A Review of the Regulatory Mechanisms of N-Myc on Cell Cycle
Abstract
1. Introduction
2. The Target Genes of N-Myc
2.1. POU5F1
2.2. PRMT1
2.3. VRK1
2.4. SKP2
2.5. PTK2
2.6. DKC1
2.7. MDM2
2.8. FOXM1
2.9. PLK1
2.10. PLAGL2
3. Another Seven N-Myc Target Genes
3.1. GLDC
3.2. TERT
3.3. PBK
3.4. SGO1
3.5. AURKB
3.6. E2F5
3.7. TEAD4
4. Discussion
| General Mode of Action | Compound | Structure | Target | Cell-Free Assay | Cell Data | Clinical Trails |
|---|---|---|---|---|---|---|
| Prevent the abnormal expression of MYCN | (+)-JQ1 | 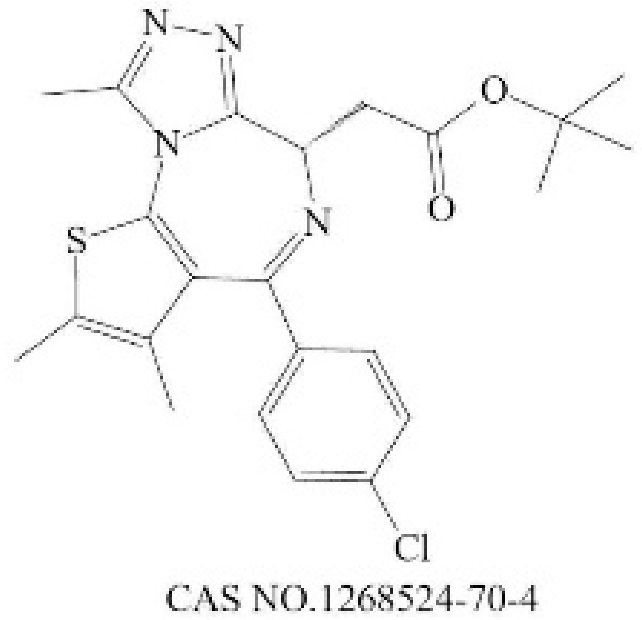 | BRD4(1/2) [224] | 77/33 (1 h) [224] | 4 (NMC 11060, 72 h) [224] | N |
| THZ1 | 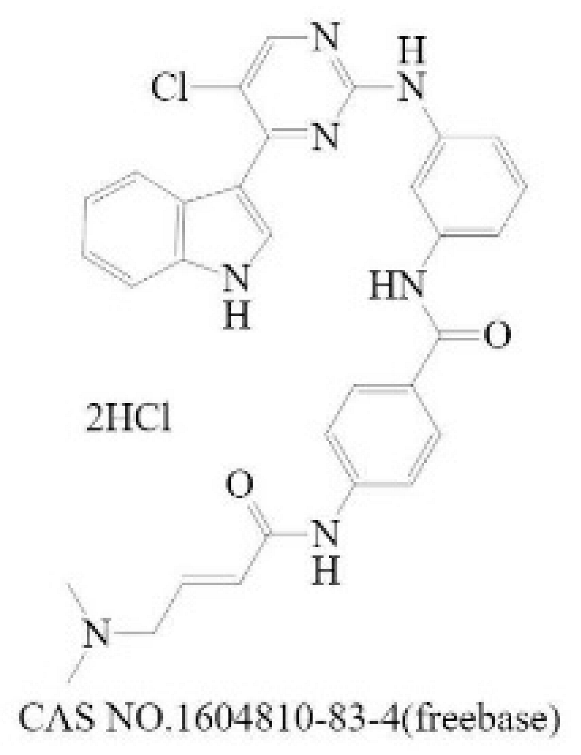 | CDK7 [225] | 3.2 (3 h) [225] | 50 (Jurkat cells, 72 h) [226] | N | |
| CYC065 | 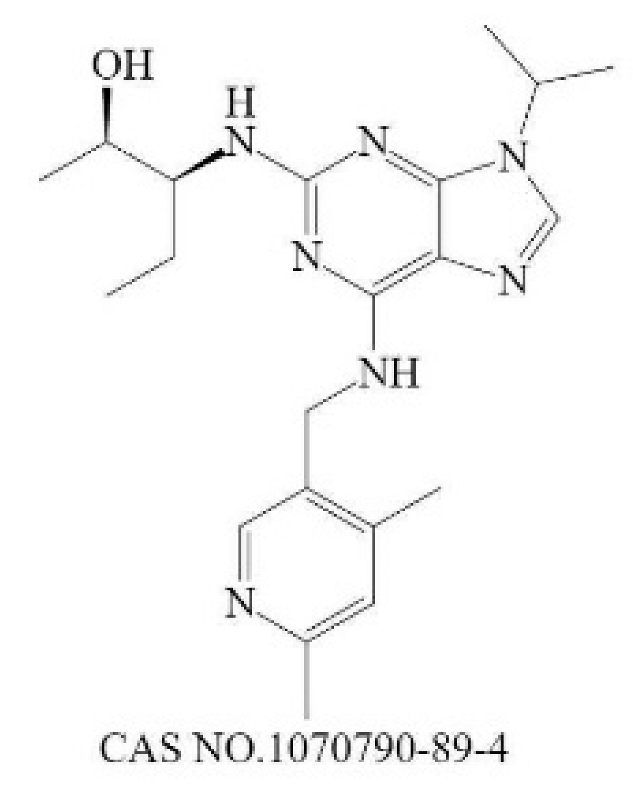 | CDK9 [227] | 26 a [227] | 370 (Hop63 24 h) [228] | Phase 1/2: Solid Tumor, Adult Lymphoma (Recruiting) [229] | |
| Promote the degradation of N-Myc | EPZ015666 |  | PRMT5 [230] | 22 (120 h) [231] | 96 (Z-138, 12 days) [231] | N |
| BI 2536 | 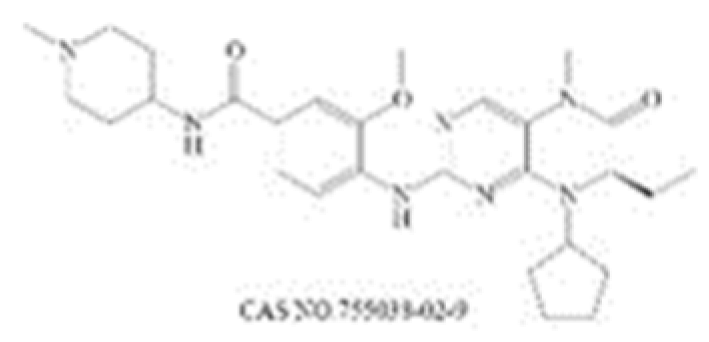 | PLK1 [232] | 0.83 (45 min) [232] | 1.78 (NALM-6, 72 h) [233] | Phase 1: NSCLC; advanced solid tumours; Pancreatic Neoplasms; Non-Hodgkin’s Lymphoma Phase 2: AML; Prostatic Neoplasms; NSCLC; SCLC; Pancreatic Cancer; Breast Cancer/Endometrial Cancer/Head and Neck Cancer/Melanoma (Skin)/Ovarian Cancer/Sarcoma [234] | |
| Inhibit the formation of heterodimers between N-Myc and MAX | MYCi361 |  | MYC [221] | 3200 (Kd) [221] | 490 (SK-NB2, 120 h) [221] | N |
| Inhibit downstream target of N-Myc | Nutlin-3 | 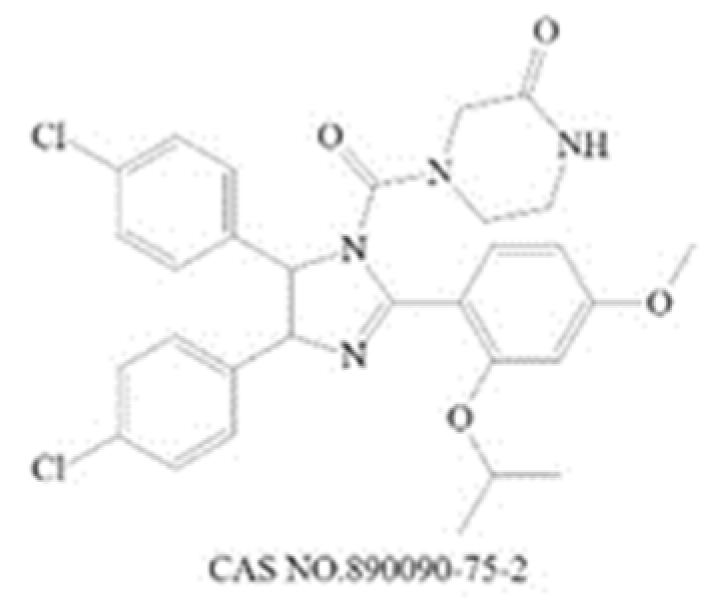 | Mdm2 [235] | 90 (1 h) [236] | 650 (U87MG,48 h) [237] | N |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef]
- Tomioka, N.; Oba, S.; Ohira, M.; Misra, A.; Fridlyand, J.; Ishii, S.; Nakamura, Y.; Isogai, E.; Hirata, T.; Yoshida, Y.; et al. Novel risk stratification of patients with neuroblastoma by genomic signature, which is independent of molecular signature. Oncogene 2008, 27, 441–449. [Google Scholar] [CrossRef]
- Cheung, N.K.; Zhang, J.; Lu, C.; Parker, M.; Bahrami, A.; Tickoo, S.K.; Heguy, A.; Pappo, A.S.; Federico, S.; Dalton, J.; et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 2012, 307, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Hertwig, F.; Roels, F.; Dreidax, D.; Gartlgruber, M.; Menon, R.; Krämer, A.; Roncaioli, J.L.; Sand, F.; Heuckmann, J.M.; et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015, 526, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.; Gastier-Foster, J.M.; Mann, M.; Naranjo, A.H.; van Ryn, C.; Bagatell, R.; Matthay, K.K.; London, W.B.; Irwin, M.S.; Shimada, H.; et al. Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: A report from the Children’s Oncology Group. Cancer 2017, 123, 4224–4235. [Google Scholar] [CrossRef] [PubMed]
- Marrano, P.; Irwin, M.S.; Thorner, P.S. Heterogeneity of MYCN amplification in neuroblastoma at diagnosis, treatment, relapse, and metastasis. Genes Chromosomes Cancer 2017, 56, 28–41. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Grady, S.; Tang, M.; Crown, J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021, 94, 102154. [Google Scholar] [CrossRef]
- Beaulieu, M.E.; Castillo, F.; Soucek, L. Structural and Biophysical Insights into the Function of the Intrinsically Disordered Myc Oncoprotein. Cells 2020, 9, 1038. [Google Scholar] [CrossRef]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef]
- Beltran, H. The N-myc Oncogene: Maximizing its Targets, Regulation, and Therapeutic Potential. Mol. Cancer Res. 2014, 12, 815–822. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.S.; Clarke, S.; Veschi, V.; Thiele, C.J. Targeting MYCN in Pediatric and Adult Cancers. Front. Oncol. 2020, 10, 623679. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, A.; Schwab, M. The mycN/max protein complex in neuroblastoma. Short review. Eur. J. Cancer 1995, 31, 516–519. [Google Scholar] [CrossRef]
- Murphy, D.M.; Buckley, P.G.; Bryan, K.; Das, S.; Alcock, L.; Foley, N.H.; Prenter, S.; Bray, I.; Watters, K.M.; Higgins, D.; et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS ONE 2009, 4, e8154. [Google Scholar] [CrossRef]
- Sun, Y.; Bell, J.L.; Carter, D.; Gherardi, S.; Poulos, R.C.; Milazzo, G.; Wong, J.W.; Al-Awar, R.; Tee, A.E.; Liu, P.Y.; et al. WDR5 Supports an N-Myc Transcriptional Complex That Drives a Protumorigenic Gene Expression Signature in Neuroblastoma. Cancer Res. 2015, 75, 5143–5154. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; AlTahan, A.M.; Hu, D.; Wang, Y.; Cheng, P.H.; Morton, C.L.; Qu, C.; Nathwani, A.C.; Shohet, J.M.; Fotsis, T.; et al. The role of histone demethylase KDM4B in Myc signaling in neuroblastoma. J. Natl. Cancer Inst. 2015, 107, djv080. [Google Scholar] [CrossRef]
- Amente, S.; Milazzo, G.; Sorrentino, M.C.; Ambrosio, S.; di Palo, G.; Lania, L.; Perini, G.; Majello, B. Lysine-specific demethylase (LSD1/KDM1A) and MYCN cooperatively repress tumor suppressor genes in neuroblastoma. Oncotarget 2015, 6, 14572–14583. [Google Scholar] [CrossRef]
- Zeid, R.; Lawlor, M.A.; Poon, E.; Reyes, J.M.; Fulciniti, M.; Lopez, M.A.; Scott, T.G.; Nabet, B.; Erb, M.A.; Winter, G.E.; et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 2018, 50, 515–523. [Google Scholar] [CrossRef]
- Agarwal, S.; Milazzo, G.; Rajapakshe, K.; Bernardi, R.; Chen, Z.; Barbieri, E.; Koster, J.; Perini, G.; Coarfa, C.; Shohet, J.M. MYCN acts as a direct co-regulator of p53 in MYCN amplified neuroblastoma. Oncotarget 2018, 9, 20323–20338. [Google Scholar] [CrossRef]
- O’Brien, E.M.; Selfe, J.L.; Martins, A.S.; Walters, Z.S.; Shipley, J.M. The long non-coding RNA MYCNOS-01 regulates MYCN protein levels and affects growth of MYCN-amplified rhabdomyosarcoma and neuroblastoma cells. BMC Cancer 2018, 18, 217. [Google Scholar] [CrossRef]
- Tchakarska, G.; Sola, B. The double dealing of cyclin D1. Cell Cycle 2020, 19, 163–178. [Google Scholar] [CrossRef]
- Tatum, N.J.; Endicott, J.A. Chatterboxes: The structural and functional diversity of cyclins. Semin. Cell Dev. Biol. 2020, 107, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Nie, L.; Wei, W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2021, 28, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Balboni, N.; Palma, A.; Aleo, E.; Sanna, P.P.; Perini, G.; Giorgi, F.M. Single-Cell Gene Network Analysis and Transcriptional Landscape of MYCN-Amplified Neuroblastoma Cell Lines. Biomolecules 2021, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Li, C.; Su, Y.; Fang, W.; Zhong, C.; Ji, W.; Zhang, Q.; Su, C. Transcription factor OCT4 promotes cell cycle progression by regulating CCND1 expression in esophageal carcinoma. Cancer Lett. 2014, 354, 77–86. [Google Scholar] [CrossRef]
- Schoenhals, M.; Kassambara, A.; de Vos, J.; Hose, D.; Moreaux, J.; Klein, B. Embryonic stem cell markers expression in cancers. Biochem. Biophys. Res. Commun. 2009, 383, 157–162. [Google Scholar] [CrossRef]
- Suenaga, Y.; Nakatani, K.; Nakagawara, A. De novo evolved gene product NCYM in the pathogenesis and clinical outcome of human neuroblastomas and other cancers. Jpn. J. Clin. Oncol. 2020, 50, 839–846. [Google Scholar] [CrossRef]
- Yang, L.; Zheng, J.; Xu, T.; Xiao, X. Downregulation of OCT4 promotes differentiation and inhibits growth of BE (2)-C human neuroblastoma I-type cells. Oncol. Rep. 2013, 29, 2191–2196. [Google Scholar] [CrossRef]
- Su, C. Survivin in survival of hepatocellular carcinoma. Cancer Lett. 2016, 379, 184–190. [Google Scholar] [CrossRef]
- Bai, M.; Yuan, M.; Liao, H.; Chen, J.; Xie, B.; Yan, D.; Xi, X.; Xu, X.; Zhang, Z.; Feng, Y. OCT4 pseudogene 5 upregulates OCT4 expression to promote proliferation by competing with miR-145 in endometrial carcinoma. Oncol. Rep. 2015, 33, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Han, S.M.; Han, S.H.; Coh, Y.R.; Jang, G.; Chan Ra, J.; Kang, S.K.; Lee, H.W.; Youn, H.Y. Enhanced proliferation and differentiation of Oct4- and Sox2-overexpressing human adipose tissue mesenchymal stem cells. Exp. Mol. Med. 2014, 46, e101. [Google Scholar] [CrossRef] [PubMed]
- Card, D.A.; Hebbar, P.B.; Li, L.; Trotter, K.W.; Komatsu, Y.; Mishina, Y.; Archer, T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008, 28, 6426–6438. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Ying, S.Y.; Leu, D.; Wu, D.T. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010, 70, 9473–9482. [Google Scholar] [CrossRef]
- Schoeftner, S.; Scarola, M.; Comisso, E.; Schneider, C.; Benetti, R. An Oct4-pRb axis, controlled by MiR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells 2013, 31, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.J.; Lin, M.; Li, C.X.; Liu, H.; Gong, C.J. A comprehensive review of the roles of E2F1 in colon cancer. Am. J. Cancer Res. 2020, 10, 757–768. [Google Scholar]
- Pennycook, B.R.; Barr, A.R. Restriction point regulation at the crossroads between quiescence and cell proliferation. FEBS Lett. 2020, 594, 2046–2060. [Google Scholar] [CrossRef] [PubMed]
- She, S.; Wei, Q.; Kang, B.; Wang, Y.J. Cell cycle and pluripotency: Convergence on octamer-binding transcription factor 4 (Review). Mol. Med. Rep. 2017, 16, 6459–6466. [Google Scholar] [CrossRef] [PubMed]
- Wierstra, I.; Alves, J. Transcription and pluripotency: Convergence on octamer-binding transcrip factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol. Chem. 2006, 387, 949–962. [Google Scholar] [CrossRef]
- Zhao, R.; Deibler, R.W.; Lerou, P.H.; Ballabeni, A.; Heffner, G.C.; Cahan, P.; Unternaehrer, J.J.; Kirschner, M.W.; Daley, G.Q. A nontranscriptional role for Oct4 in the regulation of mitotic entry. Proc. Natl. Acad. Sci. USA 2014, 111, 15768–15773. [Google Scholar] [CrossRef]
- Yu, K.R.; Yang, S.R.; Jung, J.W.; Kim, H.; Ko, K.; Han, D.W.; Park, S.B.; Choi, S.W.; Kang, S.K.; Scholer, H.; et al. CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem Cells 2012, 30, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; Su, P.F.; Huang, Y.F.; Yew, T.L.; Hung, S.C. Oct4 and Nanog Directly Regulate Dnmt1 to Maintain Self-Renewal and Undifferentiated State in Mesenchymal Stem Cells. Mol. Cell 2012, 47, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Chen, T.; He, L.; Ma, N.; Li, J.A.; Rong, Y.F.; Fang, Y.; Liu, M.; Xie, D.; Lou, W. PRMT1 promotes pancreatic cancer growth and predicts poor prognosis. Cell. Oncol. 2020, 43, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.M.; Sun, W.Z.; Fan, X.Z.; Xu, Y.L.; Cheng, M.B.; Zhang, Y. Methylation of C/EBPα by PRMT1 Inhibits Its Tumor-Suppressive Function in Breast Cancer. Cancer Res. 2019, 79, 2865–2877. [Google Scholar] [CrossRef]
- Chuang, C.Y.; Chang, C.P.; Lee, Y.J.; Lin, W.L.; Chang, W.W.; Wu, J.S.; Cheng, Y.W.; Lee, H.; Li, C. PRMT1 expression is elevated in head and neck cancer and inhibition of protein arginine methylation by adenosine dialdehyde or PRMT1 knockdown downregulates proliferation and migration of oral cancer cells. Oncol. Rep. 2017, 38, 1115–1123. [Google Scholar] [CrossRef]
- Valentijn, L.J.; Koster, J.; Haneveld, F.; Aissa, R.A.; van Sluis, P.; Broekmans, M.E.; Molenaar, J.J.; van Nes, J.; Versteeg, R. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. USA 2012, 109, 19190–19195. [Google Scholar] [CrossRef]
- Eberhardt, A.; Hansen, J.N.; Koster, J.; Lotta, L.T., Jr.; Wang, S.; Livingstone, E.; Qian, K.; Valentijn, L.J.; Zheng, Y.G.; Schor, N.F.; et al. Protein arginine methyltransferase 1 is a novel regulator of MYCN in neuroblastoma. Oncotarget 2016, 7, 63629–63639. [Google Scholar] [CrossRef]
- Hua, Z.Y.; Hansen, J.N.; He, M.; Dai, S.K.; Choi, Y.; Fulton, M.D.; Lloyd, S.M.; Szemes, M.; Sen, J.; Ding, H.F.; et al. PRMT1 promotes neuroblastoma cell survival through ATF5. Oncogenesis 2020, 9, 50. [Google Scholar] [CrossRef]
- Klerkx, E.P.; Lazo, P.A.; Askjaer, P. Emerging biological functions of the vaccinia-related kinase (VRK) family. Histol. Histopathol. 2009, 24, 749–759. [Google Scholar] [CrossRef]
- Huang, W.; Cui, X.; Chen, Y.; Shao, M.; Shao, X.; Shen, Y.; Liu, Q.; Wu, M.; Liu, J.; Ni, W.; et al. High VRK1 expression contributes to cell proliferation and survival in hepatocellular carcinoma. Pathol. Res. Pract. 2016, 212, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Ben, Z.; Gong, L.; Qiu, Y. High expression of VRK1 is related to poor prognosis in glioma. Pathol. Res. Pract. 2018, 214, 112–118. [Google Scholar] [CrossRef]
- Colmenero-Repiso, A.; Gómez-Muñoz, M.A.; Rodríguez-Prieto, I.; Amador-Álvarez, A.; Henrich, K.O.; Pascual-Vaca, D.; Okonechnikov, K.; Rivas, E.; Westermann, F.; Pardal, R.; et al. Identification of VRK1 as a New Neuroblastoma Tumor Progression Marker Regulating Cell Proliferation. Cancers 2020, 12, 3465. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.R.; Rodríguez-Pinilla, M.; Vega, F.M.; Rodríguez-Peralto, J.L.; Blanco, S.; Sevilla, A.; Valbuena, A.; Hernández, T.; van Wijnen, A.J.; Li, F.; et al. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol. Cancer Res. 2006, 4, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.H.; Park, D.Y.; Kim, W.; Kim, K.T. VRK1 phosphorylates CREB and mediates CCND1 expression. J. Cell Sci. 2008, 121, 3035–3041. [Google Scholar] [CrossRef]
- Lee, N.; Kwon, J.H.; Kim, Y.B.; Kim, S.H.; Park, S.J.; Xu, W.; Jung, H.Y.; Kim, K.T.; Wang, H.J.; Choi, K.Y. Vaccinia-related kinase 1 promotes hepatocellular carcinoma by controlling the levels of cell cycle regulators associated with G1/S transition. Oncotarget 2015, 6, 30130–30148. [Google Scholar] [CrossRef]
- Valbuena, A.; Lopez-Sanchez, I.; Lazo, P.A. Human VRK1 Is an Early Response Gene and Its Loss Causes a Block in Cell Cycle Progression. PLoS ONE 2008, 3, e1642. [Google Scholar] [CrossRef]
- Ren, Z.; Geng, J.; Xiong, C.; Li, X.; Li, Y.; Li, J.; Liu, H. Downregulation of VRK1 reduces the expression of BANF1 and suppresses the proliferative and migratory activity of esophageal cancer cells. Oncol. Lett. 2020, 20, 1163–1170. [Google Scholar] [CrossRef]
- Jamin, A.; Wicklund, A.; Wiebe, M.S. Cell- and virus-mediated regulation of the barrier-to-autointegration factor’s phosphorylation state controls its DNA binding, dimerization, subcellular localization, and antipoxviral activity. J. Virol. 2014, 88, 5342–5355. [Google Scholar] [CrossRef]
- Nichols, R.J.; Wiebe, M.S.; Traktman, P. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 2006, 17, 2451–2464. [Google Scholar] [CrossRef]
- Aihara, H.; Nakagawa, T.; Mizusaki, H.; Yoneda, M.; Kato, M.; Doiguchi, M.; Imamura, Y.; Higashi, M.; Ikura, T.; Hayashi, T.; et al. Histone H2A T120 Phosphorylation Promotes Oncogenic Transformation via Upregulation of Cyclin D1. Mol. Cell 2016, 64, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Yan, G.; Song, X.; Xie, L.; Zhou, Y.; Li, J.; Hu, X.; Li, Z.; Hu, J.; Zhang, Y.; et al. Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proc. Natl. Acad. Sci. USA 2018, 115, 4678–4683. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Zhang, L.; Duan, Y.; Zhang, M.; Wang, G.; Zhang, J.; Zhao, Z. The differential susceptibilities of MCF-7 and MDA-MB-231 cells to the cytotoxic effects of curcumin are associated with the PI3K/Akt-SKP2-Cip/Kips pathway. Cancer Cell Int. 2014, 14, 126. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Su, H.K.; Yu, Z.H.; Xi, S.Y.; Guo, C.C.; Hu, Z.Y.; Qu, Y.; Cai, H.P.; Zhao, Y.Y.; Zhao, H.F.; et al. Skp2 modulates proliferation, senescence and tumorigenesis of glioma. Cancer Cell Int. 2020, 20, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Du, L.; Ren, Y.; Liu, X.; Jiao, Q.; Cui, D.; Wen, M.; Wang, C.; Wei, G.; Wang, Y.; et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J. Exp. Clin. Cancer Res. 2019, 38, 76. [Google Scholar] [CrossRef]
- Wei, X.; Li, X.; Yan, W.; Zhang, X.; Sun, Y.; Zhang, F. SKP2 Promotes Hepatocellular Carcinoma Progression Through Nuclear AMPK-SKP2-CARM1 Signaling Transcriptionally Regulating Nutrient-Deprived Autophagy Induction. Cell. Physiol. Biochem. 2018, 47, 2484–2497. [Google Scholar] [CrossRef]
- Evans, L.; Chen, L.; Milazzo, G.; Gherardi, S.; Perini, G.; Willmore, E.; Newell, D.R.; Tweddle, D.A. SKP2 is a direct transcriptional target of MYCN and a potential therapeutic target in neuroblastoma. Cancer Lett. 2015, 363, 37–45. [Google Scholar] [CrossRef]
- Muth, D.; Ghazaryan, S.; Eckerle, I.; Beckett, E.; Pöhler, C.; Batzler, J.; Beisel, C.; Gogolin, S.; Fischer, M.; Henrich, K.O.; et al. Transcriptional repression of SKP2 is impaired in MYCN-amplified neuroblastoma. Cancer Res. 2010, 70, 3791–3802. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, C. F-box protein Skp2: A novel transcriptional target of E2F. Oncogene 2006, 25, 2615–2627. [Google Scholar] [CrossRef]
- Binne, U.K.; Classon, M.K.; Dick, F.A.; Wei, W.; Rape, M.; Kaelin, W.G.; Naar, A.M.; Dyson, N.J. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 2007, 9, 225–232. [Google Scholar] [CrossRef]
- Assoian, R.K.; Yung, Y. A reciprocal relationship between Rb and Skp2—Implications for restriction point control, signal transduction to the cell cycle and cancer. Cell Cycle 2008, 7, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Hydbring, P.; Castell, A.; Larsson, L.G. MYC Modulation around the CDK2/p27/SKP2 Axis. Genes. 2017, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Yung, Y.; Walker, J.L.; Roberts, J.M.; Assoian, R.K. A Skp2 autoinduction loop and restriction point control. J. Cell Biol. 2007, 178, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, V.; Vail, P.; Nambiar, R.; Witkiewicz, A.K.; Knudsen, E.S. Functional Determinants of Cell Cycle Plasticity and Sensitivity to CDK4/6 Inhibition. Cancer Res. 2021, 81, 1347–1360. [Google Scholar] [CrossRef]
- Kothapalli, D.; Zhao, L.; Hawthorne, E.A.; Cheng, Y.; Lee, E.; Pure, E.; Assoian, R.K. Hyaluronan and CD44 antagonize mitogen-dependent cyclin D1 expression in mesenchymal cells. J. Cell Biol. 2007, 176, 535–544. [Google Scholar] [CrossRef]
- Zhou, W.; Srinivasan, S.; Nawaz, Z.; Slingerland, J.M. ER alpha, SKP2 and E2F-1 form a feed forward loop driving late ER alpha targets and G1 cell cycle progression. Oncogene 2014, 33, 2341–2353. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.; Lunec, J.; Tweddle, D.A. Cell cycle regulation targets of MYCN identified by gene expression microarrays. Cell Cycle 2007, 6, 1249–1256. [Google Scholar] [CrossRef]
- Cox, B.D.; Natarajan, M.; Stettner, M.R.; Gladson, C.L. New concepts regarding focal adhesion kinase promotion of cell migration and proliferation. J. Cell. Biochem. 2006, 99, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Beierle, E.A.; Trujillo, A.; Nagaram, A.; Kurenova, E.V.; Finch, R.; Ma, X.; Vella, J.; Cance, W.G.; Golubovskaya, V.M. N-MYC regulates focal adhesion kinase expression in human neuroblastoma. J. Biol. Chem. 2007, 282, 12503–12516. [Google Scholar] [CrossRef]
- Stafman, L.L.; Williams, A.P.; Marayati, R.; Aye, J.M.; Markert, H.R.; Garner, E.F.; Quinn, C.H.; Lallani, S.B.; Stewart, J.E.; Yoon, K.J.; et al. Focal Adhesion Kinase Inhibition Contributes to Tumor Cell Survival and Motility in Neuroblastoma Patient-Derived Xenografts. Sci. Rep. 2019, 9, 13259. [Google Scholar] [CrossRef]
- Mitra, S.K.; Schlaepfer, D.D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 18, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Pestell, R.; Guan, J.L. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Mol. Biol. Cell 2001, 12, 4066–4077. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bian, Z.C.; Yee, K.; Chen, B.P.; Chien, S.; Guan, J.L. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol. Cell 2003, 11, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Grammer, J.R.; Nelson, M.A.; Guan, J.L.; Stewart, J.E., Jr.; Gladson, C.L. p27Kip1 and cyclin D1 are necessary for focal adhesion kinase regulation of cell cycle progression in glioblastoma cells propagated in vitro and in vivo in the scid mouse brain. J. Biol. Chem. 2005, 280, 6802–6815. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.T.; Meier, U.T. RNA-guided isomerization of uridine to pseudouridine—Pseudouridylation. RNA Biol. 2014, 11, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Pan, Y.; Jiang, R.; Hou, P.; Shan, H.; Chen, F.; Jiang, T.; Bai, J.; Zheng, J. DKC1 serves as a potential prognostic biomarker for human clear cell renal cell carcinoma and promotes its proliferation, migration and invasion via the NF-κB pathway. Oncol. Rep. 2018, 40, 968–978. [Google Scholar] [CrossRef]
- Miao, F.A.; Chu, K.; Chen, H.R.; Zhang, M.; Shi, P.C.; Bai, J.; You, Y.P. Increased DKC1 expression in glioma and its significance in tumor cell proliferation, migration and invasion. Investig. New Drugs 2019, 37, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.; Tran, S.L.; Maritz, M.F.; Liu, B.; Kong, C.F.; Purgato, S.; Yang, C.; Murray, J.; Russell, A.J.; Flemming, C.L.; et al. MYC-Driven Neuroblastomas Are Addicted to a Telomerase-Independent Function of Dyskerin. Cancer Res. 2016, 76, 3604–3617. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Huang, C.; Liu, H. Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis. PLoS ONE 2012, 7, e43147. [Google Scholar] [CrossRef]
- Bellodi, C.; Krasnykh, O.; Haynes, N.; Theodoropoulou, M.; Peng, G.; Montanaro, L.; Ruggero, D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010, 70, 6026–6035. [Google Scholar] [CrossRef]
- Yoon, A.; Peng, G.; Brandenburger, Y.; Zollo, O.; Xu, W.; Rego, E.; Ruggero, D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 2006, 312, 902–906. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Gu, L.; Zhang, H.; Zhou, M. Crosstalk between MYCN and MDM2-p53 signal pathways regulates tumor cell growth and apoptosis in neuroblastoma. Cell Cycle 2011, 10, 2994–3002. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Yen, C.C.; Hung, G.Y.; Pan, C.C.; Chen, W.M. Gene amplification and tumor grading in parosteal osteosarcoma. J. Chin. Med. Assoc. 2019, 82, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Deb, S.P.; Singh, S.; Deb, S. MDM2 overexpression, activation of signaling networks, and cell proliferation. Subcell. Biochem. 2014, 85, 215–234. [Google Scholar] [CrossRef]
- Slack, A.; Chen, Z.; Tonelli, R.; Pule, M.; Hunt, L.; Pession, A.; Shohet, J.M. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc. Natl. Acad. Sci. USA 2005, 102, 731–736. [Google Scholar] [CrossRef]
- Zhu, S.; Lee, J.S.; Guo, F.; Shin, J.; Perez-Atayde, A.R.; Kutok, J.L.; Rodig, S.J.; Neuberg, D.S.; Helman, D.; Feng, H.; et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 2012, 21, 362–373. [Google Scholar] [CrossRef]
- Brown, D.R.; Thomas, C.A.; Deb, S.P. The human oncoprotein MDM2 arrests the cell cycle: Elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998, 17, 2513–2525. [Google Scholar] [CrossRef]
- Deb, S.P. Cell cycle regulatory functions of the human oncoprotein MDM2. Mol. Cancer Res. 2003, 1, 1009–1016. [Google Scholar]
- Chen, J.; Wu, X.; Lin, J.; Levine, A.J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol. Cell. Biol. 1996, 16, 2445–2452. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.C.; Lozano, G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010, 17, 93–102. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, H.; Hu, W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim. Biophys. Sin. 2014, 46, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiong, Y.; Yarbrough, W.G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998, 92, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, J.; Schreiber-Agus, N.; Liegeois, N.J.; Silverman, A.; Alland, L.; Chin, L.; Potes, J.; Chen, K.; Orlow, I.; Lee, H.W.; et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 1998, 92, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Stott, F.J.; Bates, S.; James, M.C.; McConnell, B.B.; Starborg, M.; Brookes, S.; Palmero, I.; Ryan, K.; Hara, E.; Vousden, K.H.; et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998, 17, 5001–5014. [Google Scholar] [CrossRef]
- Yap, D.B.; Hsieh, J.K.; Chan, F.S.; Lu, X. mdm2: A bridge over the two tumour suppressors, p53 and Rb. Oncogene 1999, 18, 7681–7689. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, H.; Li, M.; Rayburn, E.R.; Agrawal, S.; Zhang, R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene 2005, 24, 7238–7247. [Google Scholar] [CrossRef]
- Bell, L.A.; Ryan, K.M. Life and death decisions by E2F-1. Cell Death Differ. 2004, 11, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Jiang, X.; Peng, X. Forkhead box M1 inhibits endothelial cell apoptosis and cell-cycle arrest through ROS generation. Int. J. Clin. Exp. Pathol. 2018, 11, 4899–4907. [Google Scholar]
- Laoukili, J.; Stahl, M.; Medema, R.H. FoxM1: At the crossroads of ageing and cancer. Biochim. Biophys. Acta 2007, 1775, 92–102. [Google Scholar] [CrossRef]
- Halasi, M.; Gartel, A.L. FOX(M1) news—It is cancer. Mol. Cancer Ther. 2013, 12, 245–254. [Google Scholar] [CrossRef]
- Vanhauwaert, S.; Decaesteker, B.; De Brouwer, S.; Leonelli, C.; Durinck, K.; Mestdagh, P.; Vandesompele, J.; Sermon, K.; Denecker, G.; Van Neste, C.; et al. In silico discovery of a FOXM1 driven embryonal signaling pathway in therapy resistant neuroblastoma tumors. Sci. Rep. 2018, 8, 17468. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, F.C.; O’Sullivan, H. FOXM1 in sarcoma: Role in cell cycle, pluripotency genes and stem cell pathways. Oncotarget 2016, 7, 42792–42804. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Jiang, L.; Wang, C.; Zhao, D.; He, W.; Zhou, K.; Liang, Y. FoxM1 Regulates Proliferation and Apoptosis of Human Neuroblastoma Cell through PI3K/AKT Pathway. Fetal Pediatr. Pathol. 2022, 41, 355–370. [Google Scholar] [CrossRef]
- Costa, R.H. FoxM1 dances with mitosis. Nat. Cell Biol. 2005, 7, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.H.; Kalinichenko, V.V.; Holterman, A.X.; Wang, X. Transcription factors in liver development, differentiation, and regeneration. Hepatology 2003, 38, 1331–1347. [Google Scholar] [CrossRef]
- Leung, T.W.; Lin, S.S.; Tsang, A.C.; Tong, C.S.; Ching, J.C.; Leung, W.Y.; Gimlich, R.; Wong, G.G.; Yao, K.M. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001, 507, 59–66. [Google Scholar] [CrossRef]
- Wang, X.; Quail, E.; Hung, N.J.; Tan, Y.; Ye, H.; Costa, R.H. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc. Natl. Acad. Sci. USA 2001, 98, 11468–11473. [Google Scholar] [CrossRef]
- Lam, E.W.; Brosens, J.J.; Gomes, A.R.; Koo, C.Y. Forkhead box proteins: Tuning forks for transcriptional harmony. Nat. Rev. Cancer 2013, 13, 482–495. [Google Scholar] [CrossRef]
- Fu, Z.; Malureanu, L.; Huang, J.; Wang, W.; Li, H.; van Deursen, J.M.; Tindall, D.J.; Chen, J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 2008, 10, 1076–1082. [Google Scholar] [CrossRef]
- Rizki, A.; Mott, J.D.; Bissell, M.J. Polo-like kinase 1 is involved in invasion through extracellular matrix. Cancer Res. 2007, 67, 11106–11110. [Google Scholar] [CrossRef]
- Bahassi, E. Polo-like kinases and DNA damage checkpoint: Beyond the traditional mitotic functions. Exp. Biol. Med. 2011, 236, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.B.; Yue, M.; Su, H.X.; Ren, P.; Jiang, J.; Li, F.; Hu, Y.F.; Du, H.N.; Liu, H.D.; Qing, G.L. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol. Cell 2016, 64, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Schmucker, S.; Sumara, I. Molecular dynamics of PLK1 during mitosis. Mol. Cell. Oncol. 2014, 1, e954507. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A.R.; Sharma, G.; Chakraborty, C.; Kim, J. PLK-1: Angel or devil for cell cycle progression. Biochim. Biophys. Acta 2016, 1865, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Casenghi, M.; Meraldi, P.; Weinhart, U.; Duncan, P.I.; Korner, R.; Nigg, E.A. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 2003, 5, 113–125. [Google Scholar] [CrossRef]
- Lee, K.; Rhee, K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 2011, 195, 1093–1101. [Google Scholar] [CrossRef]
- Mardin, B.R.; Agircan, F.G.; Lange, C.; Schiebel, E. Plk1 controls the Nek2A-PP1gamma antagonism in centrosome disjunction. Curr. Biol. 2011, 21, 1145–1151. [Google Scholar] [CrossRef]
- Roshak, A.K.; Capper, E.A.; Imburgia, C.; Fornwald, J.; Scott, G.; Marshall, L.A. The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell. Signal. 2000, 12, 405–411. [Google Scholar] [CrossRef]
- Inoue, D.; Sagata, N. The Polo-like kinase Plx1 interacts with and inhibits Myt1 after fertilization of Xenopus eggs. EMBO J. 2005, 24, 1057–1067. [Google Scholar] [CrossRef]
- Watanabe, N.; Arai, H.; Nishihara, Y.; Taniguchi, M.; Watanabe, N.; Hunter, T.; Osada, H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl. Acad. Sci. USA 2004, 101, 4419–4424. [Google Scholar] [CrossRef]
- Yuan, J.; Eckerdt, F.; Bereiter-Hahn, J.; Kurunci-Csacsko, E.; Kaufmann, M.; Strebhardt, K. Cooperative phosphorylation including the activity of polo-like kinase 1 regulates the subcellular localization of cyclin B1. Oncogene 2002, 21, 8282–8292. [Google Scholar] [CrossRef] [PubMed]
- Sumara, I.; Vorlaufer, E.; Stukenberg, P.T.; Kelm, O.; Redemann, N.; Nigg, E.A.; Peters, J.M. The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol. Cell 2002, 9, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, T.S.; Sakuno, T.; Ishiguro, K.; Iemura, S.; Natsume, T.; Kawashima, S.A.; Watanabe, Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 2006, 441, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, H. Functioning mechanisms of Shugoshin-1 in centromeric cohesion during mitosis. Essays Biochem. 2020, 64, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.V.; Loktev, A.V.; Ban, K.H.; Jackson, P.K. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol. Biol. Cell 2004, 15, 5623–5634. [Google Scholar] [CrossRef]
- Moshe, Y.; Boulaire, J.; Pagano, M.; Hershko, A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. USA 2004, 101, 7937–7942. [Google Scholar] [CrossRef]
- Nasmyth, K. Segregating sister genomes: The molecular biology of chromosome separation. Science 2002, 297, 559–565. [Google Scholar] [CrossRef]
- Hornig, N.C.; Uhlmann, F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J. 2004, 23, 3144–3153. [Google Scholar] [CrossRef]
- Kakeno, M.; Matsuzawa, K.; Matsui, T.; Akita, H.; Sugiyama, I.; Ishidate, F.; Nakano, A.; Takashima, S.; Goto, H.; Inagaki, M.; et al. Plk1 Phosphorylates CLIP-170 and Regulates Its Binding to Microtubules for Chromosome Alignment. Cell Struct. Funct. 2014, 39, 45–59. [Google Scholar] [CrossRef]
- Burkard, M.E.; Maciejowski, J.; Rodriguez-Bravo, V.; Repka, M.; Lowery, D.M.; Clauser, K.R.; Zhang, C.; Shokat, K.M.; Carr, S.A.; Yaffe, M.B.; et al. Plk1 Self-Organization and Priming Phosphorylation of HsCYK-4 at the Spindle Midzone Regulate the Onset of Division in Human Cells. PLoS Biol. 2009, 7, e1000111. [Google Scholar] [CrossRef]
- Neef, R.; Gruneberg, U.; Kopajtich, R.; Li, X.L.; Nigg, E.A.; Sillje, H.; Barr, F.A. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 2007, 9, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.K.; Ozlu, N.; Coughlin, M.; Steen, J.J.; Mitchison, T.J. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell 2012, 23, 2702–2711. [Google Scholar] [CrossRef] [PubMed]
- Wezensky, S.J.; Hanks, T.S.; Wilkison, M.J.; Ammons, M.C.; Siemsen, D.W.; Gauss, K.A. Modulation of PLAGL2 transactivation by positive cofactor 2 (PC2), a component of the ARC/Mediator complex. Gene 2010, 452, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, L.; Liu, R.; Mo, H.; Niu, Y.; Chen, T.; Wang, Y.; Han, S.; Tu, K.; Liu, Q. Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1α signaling loop promotes hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 2021, 40, 72. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Wiedemeyer, R.; Yan, H.; Quayle, S.N.; Ivanova, E.V.; Paik, J.H.; Zhang, H.; Xiao, Y.; Perry, S.R.; et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 2010, 17, 497–509. [Google Scholar] [CrossRef]
- Li, N.; Li, D.; Du, Y.; Su, C.; Yang, C.; Lin, C.; Li, X.; Hu, G. Overexpressed PLAGL2 transcriptionally activates Wnt6 and promotes cancer development in colorectal cancer. Oncol. Rep. 2019, 41, 875–884. [Google Scholar] [CrossRef]
- Landrette, S.F.; Kuo, Y.H.; Hensen, K.; van Waalwijk van Doorn-Khosrovani, S.B.; Perrat, P.N.; van de Ven, W.J.; Delwel, R.; Castilla, L.H. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood 2005, 105, 2900–2907. [Google Scholar] [CrossRef]
- Zhao, Z.; Shelton, S.D.; Oviedo, A.; Baker, A.L.; Bryant, C.P.; Omidvarnia, S.; Du, L. The PLAGL2/MYCN/miR-506-3p interplay regulates neuroblastoma cell fate and associates with neuroblastoma progression. J. Exp. Clin. Cancer Res. 2020, 39, 41. [Google Scholar] [CrossRef]
- Hanks, T.S.; Gauss, K.A. Pleomorphic adenoma gene-like 2 regulates expression of the p53 family member, p73, and induces cell cycle block and apoptosis in human promonocytic U937 cells. Apoptosis 2012, 17, 236–247. [Google Scholar] [CrossRef]
- Zheng, G.; Ning, J.; Yang, Y.C. PLAGL2 controls the stability of Pirh2, an E3 ubiquitin ligase for p53. Biochem. Biophys. Res. Commun. 2007, 364, 344–350. [Google Scholar] [CrossRef]
- Celeghin, A.; Giunco, S.; Freguja, R.; Zangrossi, M.; Nalio, S.; Dolcetti, R.; De Rossi, A. Short-term inhibition of TERT induces telomere length-independent cell cycle arrest and apoptotic response in EBV-immortalized and transformed B cells. Cell Death Dis. 2016, 7, e2562. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesh, S.; Banerjee, P.P. Telomerase reverse transcriptase regulates the expression of a key cell cycle regulator, cyclin D1. Biochem. Biophys. Res. Commun. 2006, 347, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.M.; Buckley, P.G.; Bryan, K.; Watters, K.M.; Koster, J.; van Sluis, P.; Molenaar, J.; Versteeg, R.; Stallings, R.L. Dissection of the oncogenic MYCN transcriptional network reveals a large set of clinically relevant cell cycle genes as drivers of neuroblastoma tumorigenesis. Mol. Carcinog. 2011, 50, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, W.; Wei, L.; Jin, J.; Tang, K.; Li, C.; Teh, B.T.; Chen, X. The effect of Aurora kinases on cell proliferation, cell cycle regulation and metastasis in renal cell carcinoma. Int. J. Oncol. 2012, 41, 2139–2149. [Google Scholar] [CrossRef] [PubMed]
- Giet, R.; Glover, D.M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001, 152, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Kallio, M.J.; McCleland, M.L.; Stukenberg, P.T.; Gorbsky, G.J. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 2002, 12, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Alptekin, A.; Ye, B.; Yu, Y.; Poole, C.J.; van Riggelen, J.; Zha, Y.; Ding, H.F. Glycine decarboxylase is a transcriptional target of MYCN required for neuroblastoma cell proliferation and tumorigenicity. Oncogene 2019, 38, 7504–7520. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Sun, L.; Wang, X.; Chen, J.; Jia, M.; Zou, Y.; Sa, H.; Cai, Y.; Xu, Y.; Sun, C.; et al. Identification of a new GLDC gene alternative splicing variant and its protumorigenic roles in lung cancer. Future Oncol. 2019, 15, 4127–4139. [Google Scholar] [CrossRef]
- Kume, A.K.H.; Sakakibara, T.; Ishiguro, Y.; Kure, S.; Hiraga, K. The glycine cleavage system. Molecular cloning of the chicken and human glycine decarboxylase cDNAs and some characteristics involved in the deduced protein structures. J. Biol. Chem. 1991, 26, 3323–3329. [Google Scholar] [CrossRef]
- Tibbetts, A.S.; Appling, D.R. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010, 30, 57–81. [Google Scholar] [CrossRef]
- Murakami-Tonami, Y.; Ikeda, H.; Yamagishi, R.; Inayoshi, M.; Inagaki, S.; Kishida, S.; Komata, Y.; Jan, K.; Takeuchi, I.; Kondo, Y.; et al. SGO1 is involved in the DNA damage response in MYCN-amplified neuroblastoma cells. Sci. Rep. 2016, 6, 31615. [Google Scholar] [CrossRef]
- Chen, Q.; Wan, X.; Chen, Y.; Liu, C.; Gu, M.; Wang, Z. SGO1 induces proliferation and metastasis of prostate cancer through AKT-mediated signaling pathway. Am. J. Cancer Res. 2019, 9, 2693–2705. [Google Scholar] [PubMed]
- Matsuura, S.; Kahyo, T.; Shinmura, K.; Iwaizumi, M.; Yamada, H.; Funai, K.; Kobayashi, J.; Tanahashi, M.; Niwa, H.; Ogawa, H.; et al. SGOL1 variant B induces abnormal mitosis and resistance to taxane in non-small cell lung cancers. Sci. Rep. 2013, 3, 3012. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Wang, X.Y.; Han, L.W.; Hu, D.D. LncRNA MALAT1 Regulates the Progression and Cisplatin Resistance of Ovarian Cancer Cells via Modulating miR-1271-5p/E2F5 Axis. Cancer Manag. Res. 2020, 12, 9999–10010. [Google Scholar] [CrossRef]
- Xu, X.; Cai, N.; Zhi, T.; Bao, Z.; Wang, D.; Liu, Y.; Jiang, K.; Fan, L.; Ji, J.; Liu, N. MicroRNA-1179 inhibits glioblastoma cell proliferation and cell cycle progression via directly targeting E2F transcription factor 5. Am. J. Cancer Res. 2017, 7, 1680–1692. [Google Scholar]
- Hijmans, E.M.; Voorhoeve, P.M.; Beijersbergen, R.L.; van’t Veer, L.J.; Bernards, R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol. Cell. Biol. 1995, 15, 3082–3089. [Google Scholar] [CrossRef]
- Chen, Q.; Liang, D.; Overbeek, P.A. Overexpression of E2F5/p130, but not E2F5 alone, can inhibit E2F-induced cell cycle entry in transgenic mice. Mol. Vis. 2008, 14, 602–614. [Google Scholar] [PubMed]
- Chen, D.; Cox, J.; Annam, J.; Weingart, M.; Essien, G.; Rathi, K.S.; Rokita, J.L.; Khurana, P.; Cuya, S.M.; Bosse, K.R.; et al. LIN28B promotes neuroblastoma metastasis and regulates PDZ binding kinase. Neoplasia 2020, 22, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Gartenhaus, R.B.; Eichberg, D.; Liu, Z.; Fang, H.B.; Rapoport, A.P. PBK/TOPK interacts with the DBD domain of tumor suppressor p53 and modulates expression of transcriptional targets including p21. Oncogene 2010, 29, 5464–5474. [Google Scholar] [CrossRef]
- Brooks, W.S.; Banerjee, S.; Crawford, D.F. G2E3 is a nucleo-cytoplasmic shuttling protein with DNA damage responsive localization. Exp. Cell Res. 2007, 313, 665–676. [Google Scholar] [CrossRef]
- Abe, Y.; Takeuchi, T.; Kagawa-Miki, L.; Ueda, N.; Shigemoto, K.; Yasukawa, M.; Kito, K. A mitotic kinase TOPK enhances Cdk1/cyclin B1-dependent phosphorylation of PRC1 and promotes cytokinesis. J. Mol. Biol. 2007, 370, 231–245. [Google Scholar] [CrossRef] [PubMed]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, P.; Lopez, G.; Capdevila, C.; Salvatori, B.; Yu, J.; Rodriguez-Barrueco, R.; Martinez, D.; Yarmarkovich, M.; Weichert-Leahey, N.; Abraham, B.J.; et al. Cross-Cohort Analysis Identifies a TEAD4-MYCN Positive Feedback Loop as the Core Regulatory Element of High-Risk Neuroblastoma. Cancer Discov. 2018, 8, 582–599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef]
- Min, H.L.; Kim, J.; Kim, W.H.; Jang, B.G.; Kim, M.A. Epigenetic Silencing of the Putative Tumor Suppressor Gene GLDC (Glycine Dehydrogenase) in Gastric Carcinoma. Anticancer Res. 2016, 36, 179–187. [Google Scholar]
- Haugen, B.R.; Nawaz, S.; Markham, N.; Hashizumi, T.; Shroyer, A.L.; Werness, B.; Shroyer, K.R. Telomerase activity in benign and malignant thyroid tumors. Thyroid 1997, 7, 337–342. [Google Scholar] [CrossRef]
- Liang, W.; Ye, D.; Dai, L.; Shen, Y.; Xu, J. Overexpression of hTERT extends replicative capacity of human nucleus pulposus cells, and protects against serum starvation-induced apoptosis and cell cycle arrest. J. Cell. Biochem. 2012, 113, 2112–2121. [Google Scholar] [CrossRef]
- Martínez, P.; Blasco, M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 2011, 11, 161–176. [Google Scholar] [CrossRef]
- Nandi, A.; Tidwell, M.; Karp, J.; Rapoport, A.P. Protein expression of PDZ-binding kinase is up-regulated in hematologic malignancies and strongly down-regulated during terminal differentiation of HL-60 leukemic cells. Blood Cells Mol. Dis. 2004, 32, 240–245. [Google Scholar] [CrossRef]
- Côté, S.; Simard, C.; Lemieux, R. Regulation of growth-related genes by interleukin-6 in murine myeloma cells. Cytokine 2002, 20, 113–120. [Google Scholar] [CrossRef]
- Simons-Evelyn, M.; Bailey-Dell, K.; Toretsky, J.A.; Ross, D.D.; Fenton, R.; Kalvakolanu, D.; Rapoport, A.P. PBK/TOPK is a novel mitotic kinase which is upregulated in Burkitt’s lymphoma and other highly proliferative malignant cells. Blood Cells Mol. Dis. 2001, 27, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhou, J.Y.; Ge, Y.; Matherly, L.H.; Wu, G.S. The phosphatase MKP1 is a transcriptional target of p53 involved in cell cycle regulation. J. Biol. Chem. 2003, 278, 41059–41068. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kitajima, T.S.; Tanno, Y.; Yoshida, K.; Morita, T.; Miyano, T.; Miyake, M.; Watanabe, Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2008, 10, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jia, L.; Yu, H. Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr. Biol. 2013, 23, 1927–1933. [Google Scholar] [CrossRef]
- Liu, H.; Qu, Q.; Warrington, R.; Rice, A.; Cheng, N.; Yu, H. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol. Cell 2015, 59, 426–436. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, N.; Liu, J.; Min, J.; Ma, N.; Liu, N.; Liu, Y.; Zhang, H. Lentivirus-mediated siRNA interference targeting SGO-1 inhibits human NSCLC cell growth. Tumour Biol. 2012, 33, 515–521. [Google Scholar] [CrossRef]
- Yamada, H.Y.; Yao, Y.; Wang, X.; Zhang, Y.; Huang, Y.; Dai, W.; Rao, C.V. Haploinsufficiency of SGO1 results in deregulated centrosome dynamics, enhanced chromosomal instability and colon tumorigenesis. Cell Cycle 2012, 11, 479–488. [Google Scholar] [CrossRef]
- Yang, J.; Ikezoe, T.; Nishioka, C.; Yokoyama, A. A novel treatment strategy targeting shugoshin 1 in hematological malignancies. Leuk. Res. 2013, 37, 76–82. [Google Scholar] [CrossRef]
- Vader, G.; Medema, R.H.; Lens, S.M. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J. Cell Biol. 2006, 173, 833–837. [Google Scholar] [CrossRef]
- Chieffi, P. Aurora B: A new promising therapeutic target in cancer. Intractable Rare Dis. Res. 2018, 7, 141–144. [Google Scholar] [CrossRef]
- Bogen, D.; Wei, J.S.; Azorsa, D.O.; Ormanoglu, P.; Buehler, E.; Guha, R.; Keller, J.M.; Mathews Griner, L.A.; Ferrer, M.; Song, Y.K.; et al. Aurora B kinase is a potent and selective target in MYCN-driven neuroblastoma. Oncotarget 2015, 6, 35247–35262. [Google Scholar] [CrossRef] [PubMed]
- Terada, Y.; Tatsuka, M.; Suzuki, F.; Yasuda, Y.; Fujita, S.; Otsu, M. AIM-1: A mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998, 17, 667–676. [Google Scholar] [CrossRef]
- Xie, H.; Kang, Y.; Wang, S.; Zheng, P.; Chen, Z.; Roy, S.; Zhao, C. E2f5 is a versatile transcriptional activator required for spermatogenesis and multiciliated cell differentiation in zebrafish. PLoS Genet. 2020, 16, e1008655. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, D.H.; Wan, W.Q. MYCN-induced E2F5 promotes neuroblastoma cell proliferation through regulating cell cycle progression. Biochem. Bioph. Res. Commun. 2019, 511, 35–40. [Google Scholar] [CrossRef]
- Chen, M.; Huang, B.; Zhu, L.; Chen, K.; Liu, M.; Zhong, C. Structural and Functional Overview of TEAD4 in Cancer Biology. OncoTargets Ther. 2020, 13, 9865–9874. [Google Scholar] [CrossRef]
- Liu, X.; Li, H.; Rajurkar, M.; Li, Q.; Cotton, J.L.; Ou, J.; Zhu, L.J.; Goel, H.L.; Mercurio, A.M.; Park, J.S.; et al. Tead and AP1 Coordinate Transcription and Motility. Cell Rep. 2016, 14, 1169–1180. [Google Scholar] [CrossRef]
- Park, J.A.; Cheung, N.V. Targets and Antibody Formats for Immunotherapy of Neuroblastoma. J. Clin. Oncol. 2020, 38, 1836–1848. [Google Scholar] [CrossRef] [PubMed]
- Whittle, S.B.; Smith, V.; Doherty, E.; Zhao, S.; McCarty, S.; Zage, P.E. Overview and recent advances in the treatment of neuroblastoma. Expert Rev. Anticancer Ther. 2017, 17, 369–386. [Google Scholar] [CrossRef] [PubMed]
- Ara, T.; DeClerck, Y.A. Mechanisms of invasion and metastasis in human neuroblastoma. Cancer Metastasis Rev. 2006, 25, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef]
- Mlakar, V.; Jurkovic Mlakar, S.; Lopez, G.; Maris, J.M.; Ansari, M.; Gumy-Pause, F. 11q deletion in neuroblastoma: A review of biological and clinical implications. Mol. Cancer 2017, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.X.; Huang, C.; Gao, C.; Xing, T.Y.; Liu, S.G.; Li, X.J.; Zhao, Q.; Wang, X.S.; Zhao, W.; Jin, M.; et al. MYCN amplification predicts poor prognosis based on interphase fluorescence in situ hybridization analysis of bone marrow cells in bone marrow metastases of neuroblastoma. Cancer Cell Int. 2017, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Curto, G.; der Vartanian, A.; Frarma, Y.E.; Manceau, L.; Baldi, L.; Prisco, S.; Elarouci, N.; Causeret, F.; Korenkov, D.; Rigolet, M.; et al. The PAX-FOXO1s trigger fast trans-differentiation of chick embryonic neural cells into alveolar rhabdomyosarcoma with tissue invasive properties limited by S phase entry inhibition. PLoS Genet. 2020, 16, e1009164. [Google Scholar] [CrossRef]
- Li, J.; Kretzner, L. The growth-inhibitory Ndrg1 gene is a Myc negative target in human neuroblastomas and other cell types with overexpressed N- or c-myc. Mol. Cell. Biochem. 2003, 250, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Akiba, J.; Murakami, Y.; Noda, M.; Watari, K.; Ogasawara, S.; Yoshida, T.; Kawahara, A.; Sanada, S.; Yasumoto, M.; Yamaguchi, R.; et al. N-myc downstream regulated gene1/Cap43 overexpression suppresses tumor growth by hepatic cancer cells through cell cycle arrest at the G0/G1 phase. Cancer Lett. 2011, 310, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, Z.; Sivagurunathan, S.; Mangs, H.; Chikhani, S.; Zhang, D.; Richardson, D.R. The metastasis suppressor, N-myc downstream regulated gene 1 (NDRG1), upregulates p21 via p53-independent mechanisms. Carcinogenesis 2011, 32, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, D.; Yue, F.; Zheng, M.; Kovacevic, Z.; Richardson, D.R. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J. Biol. Chem. 2012, 287, 17016–17028. [Google Scholar] [CrossRef] [PubMed]
- Fotovati, A.; Fujii, T.; Yamaguchi, M.; Kage, M.; Shirouzu, K.; Oie, S.; Basaki, Y.; Ono, M.; Yamana, H.; Kuwano, M. 17Beta-estradiol induces down-regulation of Cap43/NDRG1/Drg-1, a putative differentiation-related and metastasis suppressor gene, in human breast cancer cells. Clin. Cancer Res. 2006, 12, 3010–3018. [Google Scholar] [CrossRef] [PubMed]
- Petroni, M.; Sardina, F.; Heil, C.; Sahún-Roncero, M.; Colicchia, V.; Veschi, V.; Albini, S.; Fruci, D.; Ricci, B.; Soriani, A.; et al. The MRN complex is transcriptionally regulated by MYCN during neural cell proliferation to control replication stress. Cell Death Differ. 2016, 23, 197–206. [Google Scholar] [CrossRef]
- Stracker, T.H.; Petrini, J.H. The MRE11 complex: Starting from the ends. Nat. Rev. Mol. Cell Biol. 2011, 12, 90–103. [Google Scholar] [CrossRef]
- Jin, M.H.; Oh, D.Y. ATM in DNA repair in cancer. Pharmacol. Ther. 2019, 203, 107391. [Google Scholar] [CrossRef] [PubMed]
- Puissant, A.; Frumm, S.M.; Alexe, G.; Bassil, C.F.; Qi, J.; Chanthery, Y.H.; Nekritz, E.A.; Zeid, R.; Gustafson, W.C.; Greninger, P.; et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013, 3, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Chipumuro, E.; Marco, E.; Christensen, C.L.; Kwiatkowski, N.; Zhang, T.; Hatheway, C.M.; Abraham, B.J.; Sharma, B.; Yeung, C.; Altabef, A.; et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014, 159, 1126–1139. [Google Scholar] [CrossRef]
- Poon, E.; Liang, T.; Jamin, Y.; Walz, S.; Kwok, C.; Hakkert, A.; Barker, K.; Urban, Z.; Thway, K.; Zeid, R.; et al. Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma. J. Clin. Investig. 2020, 130, 5875–5892. [Google Scholar] [CrossRef]
- Chesler, L.; Schlieve, C.; Goldenberg, D.D.; Kenney, A.; Kim, G.; McMillan, A.; Matthay, K.K.; Rowitch, D.; Weiss, W.A. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006, 66, 8139–8146. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, S.; Goeser, F.; Schulte, J.H.; Schramm, A.; Ehemann, V.; Hero, B.; Eggert, A.; Berthold, F.; Fischer, M. Polo-like kinase 1 is a therapeutic target in high-risk neuroblastoma. Clin. Cancer Res. 2011, 17, 731–741. [Google Scholar] [CrossRef]
- Park, J.H.; Szemes, M.; Vieira, G.C.; Melegh, Z.; Malik, S.; Heesom, K.J.; Von Wallwitz-Freitas, L.; Greenhough, A.; Brown, K.W.; Zheng, Y.G.; et al. Protein arginine methyltransferase 5 is a key regulator of the MYCN oncoprotein in neuroblastoma cells. Mol. Oncol. 2015, 9, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; He, J.Z.; Mao, L.B.; Zhang, Y.Y.; Cui, W.W.; Duan, S.J.; Jiang, A.L.; Gao, Y.; Sang, Y.; Huang, G.F. EPZ015666, a selective protein arginine methyltransferase 5 (PRMT5) inhibitor with an antitumour effect in retinoblastoma. Exp. Eye Res. 2021, 202, 108286. [Google Scholar] [CrossRef]
- Müller, I.; Larsson, K.; Frenzel, A.; Oliynyk, G.; Zirath, H.; Prochownik, E.V.; Westwood, N.J.; Henriksson, M.A. Targeting of the MYCN protein with small molecule c-MYC inhibitors. PLoS ONE 2014, 9, e97285. [Google Scholar] [CrossRef]
- Zirath, H.; Frenzel, A.; Oliynyk, G.; Segerström, L.; Westermark, U.K.; Larsson, K.; Munksgaard Persson, M.; Hultenby, K.; Lehtiö, J.; Einvik, C.; et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10258–10263. [Google Scholar] [CrossRef] [PubMed]
- Han, H.Y.; Jain, A.D.; Truica, M.I.; Izquierdo-Ferrer, J.; Anker, J.F.; Lysy, B.; Sagar, V.; Luan, Y.; Chalmers, Z.R.; Unno, K.; et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 2019, 36, 83–497.e15. [Google Scholar] [CrossRef] [PubMed]
- Struntz, N.B.; Chen, A.; Deutzmann, A.; Wilson, R.M.; Stefan, E.; Evans, H.L.; Ramirez, M.A.; Liang, T.; Caballero, F.; Wildschut, M.H.E.; et al. Stabilization of the Max Homodimer with a Small Molecule Attenuates Myc-Driven Transcription. Cell Chem. Biol. 2019, 26, 711. [Google Scholar] [CrossRef] [PubMed]
- Van Maerken, T.; Speleman, F.; Vermeulen, J.; Lambertz, I.; de Clercq, S.; de Smet, E.; Yigit, N.; Coppens, V.; Philippé, J.; de Paepe, A.; et al. Small-molecule MDM2 antagonists as a new therapy concept for neuroblastoma. Cancer Res. 2006, 66, 9646–9655. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef]
- Sánchez-Martínez, C.; Gelbert, L.M.; Lallena, M.J.; de Dios, A. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorg. Med. Chem. Lett. 2015, 25, 3420–3435. [Google Scholar] [CrossRef]
- Frame, S.; Saladino, C.; MacKay, C.; Atrash, B.; Sheldrake, P.; McDonald, E.; Clarke, P.A.; Workman, P.; Blake, D.; Zheleva, D. Fadraciclib (CYC065), a novel CDK inhibitor, targets key pro-survival and oncogenic pathways in cancer. PLoS ONE 2020, 15, e0234103. [Google Scholar] [CrossRef]
- Kawakami, M.; Mustachio, L.M.; Rodriguez-Canales, J.; Mino, B.; Roszik, J.; Tong, P.; Wang, J.; Lee, J.J.; Myung, J.H.; Heymach, J.V.; et al. Next-Generation CDK2/9 Inhibitors and Anaphase Catastrophe in Lung Cancer. J. Natl. Cancer Inst. 2017, 109, djw297. [Google Scholar] [CrossRef]
- Diolaiti, D.; McFerrin, L.; Carroll, P.A.; Eisenman, R.N. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta 2015, 1849, 484–500. [Google Scholar] [CrossRef]
- Braun, C.J.; Stanciu, M.; Boutz, P.L.; Patterson, J.C.; Calligaris, D.; Higuchi, F.; Neupane, R.; Fenoglio, S.; Cahill, D.P.; Wakimoto, H.; et al. Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer Cell 2017, 32, 411–426.e11. [Google Scholar] [CrossRef]
- Chan-Penebre, E.; Kuplast, K.G.; Majer, C.R.; Boriack-Sjodin, P.A.; Wigle, T.J.; Johnston, L.D.; Rioux, N.; Munchhof, M.J.; Jin, L.; Jacques, S.L.; et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015, 11, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Steegmaier, M.; Hoffmann, M.; Baum, A.; Lenart, P.; Petronczki, M.; Krssak, M.; Gurtler, U.; Garin-Chesa, P.; Lieb, S.; Quant, J.; et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007, 17, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.C.; Pezuk, J.A.; Brassesco, M.S.; Morales, A.G.; Queiroz, R.G.; Scrideli, C.A.; Tone, L.G. PLK1 expression and BI 2536 effects in childhood acute lymphoblastic leukemia. Pediatr. Blood Cancer 2014, 61, 1227–1231. [Google Scholar] [CrossRef]
- Murray, M.J.; Nicholson, J.C.; Coleman, N. Biology of childhood germ cell tumours, focussing on the significance of microRNAs. Andrology 2015, 3, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Yu, Z.; Zhuang, C.; Wu, Y.; Guo, Z.; Li, J.; Dong, G.; Yao, J.; Sheng, C.; Miao, Z.; Zhang, W. Design, synthesis and biological evaluation of sulfamide and triazole benzodiazepines as novel p53-MDM2 inhibitors. Int. J. Mol. Sci. 2014, 15, 15741–15753. [Google Scholar] [CrossRef]
- Daniele, S.; la Pietra, V.; Barresi, E.; di Maro, S.; da Pozzo, E.; Robello, M.; la Motta, C.; Cosconati, S.; Taliani, S.; Marinelli, L.; et al. Lead Optimization of 2-Phenylindolylglyoxylyldipeptide Murine Double Minute (MDM)2/Translocator Protein (TSPO) Dual Inhibitors for the Treatment of Gliomas. J. Med. Chem. 2016, 59, 4526–4538. [Google Scholar] [CrossRef]
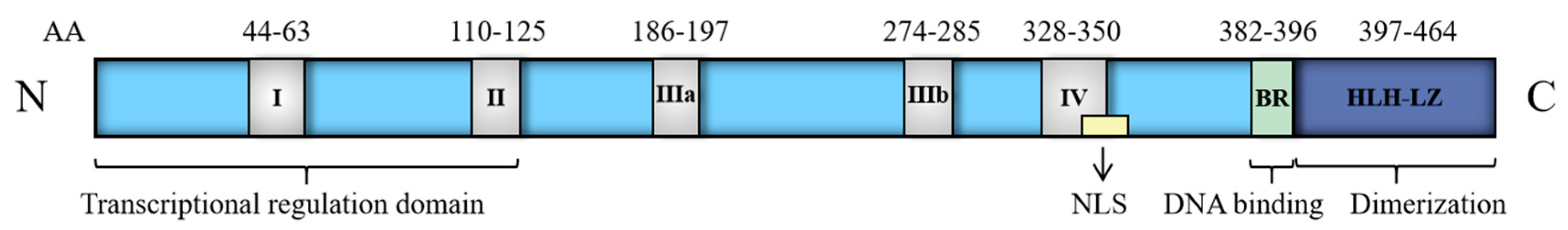
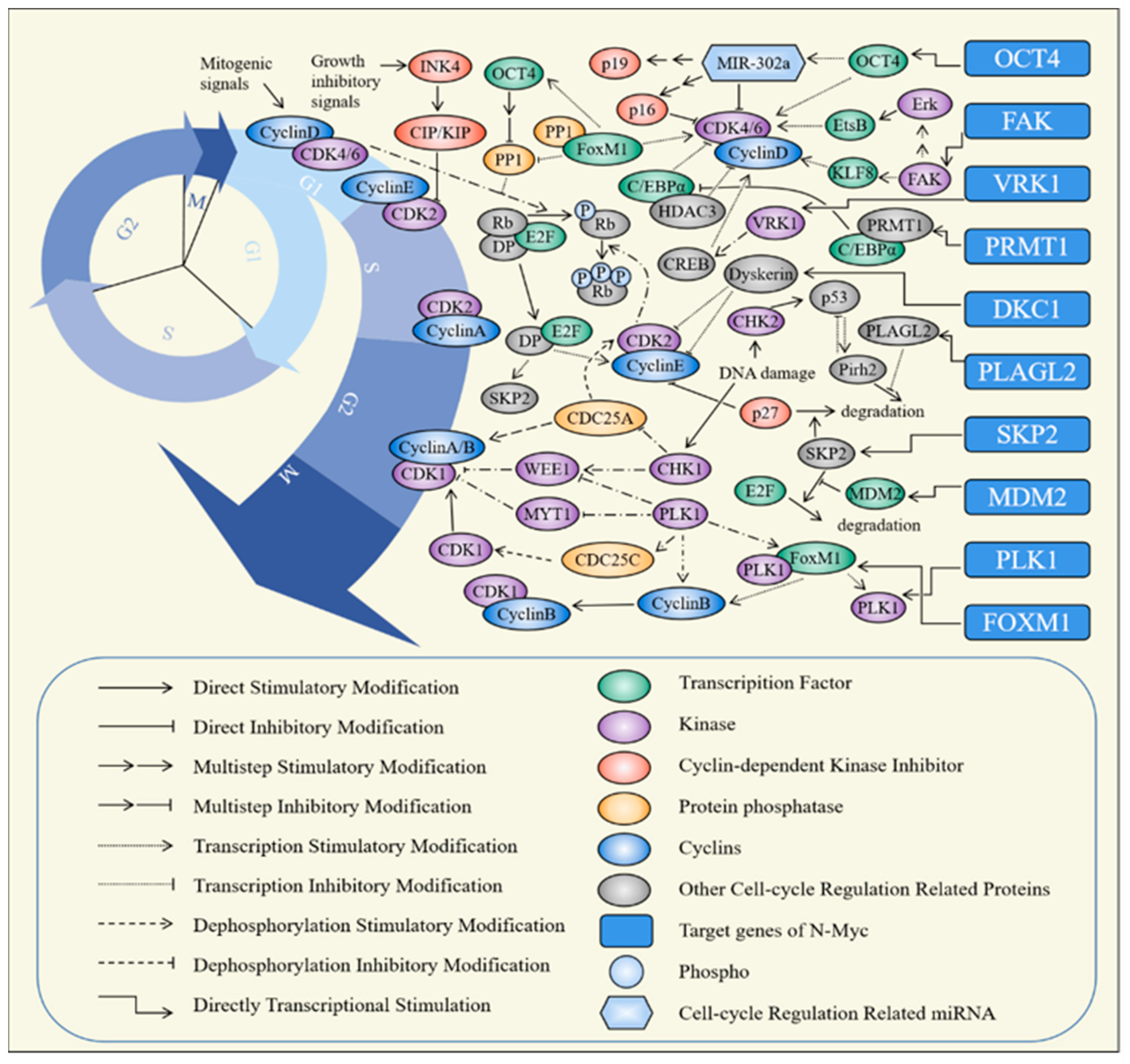
| Target Gene | Phase | Regulating Target |
|---|---|---|
| TERT [88] | S [151] | Inhibition of TERT leads to DDR and down-regulation of CCND1. The high expression of TERT significantly up-regulated the expression of CCND1 [151,152]. |
| AURKB [153] | G2/M [154], M [155,156] | Inhibition of AURKB results in down-regulation of CCNB and CDC2 and CDC25C, significantly reduces phosphorylation of histone H3, and blocks chromosome segregation and cytokinesis [154,155,156]. |
| GLDC [157] | G1 [157] | The expressions of CCNA2, CCNB1, CDK1, CDK2, CCND1, CCNE1, POLE2 and MCM5 are decreased after knocking down GLDC. GLDC can also regulate the synthesis of nucleotides and cholesterol [157,158,159,160]. |
| SGO1 [161] | G0/G1 [162], S [162], G2/M [161,162] | The expressions of CCNA, CDK2, CCND1 are decreased after knocking down SGO1. SGO1 regulate separation of sister chromatids [161,162,163]. |
| E2F5 [164] | G0/G1 [164,165], G1 [166,167] | The expressions of CDK2, CDK6 are decreased after knocking down E2F5 [164]. |
| PBK [168] | G2/M [169,170], M [171] | PBK forms a complex with Cyclin B1-CDK1 and phosphorylates PRC1 in a Cyclin B1-CDK1-dependent manner, thereby participating in the formation of the mitotic spindle and promotes cytokinesis [171]. PBK interacts with p53 containing the DBD domain to down-regulate the transactivation function of TP53. Over-expression of PBK down-regulates the expression of p21 by reducing the recruitment of p21 promoter to p53 [169,170,172]. |
| TEAD4 [173] | G0/G1 [173] | After silencing the TEAD4 gene, cyclin-dependent kinases (CDK2, CDK1, CDC25B), cyclins (CCND1), DNA replication proliferating cell nuclear antigen (PCNA), minichromosome maintenance complex component 7 (MCM7), Cdc6, checkpoint kinases (CHEK1, CHEK2, WEE1) and other proteins are inhibited [173]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.-L.; Dong, L.-L.; Jin, M.-J.; Li, Q.-Y.; Wang, X.; Jia, M.-Q.; Song, J.; Zhang, S.-Y.; Yuan, S. A Review of the Regulatory Mechanisms of N-Myc on Cell Cycle. Molecules 2023, 28, 1141. https://doi.org/10.3390/molecules28031141
Li H-L, Dong L-L, Jin M-J, Li Q-Y, Wang X, Jia M-Q, Song J, Zhang S-Y, Yuan S. A Review of the Regulatory Mechanisms of N-Myc on Cell Cycle. Molecules. 2023; 28(3):1141. https://doi.org/10.3390/molecules28031141
Chicago/Turabian StyleLi, Hong-Li, Lu-Lu Dong, Min-Jie Jin, Qian-Yu Li, Xiao Wang, Mei-Qi Jia, Jian Song, Sai-Yang Zhang, and Shuo Yuan. 2023. "A Review of the Regulatory Mechanisms of N-Myc on Cell Cycle" Molecules 28, no. 3: 1141. https://doi.org/10.3390/molecules28031141
APA StyleLi, H.-L., Dong, L.-L., Jin, M.-J., Li, Q.-Y., Wang, X., Jia, M.-Q., Song, J., Zhang, S.-Y., & Yuan, S. (2023). A Review of the Regulatory Mechanisms of N-Myc on Cell Cycle. Molecules, 28(3), 1141. https://doi.org/10.3390/molecules28031141






