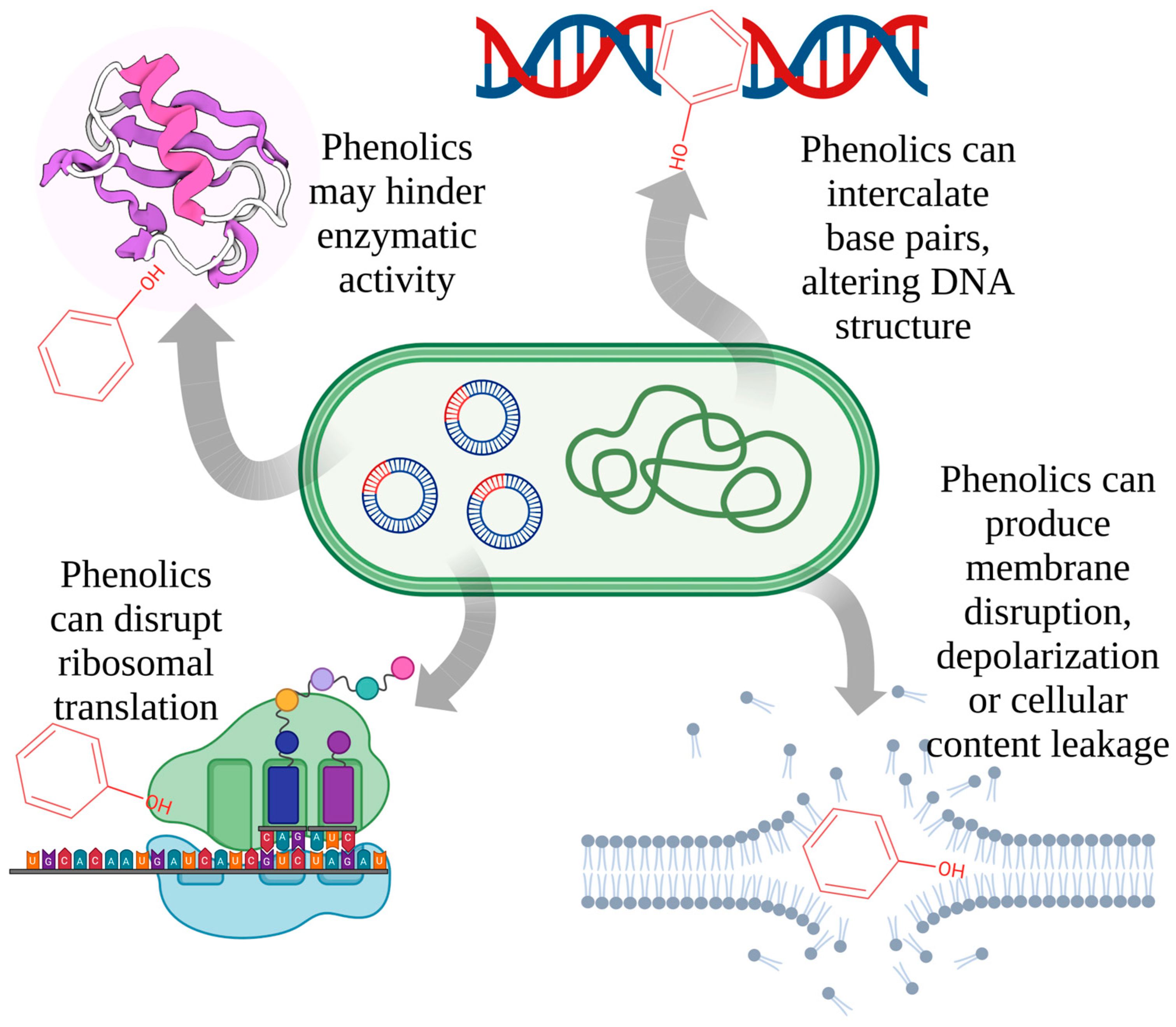
2. Antibacterial Activity
2.1. Antibacterial Activity of Phenolic Acids
2.2. Antibacterial Activity of Flavonoids
2.3. Antibacterial Activity of Lignans
2.4. Antibacterial Activity of Stilbenes
2.5. Antibacterial Activity of Tannins
2.6. Potential Mechanisms of Actions
3. Pharmacodynamics
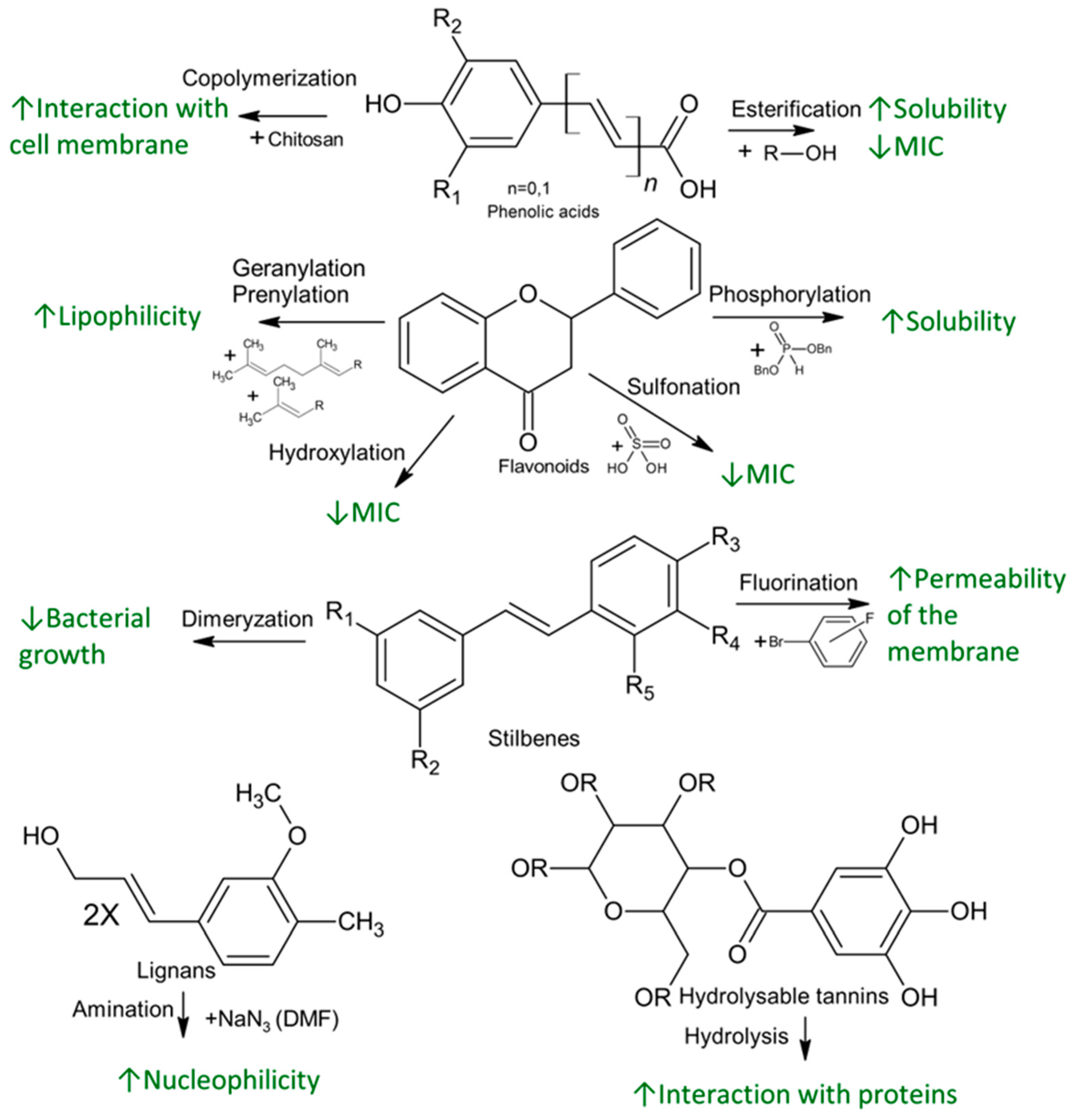
4. Chemical Functionalization and Derivatization of Natural Phenolics for Enhanced Antimicrobial Activity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rana, A.; Samtiya, M.; Dhewa, T.; Mishra, V.; Aluko, R.E. Health benefits of polyphenols: A concise review. J. Food Biochem. 2022, 46, e14264. [Google Scholar] [CrossRef] [PubMed]
- Vuolo, M.M.; Lima, V.S.; Maróstica Junior, M.R. Phenolic Compounds: Structure, Classification, and Antioxidant Power. In Bioactive Compounds: Health Benefits and Potential Applications; Woodhead Publishing: Sawston, UK, 2019; pp. 33–50. [Google Scholar]
- Olszowy, M. What is responsible for antioxidant properties of polyphenolic compounds from plants? Plant Physiol. Biochem. 2019, 144, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Bors, W.; Michel, C. Chemistry of the Antioxidant Effect of Polyphenols. Ann. New York Acad. Sci. 2002, 957, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Makarewicz, M.; Drożdż, I.; Tarko, T.; Duda-Chodak, A. The Interactions between Polyphenols and Microorganisms, Especially Gut Microbiota. Antioxidants 2021, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Belščak-Cvitanović, A.; Durgo, K.; Huđek, A.; Bačun-Družina, V.; Komes, D. Overview of polyphenols and their properties. In Polyphenols: Properties, Recovery, and Applications; Galanakis, C.M., Ed.; Woodhead Publishing: Sawston, UK, 2018; pp. 3–44. [Google Scholar] [CrossRef]
- Bouarab-Chibane, L.; Forquet, V.; Lantéri, P.; Clément, Y.; Léonard-Akkari, L.; Oulahal, N.; Degraeve, P.; Bordes, C. Antibacterial Properties of Polyphenols: Characterization and QSAR (Quantitative Structure–Activity Relationship) Models. Front. Microbiol. 2019, 10, 829. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Chen, J.; Xiao, A.; Liu, L. Antibacterial Activity of Polyphenols: Structure-Activity Relationship and Influence of Hyperglycemic Condition. Molecules 2017, 22, 1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Ni, X.; Kai, G.; Chen, X. A Review on Structure–Activity Relationship of Dietary Polyphenols Inhibiting α-Amylase. Crit. Rev. Food Sci. Nutr. 2013, 53, 497–506. [Google Scholar] [CrossRef]
- Friedman, M.; Henika, P.R.; Mandrell, R.E. Antibacterial Activities of Phenolic Benzaldehydes and Benzoic Acids against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 2003, 66, 1811–1821. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Yamauchi, S.; Masuda, K.; Nishiwaki, H.; Akiyama, K.; Maruyama, M.; Sugahara, T.; Kishida, T.; Koba, Y. Antimicrobial Activity of Stereoisomers of Butane-Type Lignans. Biosci. Biotechnol. Biochem. 2009, 73, 1806–1810. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.N.; Siji, J.V.; Nambisan, B.; Mohandas, C. Activity and synergistic interactions of stilbenes and antibiotic combinations against bacteria in vitro. World J. Microbiol. Biotechnol. 2012, 28, 3143–3150. [Google Scholar] [CrossRef]
- Farha, A.K.; Yang, Q.-Q.; Kim, G.; Li, H.-B.; Zhu, F.; Liu, H.-Y.; Gan, R.-Y.; Corke, H. Tannins as an alternative to antibiotics. Food Biosci. 2020, 38, 100751. [Google Scholar] [CrossRef]
- Rispail, N.; Morris, P.; Webb, K.J. Phenolic Compounds: Extraction and analysis. In Lotus Japonicus Handbook; Springer: Berlin/Heidelberg, Germany, 2006; pp. 349–354. [Google Scholar] [CrossRef]
- Banjarnahor, S.D.S.; Artanti, N. Antioxidant properties of flavonoids. Med. J. Indones. 2015, 23, 239–244. [Google Scholar] [CrossRef] [Green Version]
- Tsao, R. Chemistry and Biochemistry of Dietary Polyphenols. Nutrients 2010, 2, 1231–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saltveit, M.E. Synthesis and Metabolism of Phenolic Compounds. In Fruit and Vegetable Phytochemicals; Wiley-Blackwell: Oxford, UK, 2009; pp. 89–100. [Google Scholar] [CrossRef]
- de la Rosa, L.A.; Moreno-Escamilla, J.O.; Rodrigo-García, J.; Alvarez-Parrilla, E. Phenolic Compounds. In Postharvest Physiology and Biochemistry of Fruits and Vegetables; Elsevier: Amsterdam, The Netherlands, 2019; pp. 253–271. [Google Scholar]
- Zhang, Y.; Wei, J.; Qiu, Y.; Niu, C.; Song, Z.; Yuan, Y.; Yue, T. Structure-Dependent Inhibition of Stenotrophomonas maltophilia by Polyphenol and Its Impact on Cell Membrane. Front. Microbiol. 2019, 10, 2646. [Google Scholar] [CrossRef]
- Sánchez-Maldonado, A.; Schieber, A.; Gänzle, M. Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J. Appl. Microbiol. 2011, 111, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Pernin, A.; Guillier, L.; Dubois-Brissonnet, F. Inhibitory activity of phenolic acids against Listeria monocytogenes: Deciphering the mechanisms of action using three different models. Food Microbiol. 2019, 80, 18–24. [Google Scholar] [CrossRef]
- Cueva, C.; Moreno-Arribas, M.V.; Martín-Álvarez, P.J.; Bills, G.; Vicente, M.F.; Basilio, A.; Rivas, C.L.; Requena, T.; Rodríguez, J.M.; Bartolomé, B. Antimicrobial activity of phenolic acids against commensal, probiotic and pathogenic bacteria. Res. Microbiol. 2010, 161, 372–382. [Google Scholar] [CrossRef]
- Kahkeshani, N.; Farzaei, F.; Fotouhi, M.; Alavi, S.S.; Bahramsoltani, R.; Naseri, R.; Momtaz, S.; Abbasabadi, Z.; Rahimi, R.; Farzaei, M.H.; et al. Pharmacological effects of gallic acid in health and disease: A mechanistic review. Iran. J. Basic. Med. Sci. 2019, 22, 225–237. [Google Scholar] [CrossRef]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial Activity and Mode of Action of Ferulic and Gallic Acids Against Pathogenic Bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef]
- Kouassi, Y.; Shelef, L.A. Inhibition of Listeria Monocytogenes by Cinnamic Acid: Possible Interaction of the Acid with Cysteinyl Residues. J. Food Saf. 1998, 18, 231–242. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control. 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Oteiza, P.I.; Erlejman, A.G.; Verstraeten, S.V.; Keen, C.L.; Fraga, C.G. Flavonoid-membrane Interactions: A Protective Role of Flavonoids at the Membrane Surface? Clin. Dev. Immunol. 2005, 12, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Pandey, A.K. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renzetti, A.; Betts, J.W.; Fukumoto, K.; Rutherford, R.N. Antibacterial green tea catechins from a molecular perspective: Mechanisms of action and structure–activity relationships. Food Funct. 2020, 11, 9370–9396. [Google Scholar] [CrossRef] [PubMed]
- Abuga, I.; Sulaiman, S.F.; Wahab, R.A.; Ooi, K.L.; Rasad, M.S.B.A. In vitro antibacterial effect of the leaf extract of Murraya koenigii on cell membrane destruction against pathogenic bacteria and phenolic compounds identification. Eur. J. Integr. Med. 2020, 33, 101010. [Google Scholar] [CrossRef]
- Khameneh, B.; Iranshahy, M.; Soheili, V.; Bazzaz, B.S.F. Review on plant antimicrobials: A mechanistic viewpoint. Antimicrob. Resist. Infect. Control. 2019, 8, 1–28. [Google Scholar] [CrossRef]
- Rafał, I.G.; Króliczewski, B.J.; Górniak, I.; Bartoszewski, R.; Króliczewski, Á.J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2018, 18, 10471. [Google Scholar] [CrossRef] [Green Version]
- Yuan, G.; Guan, Y.; Yi, H.; Lai, S.; Sun, Y.; Cao, S. Antibacterial activity and mechanism of plant flavonoids to gram-positive bacteria predicted from their lipophilicities. Sci. Rep. 2021, 11, 10471. [Google Scholar] [CrossRef]
- Tsou, L.K.; Lara-Tejero, M.; RoseFigura, J.; Zhang, Z.J.; Wang, Y.-C.; Yount, J.S.; Lefebre, M.; Dossa, P.D.; Kato, J.; Guan, F.; et al. Antibacterial Flavonoids from Medicinal Plants Covalently Inactivate Type III Protein Secretion Substrates. J. Am. Chem. Soc. 2016, 138, 2209–2218. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-C.; Huang, C.-Y. Inhibition of Klebsiella Pneumoniae DnaB Helicase by the Flavonol Galangin. Protein J. 2011, 30, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Biharee, A.; Sharma, A.; Kumar, A.; Jaitak, V. Antimicrobial flavonoids as a potential substitute for overcoming antimicrobial resistance. Fitoterapia 2020, 146, 104720. [Google Scholar] [CrossRef] [PubMed]
- McGillick, B.E.; Kumaran, D.; Vieni, C.; Swaminathan, S. β-Hydroxyacyl-acyl Carrier Protein Dehydratase (FabZ) from Francisella tularensis and Yersinia pestis: Structure Determination, Enzymatic Characterization, and Cross-Inhibition Studies. Biochemistry 2016, 55, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ji, Y.R.; Ryoo, Z.Y.; Choi, M.-S.; Woo, E.-R.; Lee, D.G. Antibacterial Mechanism of (−)-Nortrachelogenin in Escherichia coli O157. Curr. Microbiol. 2016, 72, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ren, J.; Li, Z. Antibacterial activity and mechanism of pinoresinol from Cinnamomum Camphora leaves against food-related bacteria. Food Control. 2017, 79, 192–199. [Google Scholar] [CrossRef]
- Favela-Hernández, J.M.J.; García, A.; Garza-González, E.; Rivas-Galindo, V.M.; Camacho-Corona, M.R. Antibacterial and Antimycobacterial Lignans and Flavonoids from Larrea tridentata. Phytotherapy Res. 2012, 26, 1957–1960. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.J.M.; Yu, Y.; Champer, J.; Kim, J. Resveratrol Demonstrates Antimicrobial Effects Against Propionibacterium acnes In Vitro. Dermatol Ther (Heidelb). 2014, 4, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Hua, X.; Xue, Z.; Ma, J. Cajanin Stilbene Acid Inhibited Vancomycin-Resistant Enterococcus by Inhibiting Phosphotransferase System. Front. Pharmacol. 2020, 11, 473. [Google Scholar] [CrossRef] [Green Version]
- Chung, K.-T.; Lu, Z.; Chou, M. Mechanism of inhibition of tannic acid and related compounds on the growth of intestinal bacteria. Food Chem. Toxicol. 1998, 36, 1053–1060. [Google Scholar] [CrossRef]
- Engels, C.; Gänzle, M.G.; Schieber, A. Fractionation of Gallotannins from Mango (Mangifera indica L.) Kernels by High-Speed Counter-Current Chromatography and Determination of Their Antibacterial Activity. J. Agric. Food Chem. 2010, 58, 775–780. [Google Scholar] [CrossRef]
- Lakey-Beitia, J.; Burillo, A.M.; La Penna, G.; Hegde, M.L.; Rao, K. Polyphenols as Potential Metal Chelation Compounds Against Alzheimer’s Disease. J. Alzheimer’s Dis. 2021, 82, S335–S357. [Google Scholar] [CrossRef] [PubMed]
- Tintino, S.R.; Morais-Tintino, C.D.; Campina, F.F.; Costa, M.D.S.; Menezes, I.R.; de Matos, Y.M.L.; Calixto-Júnior, J.T.; Pereira, P.S.; Siqueira-Junior, J.P.; Leal-Balbino, T.C.; et al. Tannic acid affects the phenotype of Staphylococcus aureus resistant to tetracycline and erythromycin by inhibition of efflux pumps. Bioorganic Chem. 2017, 74, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Olchowik-Grabarek, E.; Sękowski, S.; Kwiatek, A.; Płaczkiewicz, J.; Abdulladjanova, N.; Shlyonsky, V.; Swiecicka, I.; Zamaraeva, M. The Structural Changes in the Membranes of Staphylococcus aureus Caused by Hydrolysable Tannins Witness Their Antibacterial Activity. Membranes 2022, 12, 1124. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, C.; Song, L.; Li, T.; Cui, S.; Zhang, L.; Jia, Y. Antimicrobial activity and mechanism of Larch bark procyanidins against Staphylococcus aureus. Acta Biochim. et Biophys. Sin. 2017, 49, 1058–1066. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Efenberger-Szmechtyk, M.; Nowak, A.; Czyzowska, A. Plant extracts rich in polyphenols: Antibacterial agents and natural preservatives for meat and meat products. Crit. Rev. Food Sci. Nutr. 2021, 61, 149–178. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, J.; Zhang, J.; Xu, D.; Li, Y.; Liu, Y.; Zhang, X.; Zhang, R.; Wu, Z.; Weng, P. Antimicrobial Effect of Tea Polyphenols against Foodborne Pathogens: A Review. J. Food Prot. 2021, 84, 1801–1808. [Google Scholar] [CrossRef]
- Guo, L.; Sun, Q.; Gong, S.; Bi, X.; Jiang, W.; Xue, W.; Fei, P. Antimicrobial Activity and Action Approach of the Olive Oil Polyphenol Extract Against Listeria monocytogenes. Front. Microbiol. 2019, 10, 1586. [Google Scholar] [CrossRef] [Green Version]
- Piekarska-Radzik, L.; Klewicka, E. Mutual influence of polyphenols and Lactobacillus spp. bacteria in food: A review. Eur. Food Res. Technol. 2021, 247, 9–24. [Google Scholar] [CrossRef]
- Lin, S.; Wang, Z.; Lam, K.-L.; Zeng, S.; Tan, B.K.; Hu, J. Role of intestinal microecology in the regulation of energy metabolism by dietary polyphenols and their metabolites. Food Nutr. Res. 2019, 63, 1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Othman, L.; Sleiman, A.; Abdel-Massih, R.M. Antimicrobial Activity of Polyphenols and Alkaloids in Middle Eastern Plants. Front. Microbiol. 2019, 10, 911. [Google Scholar] [CrossRef] [PubMed]
- Kardum, N.; Glibetic, M. Polyphenols and Their Interactions with Other Dietary Compounds: Implications for Human Health. 2018, 84, 103–144. [CrossRef]
- Röttig, A.; Steinbüchel, A. Acyltransferases in Bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 277–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, H.; Subasi, B.G.; Celebioglu, H.U.; Ozdal, T.; Capanoglu, E. Chemistry of Protein-Phenolic Interactions Toward the Microbiota and Microbial Infections. Front. Nutr. 2022, 9, 914118. [Google Scholar] [CrossRef]
- Hossain, M.A.; Lee, S.J.; Park, N.H.; Mechesso, A.F.; Birhanu, B.T.; Kang, J.; Reza, A.; Suh, J.W.; Park, S.C. Impact of phenolic compounds in the acyl homoserine lactone-mediated quorum sensing regulatory pathways. Sci. Rep. 2017, 7, 10618. [Google Scholar] [CrossRef] [Green Version]
- Alvarado-Martinez, Z.; Bravo, P.; Kennedy, N.-F.; Krishna, M.; Hussain, S.; Young, A.C.; Biswas, D. Antimicrobial and Antivirulence Impacts of Phenolics on Salmonella Enterica Serovar Typhimurium. Antibiotics 2020, 9, 668. [Google Scholar] [CrossRef]
- Tapiero, H.; Tew, K.; Ba, G.N.; Mathé, G. Polyphenols: Do they play a role in the prevention of human pathologies? Biomed. Pharmacother. 2002, 56, 200–207. [Google Scholar] [CrossRef]
- Teng, H.; Chen, L. Polyphenols and bioavailability: An update. Crit. Rev. Food Sci. Nutr. 2019, 59, 2040–2051. [Google Scholar] [CrossRef]
- D’Archivio, M.; Filesi, C.; Varì, R.; Scazzocchio, B.; Masella, R. Bioavailability of the Polyphenols: Status and Controversies. Int. J. Mol. Sci. 2010, 11, 1321–1342. [Google Scholar] [CrossRef]
- Tarko, T.; Duda-Chodak, A.; Zajac, N. Digestion and absorption of phenolic compounds assessed by in vitro simulation methods. A review. Rocz. Panstw. Zakl. Hig. 2013, 64, 79–84. [Google Scholar]
- Balzerani, F.; Hinojosa-Nogueira, D.; Cendoya, X.; Blasco, T.; Pérez-Burillo, S.; Apaolaza, I.; Francino, M.P.; Rufián-Henares, J.; Planes, F.J. Prediction of degradation pathways of phenolic compounds in the human gut microbiota through enzyme promiscuity methods. NPJ Syst. Biol. Appl. 2022, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Avila, J.A.; Wall-Medrano, A.; Velderrain-Rodríguez, G.; Chen, C.-Y.O.; Salazar-López, N.J.; Robles-Sánchez, M.; González-Aguilar, G.A. Gastrointestinal interactions, absorption, splanchnic metabolism and pharmacokinetics of orally ingested phenolic compounds. Food Funct. 2017, 8, 15–38. [Google Scholar] [CrossRef] [PubMed]
- Merkl, R.; Hrádková, I.; Filip, V.; Šmidrkal, J. Antimicrobial and antioxidant properties of phenolic acids alkyl esters. Czech J. Food Sci. 2010, 28, 275–279. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xie, M.; Ma, G.; Fang, Y.; Yang, W.; Ma, N.; Fang, D.; Hu, Q.; Pei, F. The antioxidant and antimicrobial activities of different phenolic acids grafted onto chitosan. Carbohydr. Polym. 2019, 225, 115238. [Google Scholar] [CrossRef]
- Osorio, M.; Carvajal, M.; Vergara, A.; Butassi, E.; Zacchino, S.; Mascayano, C.; Montoya, M.; Mejías, S.; Martín, M.; Vásquez-Martínez, Y. Prenylated Flavonoids with Potential Antimicrobial Activity: Synthesis, Biological Activity, and In Silico Study. Int. J. Mol. Sci. 2021, 22, 5472. [Google Scholar] [CrossRef]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Shah, S.A.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial Effects of Flavonoids and Their Structure-Activity Relationship Study: A Comparative Interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef]
- Osonga, F.J.; Onyango, J.O.; Mwilu, S.K.; Noah, N.M.; Schulte, J.; An, M.; Sadik, O.A. Synthesis and characterization of novel flavonoid derivatives via sequential phosphorylation of quercetin. Tetrahedron Lett. 2017, 58, 1474–1479. [Google Scholar] [CrossRef]
- Osonga, F.J.; Akgul, A.; Miller, R.M.; Eshun, G.B.; Yazgan, I.; Akgul, A.; Sadik, O.A. Antimicrobial Activity of a New Class of Phosphorylated and Modified Flavonoids. ACS Omega 2019, 4, 12865–12871. [Google Scholar] [CrossRef] [Green Version]
- Kopacz, M. Quercetin- and Morinsulfonates as Analytical Reagents. J. Anal. Chem. 2003, 58, 225–229. [Google Scholar] [CrossRef]
- Supriya, C.; Sivareddy, C.; Basaveswarao, M.; Kalyani, C.; Deviswetha, V. Evaluation of Anti Bacterial Activity and Anti-Inflammatory Activity of Diastase Conjugated Naringin. IOSR J. Pharm. 2017, 7, 46–52. [Google Scholar]
- Tago, R.; Yamauchi, S.; Maruyama, M.; Akiyama, K.; Sugahara, T.; Kishida, T.; Koba, Y. Structure-Antibacterial Activity Relationship for 9-O,9′-O-Demethyl (+)-virgatusin. Biosci. Biotechnol. Biochem. 2008, 72, 1032–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, S.; Horbach, R.; Deising, H.B.; Siewert, B.; Csuk, R. Synthesis and antimicrobial activity of (E) stilbene derivatives. Bioorganic Med. Chem. 2011, 19, 5155–5166. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chauhan, N.; Koli, M.; Nayak, S.K.; Subramanian, M. Dimer stilbene, a resveratrol analogue exhibits synergy with antibiotics that target protein synthesis in eradicating Staphylococcus aureus infection. Biochimie 2022, 201, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Silva, J.; Kim, M.; Jung, Y. Enhanced antioxidant capacity and antimicrobial activity of tannic acid by thermal processing. Food Chem. 2010, 118, 740–746. [Google Scholar] [CrossRef]
- Widsten, P.; Heathcote, C.; Kandelbauer, A.; Guebitz, G.; Nyanhongo, G.S.; Prasetyo, E.N.; Kudanga, T. Enzymatic surface functionalisation of lignocellulosic materials with tannins for enhancing antibacterial properties. Process. Biochem. 2010, 45, 1072–1081. [Google Scholar] [CrossRef]
| Polyphenols | General Structure | Representative Compounds | Antibacterial Activity |
|---|---|---|---|
| Phenolic acids |  |
| Antibacterial effect increases significantly with pH values [10]. |
| a. Benzoic acid derivatives | |||
| b. Derivates of cinnamic acid |  |
| |
| Flavonoids |  |
| Flavonoids act against bacteria such as S. aureus and P. aeruginosa with a very low minimum inhibitory concentration (MIC) value (0.062 µg/mL) [11]. |
| Lignans | 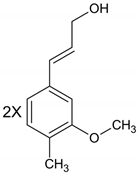 |
| Due to structural properties, antibacterial activity of lignans is influenced by the stereochemistry of molecules [12]. |
| Stilbenes | 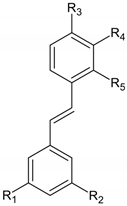 |
| In combination with antibiotics, some stilbenes can be useful in treating infections caused by multidrug-resistance bacteria [13]. |
| Tannins |  |
| Tannin compounds act against bacteria, causing disintegration of bacterial colonies, by interfering with the bacterial cell wall and inhibiting fatty acid biosynthesis pathways [14]. |
| a. Hydrolysable tannins | |||
| b. Nonhydrolysable tannins (Condensed tannins) |  | ||
| c. Pseudotannins | 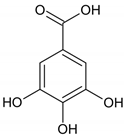 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobiuc, A.; Pavăl, N.-E.; Mangalagiu, I.I.; Gheorghiță, R.; Teliban, G.-C.; Amăriucăi-Mantu, D.; Stoleru, V. Future Antimicrobials: Natural and Functionalized Phenolics. Molecules 2023, 28, 1114. https://doi.org/10.3390/molecules28031114
Lobiuc A, Pavăl N-E, Mangalagiu II, Gheorghiță R, Teliban G-C, Amăriucăi-Mantu D, Stoleru V. Future Antimicrobials: Natural and Functionalized Phenolics. Molecules. 2023; 28(3):1114. https://doi.org/10.3390/molecules28031114
Chicago/Turabian StyleLobiuc, Andrei, Naomi-Eunicia Pavăl, Ionel I. Mangalagiu, Roxana Gheorghiță, Gabriel-Ciprian Teliban, Dorina Amăriucăi-Mantu, and Vasile Stoleru. 2023. "Future Antimicrobials: Natural and Functionalized Phenolics" Molecules 28, no. 3: 1114. https://doi.org/10.3390/molecules28031114
APA StyleLobiuc, A., Pavăl, N.-E., Mangalagiu, I. I., Gheorghiță, R., Teliban, G.-C., Amăriucăi-Mantu, D., & Stoleru, V. (2023). Future Antimicrobials: Natural and Functionalized Phenolics. Molecules, 28(3), 1114. https://doi.org/10.3390/molecules28031114






.png)


