Abstract
Globally, dental caries is one of the most common non-communicable diseases for patients of all ages; Streptococcus mutans (S. mutans) is its principal pathogen. Lactobacillus paracasei (L. paracasei) shows excellent anti-pathogens and immune-regulation functions in the host. The aim of this study is to evaluate the effects of L. paracasei ET-22 on the formation of S. mutans biofilms. The living bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 were prepared using the same number of bacteria. In vitro, they were added into artificial-saliva medium, and used to coculture with the S. mutans. Results showed that the living bacteria and secretions of L. paracasei ET-22 inhibited biofilm-growth, the synthesis of water-soluble polysaccharide and water-insoluble polysaccharide, and virulence-gene-expression levels related to the formation of S. mutans biofilms. Surprisingly, the heat-killed L. paracasei ET-22, which is a postbiotic, also showed a similar regulation function. Non-targeted metabonomics technology was used to identify multiple potential active-substances in the postbiotics of L. paracasei ET-22 that inhibit the formation of S. mutans biofilms, including phenyllactic acid, zidovudine monophosphate, and citrulline. In conclusion, live bacteria and its postbiotics of L. paracasei ET-22 all have inhibitory effects on the formation of S. mutans biofilm. The postbiotics of L. paracasei ET-22 may be a promising biological anticariogenic-agent.
1. Introduction
For patients of all ages, dental caries is one of the most common non-communicable diseases worldwide [1]. It is a multifactorial chronic disease caused by chronic biofilms (plaques) containing cariogenic microorganisms. These microorganisms and their virulence products collectively cause the demineralization and progressive destruction of enamel [1,2].
As a common pathogen of oral microbiota, Streptococcus mutans (S. mutans) has long been recognized as the primary microorganism that causes dental caries [3]. The biofilm is a highly dynamic and structured community of microbial cells integrated into a self-produced extracellular polymer matrix [4,5]. A certain number of S. mutans and other bacteria can coordinate the formation of three-dimensional cariogenic biofilms by producing extracellular polysaccharides (EPS) in the extracellular matrix, creating a microenvironment rich in pathogens and fermentable dietary carbohydrates [2,6]. In this microenvironment, metabolites such as acids produced by S. mutans accumulate in the biofilms and at the interface between the biofilms and the tooth surface, causing enamel demineralization and then the dental caries [7].
Although most dental plaques can be removed, residual dental plaques are inevitable [1]. At present, the main means of preventing dental caries are the administration of fluoride and avidin and mechanical removal [8,9]. However, each of these therapies has its limitations. Mechanical removal by brushing and flossing might be considered the first line of prevention, as it is mainly used in the daily maintenance of oral hygiene [10]. However, strongly adherent biofilms may not be cleared out completely. Furthermore, some dead spaces cannot be touched. Even though fluoride can affect the glycolytic activity of Streptococcus, its ability to protect against dental caries is insufficient and it has little impact on microorganisms’ ability to produce acid [11]. As for avidin, although it effectively prevents dental caries in in vitro and in vivo experiments, its excessive use changes the microbiota of the digestive tract and then increases the drug resistance of pathogens [12]. Therefore, more convenient, efficient, and safe methods are needed to inhibit the formation of dental plaques.
Probiotics maintain oral health by promoting a microbial balance [9]. Probiotics can prevent caries by producing metabolites, such as biosurfactants, bacteriocin, and EPS, inhibiting adhesion and colonization, and downregulating the expressions of virulence genes related to the biofilm formation of cariogenic pathogens [13]. Lactobacillus is a symbiotic microorganism found in the human oral microbiota, and has a strong impact on Streptococcus in the oral cavity [14]. Meanwhile, long-term studies also found that heat-killed probiotics, known as postbiotics, also show beneficial effects [15]. This may be another pathway for using supplemented probiotics to perform regulated functions. Moreover, the effect of postbiotics may be more stable and superior in some application scenarios [16]. In Lactobacillus, L. paracasei, L. plantarum, L. alivarius, and L. rhamnosus are the most used species for digestive-system health [14]. However, the effect mechanisms of Lactobacillus on oral S. mutans have not been fully clarified. LC-MS/MS is an important technique for analyzing the functional substances of postbiotics. In addition, within the same species, bacterial bioactivity is strain specific. Therefore, the mechanisms of regulation may be disparate and worth interrogating further.
The purpose of this study was to evaluate the effects of L. paracasei ET-22 on cariogenic biofilms and investigate the potential mechanisms of L. paracasei ET-22 in the intervention of cariogenic biofilms. In vitro experiments were used to conduct preliminary investigations of the effects of live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 on the formation of S. mutans biofilms and functional mechanisms.
2. Results
2.1. Effects of L. paracasei ET-22 on the Biofilm Formation and EPS Production of S. mutans
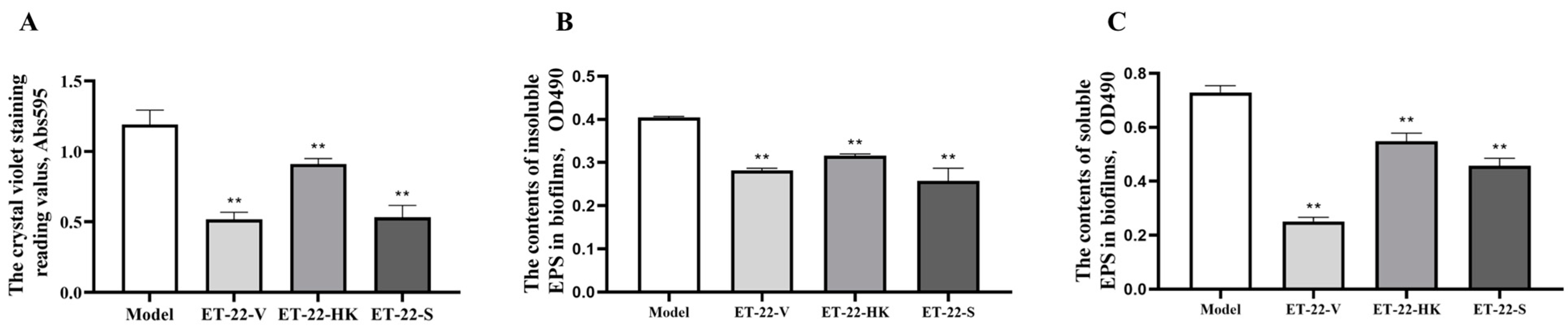
To assess the effects of L. paracasei ET-22 on S. mutans biofilms, the live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 were used in a co-culture with S. mutans. An equal volume of bacterial solutions with a concentration of 5.5 × 108 CFU/mL was used to prepare the live bacteria (ET-22-V), heat-killed bacteria (ET-22-HK), and secretions (ET-22-S) of L. paracasei ET-22. The single pathogen S. mutans (Model) was cultured as a control. After a 24 h co-culture with S. mutans, the ET-22-V and ET-22-S groups all showed decreased crystal-violet-staining reading values (p < 0.01, Figure 1A). Surprisingly, we found that the ET-22-HK group also showed a reduced crystal-violet-staining reading value (p < 0.01, Figure 1A).
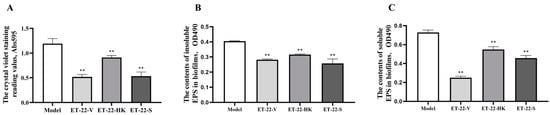
Figure 1.
Effects of L. paracasei ET-22 on formation of S. mutans biofilms. (A). The effects of live bacteria (ET-22-V), heat-killed bacteria (ET-22-HK), and secretions (ET-22-S) of L. paracasei ET-22 on the formation levels of S. mutans biofilms. The single pathogen S. mutans (Model) was cultured as a control. Means ± SEM are shown (n = 6). ** p < 0.01, vs. Model group. (B). The effects of live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 on the contents of insoluble extracellular polysaccharides (EPS) in S. mutans biofilms. Means ± SEM are shown (n = 6). ** p < 0.01, vs. Model group. (C). The effects of live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 on the contents of soluble EPS in S. mutans biofilms. Means ± SEM are shown (n = 6). ** p < 0.01, vs. Model group.
Based on the inhibitory effects of the live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 on S. mutans biofilms, we further analyzed the EPS contents of S. mutans biofilms. Compared with the Model group, the ET-22-V, ET-22-HK, and ET-22-S groups all showed significantly decreased contents of insoluble EPS in S. mutans biofilms (p < 0.01, Figure 1B). Meanwhile, the ET-22-V, ET-22-HK, and ET-22-S groups all showed significantly reduced contents of soluble EPS in S. mutans biofilms compared to the Model group (p < 0.01, Figure 1C).
2.2. Biofilm Microstructure Is Changed by L. paracasei ET-22
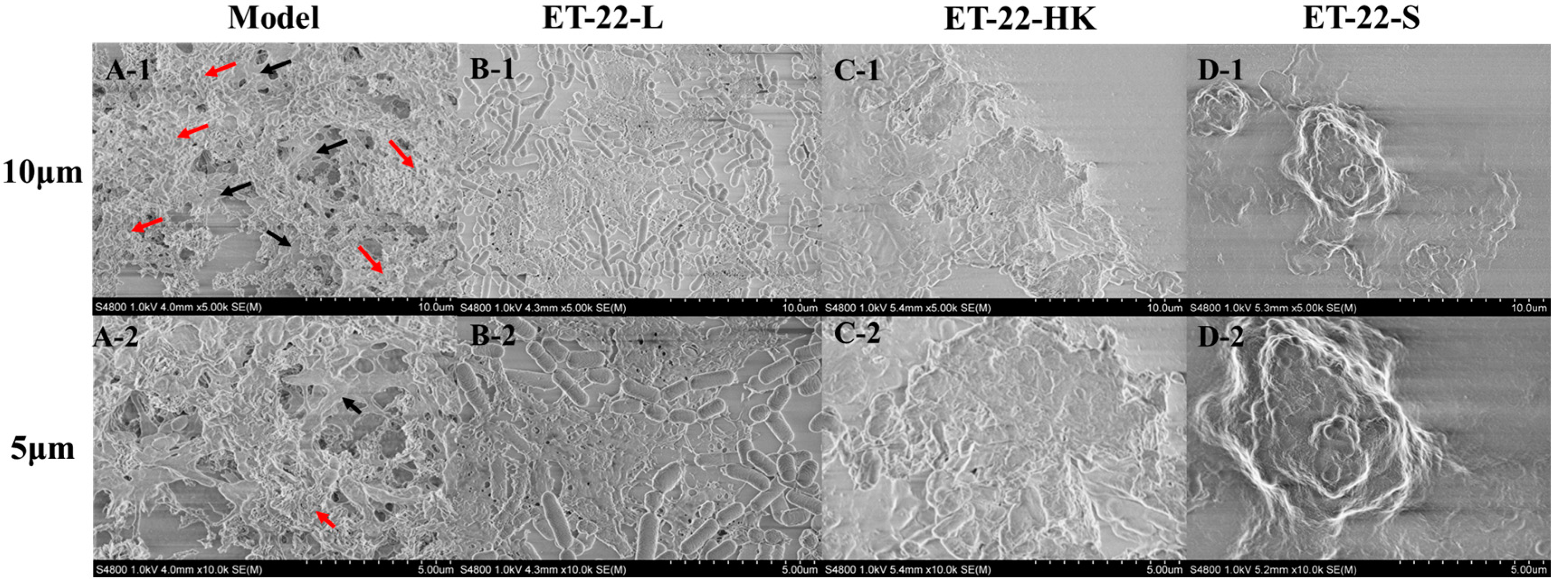
To further verify the bacteria’s inhibitory effects for S. mutans biofilms, we viewed its microstructure using scanning electron microscopy (SEM) under the 10 μm or 5 μm scales. According to the representative SEM images, the Model group showed a stable, multilayer, and reticular biofilm, in which the S. mutans was tightly wrapped with a cauliflower-like structure (Figure 2(A-1)). A small number of bacteria were exposed, and closely adhered to the “flower stalk” (black arrow) and “corolla” (red arrow) positions (Figure 2(A-2)). The ET-22-V, ET-22-HK and ET-22-S groups all showed obviously changed biofilm microstructures (Figure 2B–D). In the ET-22-V group, the multi-layer and compact cauliflower-like structure disappeared. Instead, a single-layer and loose biofilm-microstructure was exhibited, in which the bacilliform L. paracasei ET-22 and spherical S. mutans were completely exposed on the biofilms. The bindings of these two strains with the biofilms were loose (Figure 2B). Similarly, the ET-22-HK and ET-22-S groups also presented a lack of cauliflower-like structures (Figure 2C,D). In the ET-22-HK group, the heat-killed L. paracasei ET-22 and spherical S. mutans were tightly wrapped into a single-layer sheet structure (Figure 2C). The ET-22-S group showed a compact and discontinuous biofilm with a certain thickness, in which the S. mutans were tightly wrapped (Figure 2D).
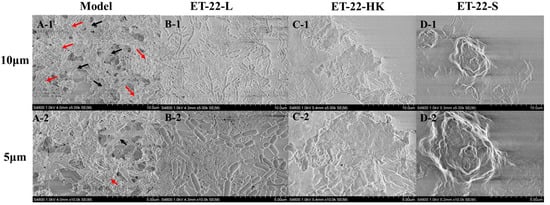
Figure 2.
Biofilm microstructure of S. mutans after the co-culture with live bacteria (ET-22-V), heat-killed bacteria (ET-22-HK), and secretions (ET-22-S) of L. paracasei ET-22, produced using scanning electron microscopy. (A-1,A-2) are typical images of S. mutans biofilms in the Model group under a 10 μm (A-1) or 5 μm (A-2) scale. The red arrow points to the “corolla” of the cauliflower-like structure. The black arrow points to the “flower stalk” of the cauliflower-like structure. (B-1,B-2) are representative images of S. mutans biofilms after the co-culture with live L. paracasei ET-22 under a 10 μm (B-1) or 5 μm (B-2) scale. (C-1,C-2) are representative images of S. mutans biofilms after the co-treatment with heat-killed L. paracasei ET-22 under a 10 μm (C-1) or 5 μm (C-2) scale. (D-1,D-2) are representative images of S. mutans biofilms after the co-treatment with secretions of L. paracasei ET-22 under a 10 μm (D-1) or 5 μm (D-2) scale.
2.3. Biofilm Thickness Is Changed by L. paracasei ET-22
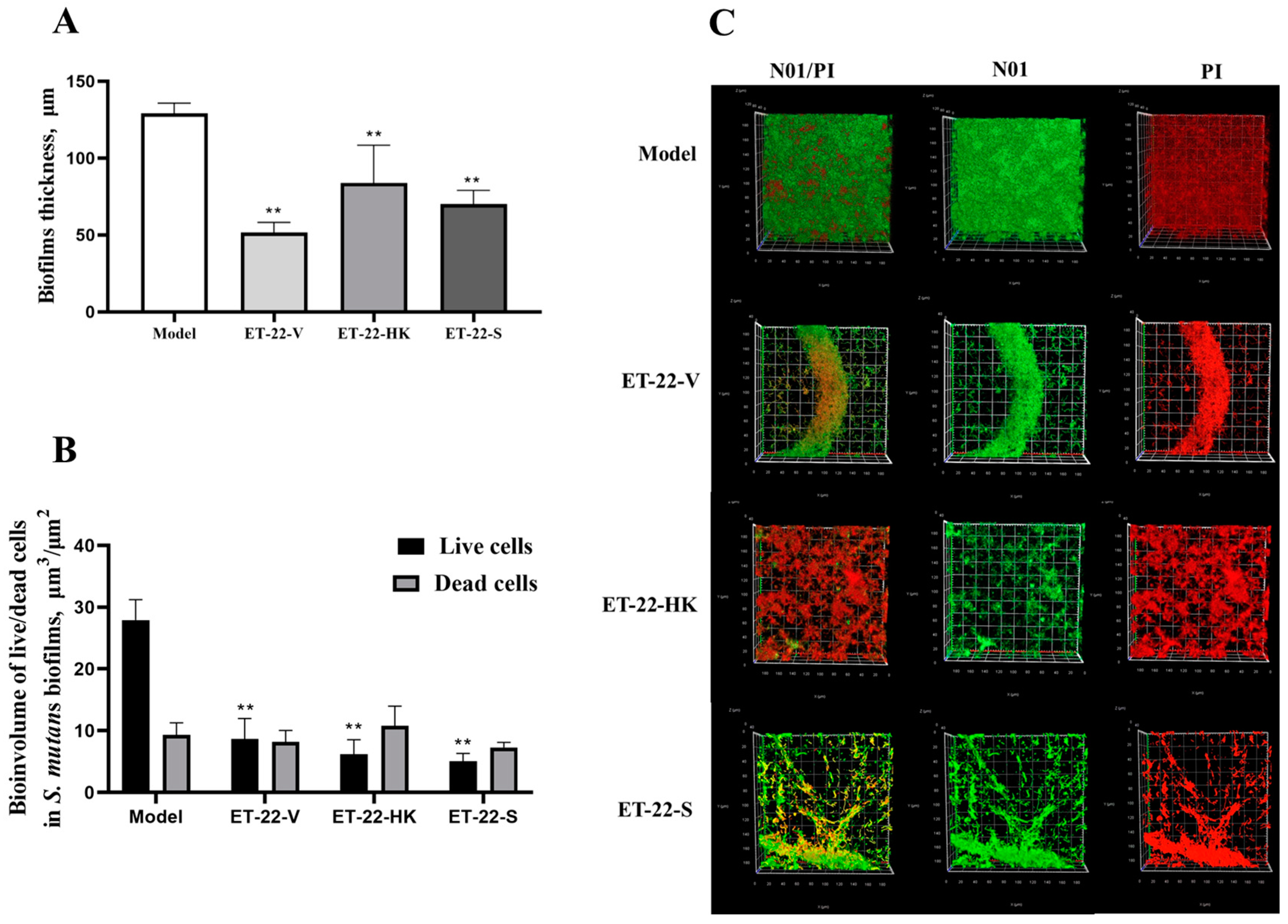
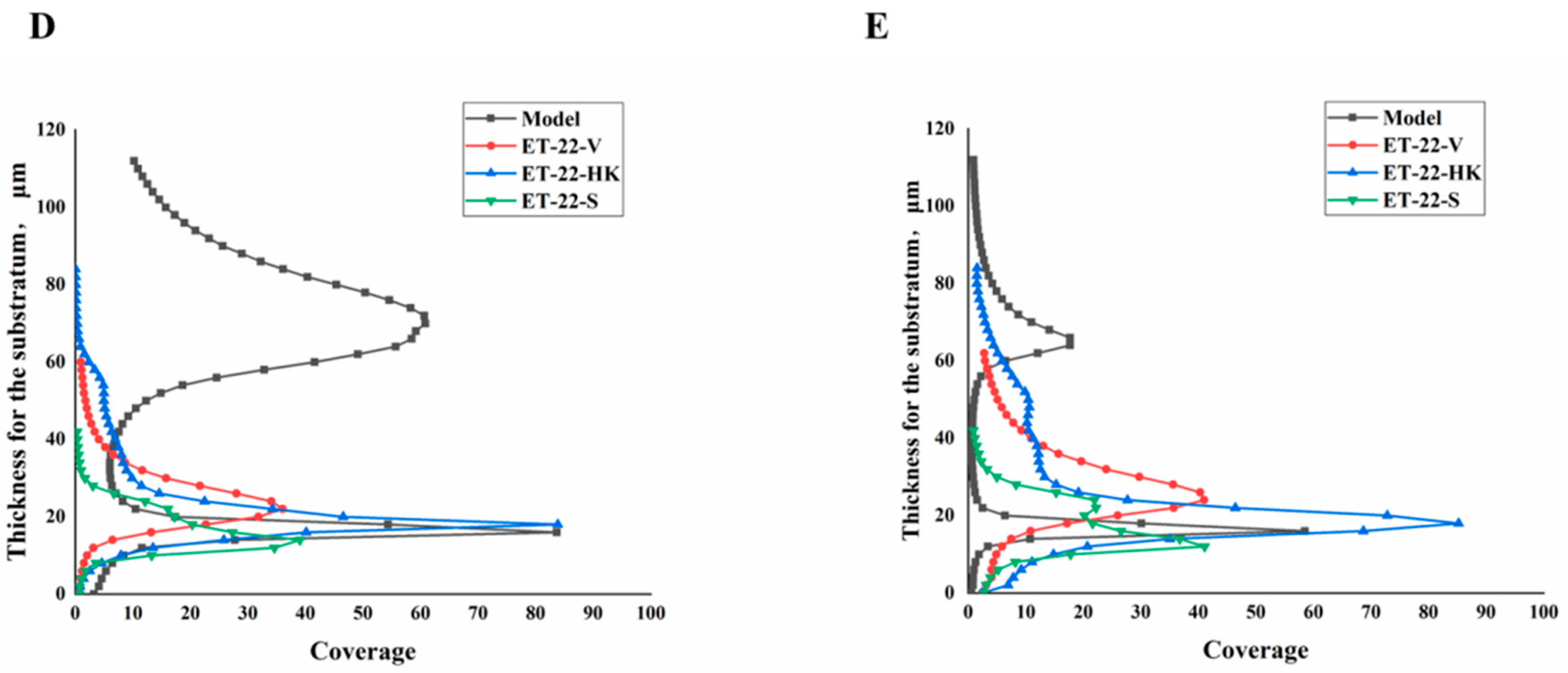
We further detected the biofilm thickness using confocal laser scanning microscopy (CLSM). Using the COMSTAT 2 analysis for CLSM views, the biofilm thicknesses are displayed. As shown in Figure 3A, compared with the Model group, the thicknesses of the S. mutans biofilms were significantly lower in the ET-22-V, ET-22-HK, and ET-22-S groups (p < 0.01, Figure 3A). After being stained with N01 and propidium iodide (PI), the live bacteria (green) and dead bacteria (red) in the biofilms were viewed (Figure 3B). Based on the COMSTAT 2 analysis, living and dead cells were respectively quantified. After the co-culture, the biomasses of live bacteria in the biofilms of the ET-22-V, ET-22-HK, and ET-22-S groups were remarkably lower than those in the Model group (p < 0.01, Figure 3C). However, there was no significant difference in the biomasses of dead bacteria among these groups (p > 0.05, Figure 3C). The vertical biofilm-coverage was checked. In the Model group, both the living bacteria and the dead bacteria in the biofilms gathered mainly at the 50–115 μm and 15–40 μm positions, above the bottom of the hydroxyapatite discs (Figure 3D,E). By contrast, the living bacteria and dead bacteria in the biofilms of the ET-22-V, ET-22-HK, and ET-22-S groups mainly gathered at the 15–40 μm position (Figure 3D,E).
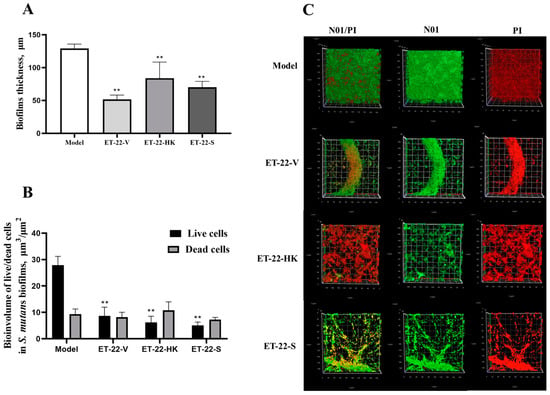
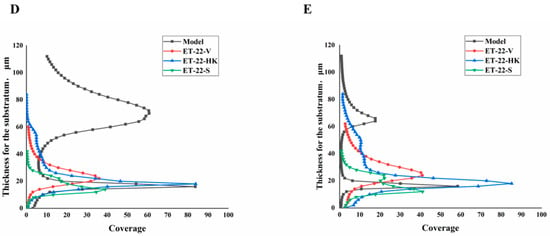
Figure 3.
Effects of L. paracasei ET-22 on the thickness and biomasses of S. mutans biofilms. (A) Changes in thickness of the S. mutans biofilms after the co-culture with live bacteria (ET-22-V), heat-killed bacteria (ET-22-HK), and secretions (ET-22-S) of L. paracasei ET-22. Means ± SEM are shown (n = 6). ** p < 0.01, vs. Model group. (B) The staining results of live and dead bacteria in S. mutans biofilms viewed using confocal laser scanning microscopy. (C) The biomasses of live bacteria (green) and dead bacteria (red) in S. mutans biofilms of the ET-22-V, ET-22-HK, and ET-22-S groups. N01 was used for the nuclear staining of live bacteria. Propidium iodide (PI) was used for the nuclear staining of dead bacteria. Means ± SEM are shown (n = 6). ** p < 0.01, vs. live bacteria in the Model group. (D,E), the gathering positions of living bacteria (D) and dead bacteria (E) above the bottom of hydroxyapatite discs.
2.4. The Expression of Virulence Genes and the Quorum-Sensing System Are Regulated by L. paracasei ET-22
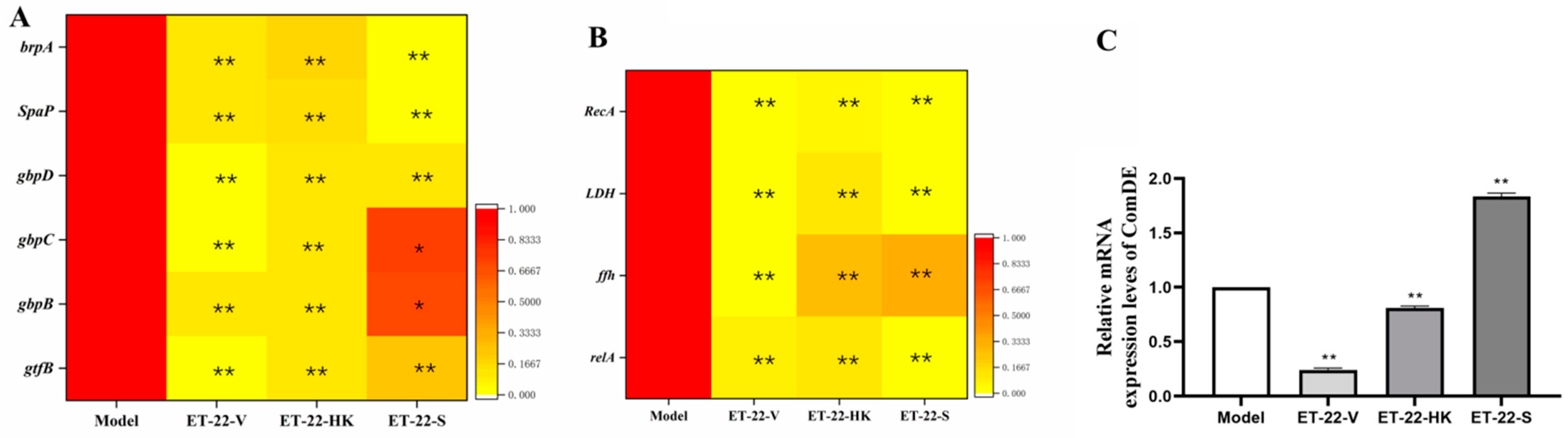
In order to investigate the mechanisms that inhibit the formation of S. mutans biofilms, the gene-expression levels of virulence-factors and quorum-sensing (QS) systems were checked. brpA is involved in the formation of biofilms and the stress tolerance of S. mutans. SpaP is a non-sucrose-dependent adhesion gene. Its protein SPAP mediates the initial adhesion of S. mutans to teeth. S. mutans also synthesizes some surface-associated glucan-binding proteins (GBP), including gbpA, gbpB, gbpC, and gbpD, and glucosyltransferases including gtfB, gtfC, and gtfD, contributing to the formation of biofilms. After the cotreatment, the ET-22-V, ET-22-HK, and ET-22-S groups all presented significantly decreased gene-expression-levels of brpA, SpaP, gbpB, gbpC, gbpD, and gtfB, compared to the Model group (p < 0.05, Figure 4A).
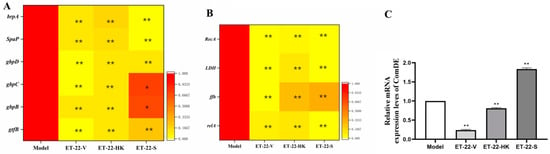
Figure 4.
Effects of L. paracasei ET-22 on gene expressions involved in the adhesive, acid-resistant, acidogenic, and quorum-sensing (QS) qualities of S. mutans. (A) A heatmap of the relative gene-expression-levels of proteins involving adhesions and the formation of the S. mutans biofilm, including brpA, SpaP, gbpD, gbpC, gbpB, and gtfB. Means ± SEM are shown (n = 6). * p < 0.05, ** p < 0.01, vs. Model group. (B) A heatmap of the relative gene-expression-levels of proteins involved in the acid production and acid tolerance of S. mutans, including RecA, LDH, ffh, and relA. Means ± SEM are shown (n = 6). ** p < 0.01, vs. Model group. (C) The gene-expression-levels of comDE in the QS system of S. mutans. Means ± SEM are shown (n = 6). ** p < 0.01, vs. Model group.
Lactate dehydrogenase (LDH), rela, RecA, and ffh are all involved in the acid production and acid tolerance of S. mutans. Compared with the Model group, the gene expressions of LDH, rela, RecA, and ffh were significantly reduced in the ET-22-V, ET-22-HK, and ET-22-S groups (p < 0.01, Figure 4B). As a component of the QS system, ComDE can regulate the intraspecific communications of S. mutans in biofilms by stimulating the competitive signaling-peptides. Compared with the Model group, the ET-22-V and ET-22-HK groups both showed similarly reduced gene-expression-levels of ComDE (p < 0.01, Figure 4C). On the contrary, the ET-22-S group showed a remarkably increased gene-expression-level of ComDE (p < 0.01, Figure 4C).
2.5. The Bioactive Substances for Resisting Formations of S. mutans Biofilms in Heat-Killed Bacteria and Secretions of L. paracasei ET-22
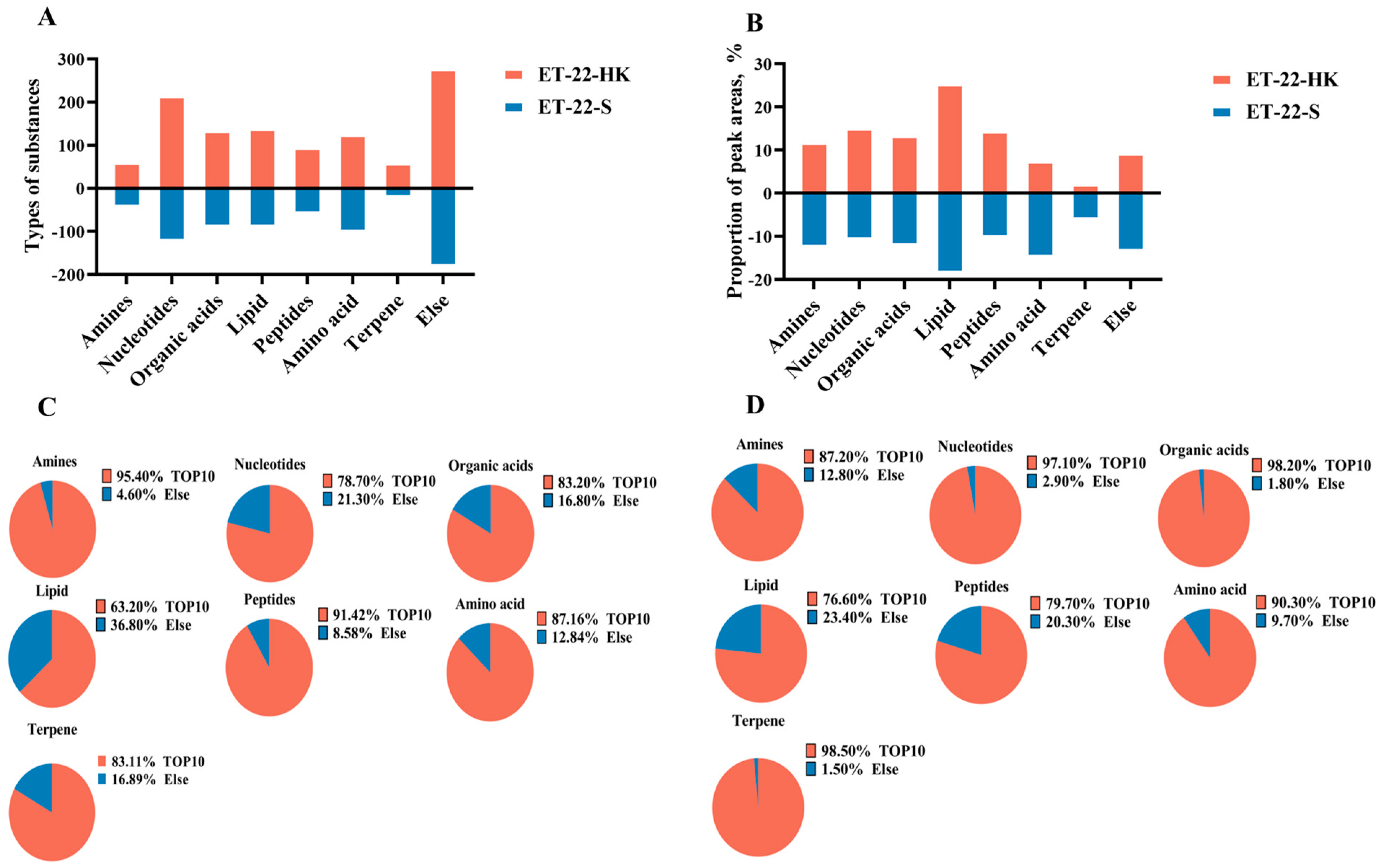
In order to investigate the potential bioactive substances that resist the formation of S. mutans biofilms, we analyzed the heat-killed bacteria and secretions of L. paracasei ET-22 in detail, using non-targeted metabonomics, whereby LC-MS/MS analysis was performed. This is a novel approach to confirming functional substances. Non-targeted metabonomics involves the comprehensive and systematic analysis of organic substances. An unbiased metabolomics analysis can contribute to the discovery of potential biomarkers. LC-MS/MS can provide more debris information that is needed for qualitative analysis, and it can reduce background noise. In addition, LC-MS/MS ensures that the spectrogram of trace components is free from the interference of abundant substances, which greatly improves the sensitivity of the detection. Therefore, in the present research, LC-MS/MS analysis is a more advantageous method of searching for possible functional substances in heat-killed bacteria and secretions of L. paracasei ET-22. As the results show, large amounts of organic substances were identified, including amines, nucleotides, organic acids, lipids, peptides, amino acids, terpene, and others in heat-killed bacteria (ET-22-HK) and secretions (ET-22-S) of L. paracasei ET-22. Of these, the nucleotides contained the most types of substances: more than 200 in ET-22-HK and 100 in ET-22-S (Figure 5A). Based on the relative ratio of peak areas, lipids represented the highest percentage, accounting for more than 20% in ET-22-HK and 10% in ET-22-S (Figure 5B). In turn, the contents of nucleotides, peptides, organic acids, and amines were relatively low, but exceeded 10% (Figure 5B). The amino acids, terpenes, and others accounted for less than 10% in ET-22-HK (Figure 5B). In each type of organic substance from ET-22-S, the top 10 types of substances made up more than 60% in the peak areas (Figure 5C). In ET-22-HK, the top 10 types of substance in each kind of organic substance accounted for more than 70% (Figure 5D).
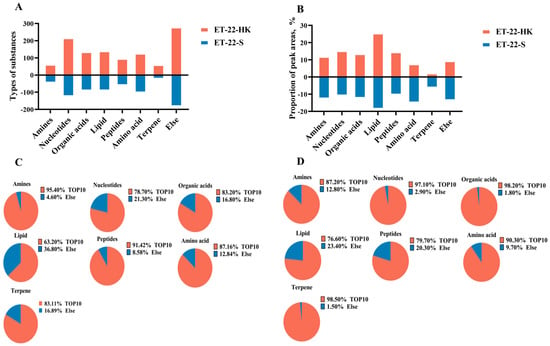
Figure 5.
The main organic substances in heat-killed bacteria and secretions of L. paracasei ET-22. (A) The number of substance types in each kind of organic substance from heat-killed bacteria (ET-22-HK) and secretions (ET-22-S) of L. paracasei ET-22. (B) The proportion of relative peak areas for each kind of organic substance. (C) The proportion of peak areas for the top 10 substance types in each kind of organic substance from ET-22-HK. (D) The proportion of peak areas for the top 10 substance types in each kind of organic substance from ET-22-S.
Based on the debris information, the specific substances need to be further identified. By checking against the HMDB database (http://www.hmdb.ca/ (accessed on 3 January 2023)), Metlin (https://metlin.scripps.edu/ (accessed on 3 January 2023)), and the Majorbio cloud platform (https://cloud.majorbio.com (accessed on 3 January 2023)), the top 10 types of substances from ET-22-HK classified as organic acids, nucleotides, terpenes, peptides, amino acids, lipids, and amines were identified. The metabolite names, formulas, and relative ratio of peak areas of these substances are shown in Table 1. Similarly, the top 10 types of substances in each kind of the above organic substances from ET-22-S are listed in Table 2. For each kind of organic substance, some of the substances in the top 10 were the same for ET-22-HK and for ET-22-S. In the organic acids, 9,10-Epoxy-18-hydroxy-octadecanoic acid, D-2-Hydroxyglutaric acid, 3-Aminopentanedioic acid, 6-Hydroxyhexanoic acid, and phenyllactic acid all existed in abundance in ET-22-HK and ET-22-S (Table 1 and Table 2). ET-22-HK and ET-22-S also contained L-Lysine, L-Tyrosine, and L-Leucine (Table 1 and Table 2). In the lipids, most substances that included PG(i-12:0/i-19:0), PA(20:4(8Z,11Z,14Z,17Z)/PGF1alpha), SM(d16:2(4E,8Z)/20:5(6E,8Z,11Z,14Z,17Z)-OH(5)), PA(TXB2/20:0), PA(PGE1/20:1(11Z)), PE(20:5(5Z,8Z,11Z,14Z,17Z)/18:2(9Z,12Z)), PI(18:0/18:2(9Z,12Z)), PA(TXB2/22:1(13Z)) and PG(a-21:0/20:4(7E,9E,11Z,13E)-3OH(5S,6R,15S)) in the top 10 coexisted in ET-22-HK and ET-22-S (Table 1 and Table 2). In the amines, N1,N12-Diacetylspermine, N-Acetylcadaverine, N-[4-[Acetyl(3-aminopropyl)amino]butyl]-N-(3-aminopropyl)acetamide, N1,N8-Diacetylspermidine, N-Cyclohexylformamide, and 2-Phenylacetamide were all abundant in ET-22-HK and ET-22-S (Table 1 and Table 2). However, only hypoxanthine and xanthine were common in the nucleotides (Table 1 and Table 2). Moreover, there was no common substance in the top 10 of terpenes and peptides between ET-22-HK and ET-22-S (Table 1 and Table 2).

Table 1.
Major compositions in ET-22-HK by non-targeted metabonomics 1.

Table 2.
Major compositions in ET-22-S by non-targeted metabonomics 1.
Other substances in the top 10 of organic acids, nucleotides, amino acids, lipids, and amines were all characteristic. For the organic acids, 9-Hydroxylinoleic acid, 2-Hydroxyadipic acid, DL-2-hydroxy stearic acid, behenic acid, and phytanic acid were peculiarly presented in ET-22-HK (Table 1). The 3-(4-Hydroxyphenyl)lactate, citramalic acid, acexamic acid, 12-hydroxyheptadecanoic acid, and azelaic acid were characteristic in ET-22-S (Table 2). In the nucleotides, ET-22-HK possessed abundant adenosine 3′-monophosphate, ADP-ribose, pseudouridine 5′-phosphate, adenosine monophosphate, guanidylic acid (guanosine monophosphate), zidovudine monophosphate, vidarabine, and 5′-Methylthioadenosine (Table 1). Notably, ET-22-S contained adenine, deoxyribose, FAPy-adenine, 7-Methylguanine, morph, oxypurinol, 5-Methyldeoxycytidine, and 1-(2-Hydroxyethyloxymethyl)-6-phenyl thiothymine (Table 2). In the amino acids, L-Carnitine, L-4-Hydroxyglutamate semialdehyde, citrulline, L-prolinamide, L-Glutamic Acid, cholylmethionine and N-Undecanoylglycine were particularly abundant in ET-22-HK (Table 1). ET-22-S contained abundant phenyl-Alanine, homocysteine, gamma-Glutamylvaline, tyrosine lactate, L-Phenylalanine, N-Acetyl-DL-Leucine, and N-Acetyl-DL-Phenylalanine (Table 2).
3. Discussion
3.1. Live L. paracasei ET-22 and Its Postbiotics Inhibit the Formation of S. mutans Biofilms
A biofilm is a collective lifestyle of microorganisms, and has many emergent characteristics, including interface aggregation, surface adhesion, water retention, nutrient absorption and retention, enhanced resistance, and collective cooperation. Once formed, the bacterial biofilm becomes a shelter for bacteria, which significantly enhances the resistance of bacteria to antimicrobial agents and their ability to escape from the host immune-system, leading to refractory and recurrent infections. In the oral cavity, caries is caused by oral biofilms formed by pathogenic S. mutans on the initial proteinaceous coating of the dental matrix [17]. Therefore, inhibiting the formation of the bacterial biofilms that cause dental caries, especially those of S. mutans, is important for the health of teeth. In view of the negative effects of antimicrobial agents, the biocontrol method has been recognized as a promising substitute. To verify whether the L. paracasei ET-22 can prevent the occurrence of dental caries in the oral cavity, the present research assessed the effects of L. paracasei ET-22 on the S. mutans biofilms, by co-culturing with S. mutans. To simulate the oral environment, the whole experiment used an artificial-saliva medium supplemented into 1% sucrose to culture S. mutans. After a 24 h co-culture, the live bacteria and secretions of L. paracasei ET-22 clearly showed significant inhibitory effects on the formation of S. mutans biofilms. This indicates that live L. paracasei ET-22 can inhibit the formation of S. mutans biofilms, by secreting metabolites. An unexpected phenomenon that emerged was that heat-killed L. paracasei ET-22 also showed a strong inhibition-effect on the formation of S. mutans biofilms, which had rarely been proved in previous studies. According to recently published research, postbiotics, productions from heat-killed probiotics, may play a stable and efficient role in protecting the health of the digestive tract [18]. Due to their colonization efficiency and stress resistance, live probiotics are less effective than postbiotics in some species such as Akkermansia muciniphila, which is used in the treatment of metabolic diseases [16]. Therefore, it can be inferred that the postbiotics of L. paracasei ET-22 may also inhibit the formation of S. mutans biofilms to protect patients from dental caries.
3.2. Live L. paracasei ET-22 and Its Postbiotics Inhibit the Formation of S. mutans Biofilms by Blocking the Initial Adhesion
EPS are the main components of oral biofilms [19,20]. By connecting with extracellular proteins, eDNA, and lipids, together they form biofilm matrices, which are conducive to bacterial colonization, biofilm formation and maintenance, and pathogenic realization [20]. S. mutans encodes several surface-associated GBP and glucosyltransferases, which serve as an integrated scaffold for biofilm formation by promoting the accumulation of local bacteria and forming a polymeric matrix [4,8,21]. To research the mechanisms whereby L. paracasei ET-22 inhibits the formation of S. mutans biofilms, we evaluated the effects of live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 on the gene expressions of brpA, gtfB, gbpB, gbpC, and gbpD of S. mutans. Among them, the water-insoluble glucan synthesized by gtfB, a glucosyltransferase of the GTF cluster encoded by the gtfB gene, was identified as the main component of EPS in S. mutans biofilms. In addition, brpA is involved in the biofilm formation and stress tolerance of S. mutans. The mutant strain lacking the brpA gene has a limited ability to grow and accumulate on solid surfaces [21]. Interestingly, our findings revealed that live L. paracasei ET-22 and its postbiotics not only reduced the gene expression of gtfB, but also reduced the gene-expression levels of gbpB-D in the GBP cluster and brpA. Similarly, previous studies have shown that Lactobacillus decreases the gene expression of brpA, the GBP cluster, and glucosyltransferase genes of the GTF cluster in S. mutans. One study found reduced biofilm-formation and lower gene-expression of gbpB and gtfB in S. mutans after co-culture with L. casei [22]. Another study also observed that, after a coculture with gellan containing L. paracasei 28.4, the S. mutans showed decreased biofilm-formation, accompanied by reduced gene-expression of brpA and gtfB-D [23]. These data suggest that live L. paracasei ET-22 and its postbiotics may inhibit the production of EPS by inhibiting the expression of the GTF cluster and brpA to defend against the formation of S. mutans biofilms. Secretions of L. paracasei ET-22 also showed inhibitory effects on the gene expression of brpA, gtfB, gbpB, gbpC, and gbpD of S. mutans, thereby blocking the construction of the biofilm structure. This indicates that suppressing the production of EPS by secretions is one of the pathways by which live L. paracasei ET-22 limits the formation of S. mutans biofilms. In line with our results, other studies also found that secretions of probiotics mainly change the biofilm structure by changing the expression of virulence genes related to the formation of S. mutans biofilms [17].
Adhesion is a prerequisite for the formation of biofilms. We aimed to determine whether L. paracasei ET-22 could affect the adhesion of S. mutans. The initial adhesion of S. mutans is achieved when it combines with the membrane receptors on the tooth surface via sucrose-independent weak interactions, which are mediated by long-range forces, i.e., van der Waals forces and Coulomb interactions, and short-range forces, including electrostatic interactions and hydrophobic interactions [24,25,26]. SPAP, a non-sucrose-dependent adhesion protein, is initially and primarily involved in promoting the colonization of S. mutans on tooth surfaces by specific interactions with salivary agglutinins [27]. Some studies have shown that SPAP-deficient S. mutans mutants showed a decreased ability to bind to salivary components and the cell matrix, as well as decreased co-aggregation-activity [28]. In this study, the SpaP expression of S. mutans was significantly downregulated by live L. paracasei ET-22 and its postbiotics. Meanwhile, they both reduced the initial adhesion-levels of S. mutans to hydroxyapatite. Consistently with these results, the CSLM results also showed the inhibitory effects of live L. paracasei ET-22 and its postbiotics on the formation of S. mutans biofilms. Therefore, it can be speculated that L. paracasei ET-22 and its postbiotics may inhibit the initial adhesion of S. mutans by changing the weak interactions between S. mutans and the tooth surface. A previous report found that postbiotics of L. paracasei may change the weak interactions during the initial adhesion of S. mutans [29]. This is in line with the findings of this study.
3.3. Live L. paracasei ET-22 and Its Postbiotics Inhibit S. mutans Biofilms by Interfering with the QS System and the Expression of Virulence Factors
Acid production and acid tolerance are considered the main virulence characteristics of S. mutans [3]. The inhibition of acid production and the regulation of acid sensitivity of S. mutans are important means of preventing dental caries [6,30,31,32]. LDH is an important acid-producing gene of S. mutans involved in the process of carbohydrate metabolism. It plays an important role in the process of tooth demineralization [13]. Some proteins, such as recA and ffh, could induce the DNA-repair mechanism of S. mutans to enhance the acid-tolerance capacity [33]. In addition, relA can improve tolerance of S. mutans to environmental stress, through the synthesis of (p)ppGpp [34]. Our study found that the transcriptions of LDH, ffh, recA, and relA were downregulated by L. paracasei ET-22 and its postbiotics, which may contribute to the inhibition of the formation of S. mutans biofilms by decreasing its acid-producing and acid-tolerance functions. Similarly, another study showed that the presence of L. rhamnosus significantly reduced the expression of LDH and then effectively modulated the formation of cariogenic biofilms [35]. To sum up, this evidence suggests that live L. paracasei ET-22 and its postbiotics may inhibit the formation of S. mutans biofilms by decreasing acid-production and acid-tolerance functions.
In the process of biofilm formation, various bacterial species communicate with others through the QS system in biofilms [3]. The ComCDE system is the most common intraspecific communication approach of QS systems in S. mutans biofilms, in which ComDE and comX can regulate the communications of S. mutans by competitive signaling-peptides [36]. The inactivation of the ComDE pathway may result in defective S. mutans biofilms [36,37]. The reduction in the gene expression of ComDE suppresses the expression of cariogenic virulence-factors in S. mutans [3,36]. Suppressed expression of ComDE and cariogenic LDH, ffh, recA, and relA genes indicate that live L. paracasei ET-22 and its postbiotics inhibit the expression of cariogenic virulence-factors by suppressing the ComDE pathway. However, the secretions of L. paracasei ET-22 showed the opposite effect on the gene expression of ComDE. The reason for this is unclear, and requires further research. Meanwhile, this finding also suggests that the bodies, but not the secretions, of L. paracasei ET-22 interfere with the QS system
3.4. The Inhibitory Effect of Live L. paracasei ET-22 and Its Postbiotics on S. mutans Biofilms Is Mediated by Multiple Components
Further research was required to determine which specific substances interfere with the QS system. Non-targeted LC-MS/MS analysis was used to further investigate the bioactive substances of the bodies and postbiotics of L. paracasei ET-22. Large numbers of organic substances were identified. Multiple organic acids can inhibit the formation of bacterial biofilms. On the one hand, an acidic environment may be detrimental for the survival and physiological activity of pathogens. On the other hand, some organic acids can inhibit the formation of biofilms through specific mechanisms. In the common organic acids of the postbiotics and secretions of L. paracasei ET-22, abundant phenyllactic acid was reported to show broad-spectrum inhibition of the biofilm formation of pathogenic bacteria, such as Actinobacillus actinomycetemcomitans, Listeria monocytogenes, Staphylococcus aureus, and Salmonella enteritidis [38]. Several studies have found that the suppressed effects were all involved in the transcriptional inhibitions of genes controlling the syntheses and secretions of exopolysaccharides, extracellular proteins, and virulence factors [39,40,41,42]. Another study further confirmed that phenyllactic acid played an inhibitive role on the QS system, including the Rhl and PQS QS systems, to suppress the gene expression of relative virulence-factors and biofilm development [43]. In agreement with previous findings, the live L. paracasei ET-22 and its postbiotics may inhibit the ComDE QS pathway in restraining the S. mutans biofilms partly by phenyllactic acids. In the characteristic nucleotides of postbiotics from L. paracasei ET-22, zidovudine has been reported to show strong activity against the biofilms of S. Typhimurium and E. coli [44]. Therefore, zidovudine monophosphate may also be a functional substance in the postbiotics of L. paracasei ET-22 that limits the development of S. mutans biofilms. However, the regulated pathway is unclear. Citrullination mediated by PPAD can constrain the formation of oral P. gingivalis biofilms, which is involved in the occurrence of periodontitis and other oral diseases [45]. In this process, citrulline is used to modify the arginine residues of gingipain-derived adhesin proteins [45]. Similarly, abundant citrulline in postbiotics of L. paracasei ET-22 may also be involved in inhibiting the development of S. mutans biofilms by the PPAD-citrullination pathway, as shown in the present study. Unlike the direct effects of the above functional substances, the characteristic metabolite N-undecanoylglycine can inhibit pathogenic bacteria and their physiological activities, through host immunity. Bacterial N-undecanoylglycine can be sensed by epithelial cells and activate the antimicrobial immunity of mucosa, such as in intestinal Tuft-2 cells [46]. Therefore, N-undecanoylglycine in the postbiotics of L. paracasei ET-22 may stimulate the oral mucosa to inhibit the development of S. mutans and its biofilms. However, the effects of these potent functional substances still need further confirmation. Additionally, there are still many common and specific substances in the postbiotics and secretions of L. paracasei ET-22, which may contain another substance that inhibits S. mutans biofilms or interferes with the QS system. The current research evidence is still insufficient. It should be noted that the research on postbiotics of L. paracasei ET-22 and its active substances still need further confirmation through animal experiments.
Dental caries is a result of biofilms formed on the teeth surface by S. mutans. Different Lactobacillus strains have different effects on the formation of S. mutans biofilms [47]. This study found that live bacteria, heat-killed bacteria, the ComDE QS pathway and secretions of L. paracasei ET-22 all inhibited the formation of S. mutans biofilms. They changed the weak-interaction forces, initial adhesions, and structure of S. mutans biofilms by competing with salivary membranes, reducing the expression of adhesion proteins, EPS, and virulence factors, and interfering with the ComDE QS pathway. The results from the present study provide new evidence for the potential efficacy of postbiotics in the prevention of dental caries. The postbiotics of L. paracasei ET-22 have strong functional activity as live bacteria, and might broaden the application of L. paracasei ET-22 for oral health. According to our results, considering the colonization efficiency of live bacteria, postbiotics of L. paracasei ET-22 may be a more stable product for use in protection against dental decay induced by S. mutans. Moreover, the application of postbiotics derived from L. paracasei ET-22 could reduce the potential risks brought about by live bacteria. Therefore, postbiotics of L. paracasei may be candidates for the prevention of dental caries.
4. Materials and Methods
4.1. Bacterial Strains and Culture Medium
The L. paracasei ET-22 was obtained from the Sanhe Fucheng Biotechnology Co. Ltd. (Sanhe, China). The S. mutans (CICC 10387) was acquired from the China Center of Industrial Culture Collection (Beijing, China). L. paracasei and S. mutans were cultured with commercial deMan, MRS, and BHI mediums (Beijing Land Bridge Technology Co., Beijing, China), at 37 °C. Before the experiments, all strains were stored at −80 °C.
4.2. Preparation of Live Bacteria, Heat-Killed Bacteria, and Secretions of L. paracasei ET-22
L. paracasei ET-22 was cultured overnight and then centrifuged at 10,000× g for 10 min. After being washed with PBS three times, the bacterial solution was resuspended and adjusted to 5.5 × 108 CFU/mL with PBS with a spectrophotometer value at OD600. After the culture on MRS agar medium (Beijing Land Bridge Technology Co., China), the bacterial concentration was further confirmed by viability count. Using this method, the live L. paracasei ET-22 was prepared. Using a similar procedure, bacterial solutions adjusted to 5.5 × 108 CFU/mL were inactivated by a water bath at 70 °C for 1 h, following the method used in a previous study [48]. Then, the heat-killed L. paracasei ET-22 was acquired. According to a previously reported method, the live bacteria adjusted to 5.5 × 108 CFU/mL were washed and resuspended with PBS (bacterial sludge (v):PBS (v) = 1:3) [49]. After being stirred at 60 g at room temperature for 2 h and then centrifuged at 10,000× g for 10 min, the supernatant was filtered with the 0.22 μm sterile filter membranes. The filtrate comprised the secretions of L. paracasei ET-22. The live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 were stored at 4 °C.
4.3. Treatment for the Formation of S. mutans Biofilms
First, 1.9 mL artificial-saliva medium (Solarbio, Beijing, China) supplemented with 1% sucrose and hydroxyapatite discs (Clarkson Chromatography Products, Inc., South Williamsport, PA, USA) was added into sterile 24-well polystyrene culture plates (Solarbio, China) [50]. The hydroxyapatite discs were used to support the biofilm formation. The S. mutans were inoculated into the culture system to reach the 5 × 108 CFU/mL. To investigate the effects of L. paracasei ET-22 on the formation of S. mutans biofilms, we used live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 to coculture with S. mutans. As Table 1 shows, a single S. mutans culture (Model group) was used as a negative control. The live bacteria (ET-22-L), heat-killed bacteria (ET-22-HK), and secretions (ET-22-S) of L. paracasei ET-22 were used to coculture with S. mutans. In each group, 50 μL PBS or live bacteria, heat-killed bacteria, and secretions of L. paracasei ET-22 were added into the culture system to coculture with S. mutans. After anaerobic culture for 24 h, the formation of S. mutans biofilms was assessed. The anaerobic-culture process was finished in anaerobic-culture bags (Hopebio, Yancheng, China), and the residual oxygen inside the bag was cleared, using an AnaeroPack (Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan). Group setting and culture conditions are as described in Table 3.

Table 3.
Group setting and culture of biofilm formation.
To assess the effects of the above treatments on the formation of S. mutans biofilms, the formation levels were analyzed using the crystal-violet staining as previously described [51]. In this method, the culture medium was cleared away. Then the biofilms were washed with PBS. After finishing the staining, the results were read with ELIASA (Perkin Elmer, PE Victor X3, Waltham, MA, USA) at Abs595nm.
4.4. Extracellular-Polysaccharide Production in Biofilms
The extracellular polysaccharide (EPS) productions of the S. mutans biofilms were measured using a previously described method [19,20]. Briefly, the resuspended biofilms were centrifuged in ultrapure water at 10,000× g for 10 min. After repeating three times, the collected supernatants were transferred into a new centrifuge tube with three times the volume of absolute ethanol. The solutions were mixed and then placed at −20 °C for 30 min to precipitate soluble polysaccharides. After centrifugation by 10,000× g for 10 min and washing with 70% alcohol, the soluble polysaccharides were resuspended and resolved with 5 mL NaOH solution at 1 M. Subsequently, 2 mL 5% phenol (m:v) and 5 mL of concentrated sulfuric acid were added, and the samples were treated away from light at room temperature for 10 min. The dissolved substances were soluble EPS. Having been vortexed for 30 s and placed into a water bath at 25 °C for 20 min, the samples’ absorbance was read at 490 nm to determine the soluble EPS contents.
After being repeatedly dissolved, the precipitates from the biofilms in ultrapure water were resuspended with 5 mL NaOH solution at 1 M and then shaken for 15 min. Subsequently, the solutions were centrifuged at 10,000× g for 10 min. The dissolved substances were insoluble EPS. The remainder of the operation was the same as the method used for soluble-EPS determination. After the process of precipitation, dissolution, reaction with phenol and sulfuric acid, a water bath, and reading values at 490 nm, the contents of insoluble EPS were acquired.
4.5. Biofilm Microstructure Observed Using Scanning Electron Microscopy
Scanning electron microscopy (SEM) was applied to observe the structure of biofilms as previously described [52]. Based on the culture method of S. mutans biofilms, glass slides were placed below the hydroxyapatite discs in the wells of 24-well polystyrene culture plates. After anaerobic culture for 24 h, the liquid culture medium was gently discarded, and PBS was used to wash the S. mutans biofilms 3 times. The S. mutans biofilms on glass slides were fixed at 4 °C overnight with PBS containing 2.5% glutaraldehyde. Glass slides were taken out and dehydrated with gradient-increased alcohol solutions from 30% (v:v) to 100% (v:v). Subsequently, the glass slides were dried in an oven at 37 °C for 24 h. After being sprayed gold, the S. mutans biofilms were observed with S-4800 SEM (HITACHI, Tokyo, Japan).
4.6. Detection of Biofilm Thickness with Confocal Laser Scanning Microscopy
Using the same method described in 4.5, S. mutans biofilms formed on hydroxyapatite discs. Live bacteria and dead bacteria in the biofilms were labeled with 2% N01 (m:v) (BBcellProbe®, Shanghai, China) and 5% PI (m:v) (BBcellProbe®, China), respectively [53]. After being washed with normal saline three times, the biofilms were stained with 500 μL 2% N01 in PBS and incubated at 25 °C for 15 min. Subsequently, the biofilms were washed twice with normal saline. Then, 500 μL 5% PI was added to incubate the biofilms at 25 °C for 15 min. Confocal imaging was performed using an LSM900 instrument (Leica, Wetzlar, Germany), in which the total magnification was 200× and the magnification of the oil lens was 630×. During the image acquisition, >600 nm was set for PI and 488–525 nm was used for N01. Moreover, the thickness of the biofilm was determined, using a 5 μm thick (8-bit, 1024 × 1024 pixels) Z-slice method. The biovolume of the biofilm and the layer-coverage distribution were quantified from the confocal stacks, using the image-processing software COMSTAT [54].
4.7. RT-qPCR for Gene-Expression Levels
After finishing the treatment, the RNAs were extracted and purified using the optimized protocol described by Kuhnert et al. [28]. Extracted crude RNAs were enzyme-catalyzed to remove contaminated genomic DNAs. Agarose gel electrophoresis was used to verify the integrity of the RNAs. Purified RNAs were reverse transcribed to cDNAs, using the PrimeScriptTM 1st Strand cDNA Synthesis Kits (Takara, Kusatsu, Japan). A quantitative real-time reverse-transcription polymerase-chain-reaction (qRT-PCR) was performed using the Applied Biosystems StepOneTM system (Thermo Fisher, Waltham, MA, USA), in which the PowerUpTM SYBRTM Green Master Mix (Thermo Fisher, USA) was used. Primers for target genes were designed for initial adhesion-protein production, glucan production, glucan-protein production, acid tolerance, and quorum sensing in S. mutans. The sequences are shown in Table 4. Primers were synthesized by Tsingke Biotechnology Co. Ltd. (Beijing, China). PCR reactions were performed as previously described [55]. The cycling conditions were as follows: one cycle at 95 °C for 5 min; 40 cycles for denaturation at 95 °C for 30 s, annealing at 52–58 °C (depending on the primers used) for 30 s; and extension- and fluorescent-data collection at 72 °C for 30 s. Dissociation curves were generated, followed by the cycling reactions. The expressions levels of the target genes were normalized to 16S rRNA [56]. Data were calculated using the 2−ΔΔCT method [55].

Table 4.
The table of primer sequences.
4.8. Active-Substances Determinations in Heat-Killed L. paracasei ET-22 by Non-Targeted Metabonomics
Here, 50 mg solid heat-killed L. paracasei ET-22 dried by freezing were accurately weighed, and the organic substances were extracted using a 400 µL 80% methanol aqueous solution (v:v) supplemented with 0.02 mg/mL L-2-chlorophenylalanin as an internal standard. The mixture was processed using a tissue crusher Wonbio-96c (Shanghai Wanbo Biotechnology Co., Ltd., Shanghai, China) at 50 Hz for 6 min at −10 °C followed by ultrasound treatment at 40 kHz for 30 min, at 5 °C. After being placed at −20 °C for 30 min to precipitate proteins, and a centrifugation by 13,000× g at 4 °C for 15 min, the supernatant was transferred for non-targeted LC-MS/MS analysis. A pooled quality-control sample was prepared by mixing equal volumes of all samples. The quality-control and analytic samples were tested in the same manner. Briefly, samples were separated by an HSS T3 column (100 mm × 2.1 mm, i.d., 1.8 μm) and then entered for mass-spectrometry detection. The mobile phases consisted of a 0.1% formic-acid aqueous solution (v:v): acetonitrile = 95:5 (v:v, solvent A) and 0.1% formic acid in acetonitrile (v:v): isopropanol: water = 47.5:47.5: 5 (v:v, solvent B). The injection volume of the sample was 2 µL and the flow rate was set to 0.4 mL/min. The column temperature was maintained at 40 °C. During the period of analysis, all of these samples were stored at 4 °C. The mass spectrometric data were collected using a Thermo UHPLC-Q Exactive HF-X Mass Spectrometer (Thermo Scientific high-resolution) equipped with an electrospray-ionization source operating in either the positive- or negative-ion mode. The optimal conditions were set as follows: heater temperature at 425 °C; capillary temperature at 325 °C; sheath-gas flow rate at 50 arb; aux-gas flow rate at 13 arb; ion-spray voltage floating (ISVF), −3500 V in the negative mode and 3500 V in the positive mode; normalized collision energy, 20–40–60 V rolling for MS/MS. The full MS-resolution was 60,000, and the MS/MS resolution was 7500. Data acquisition was performed using the data-dependent acquisition (DDA) mode. The detection was carried out over a mass range of 70–1050 m/z. The raw data of LC-MS/MS were preprocessed using the Progenesis QI software (Waters Corporation, Milford, MA, USA), and a three-dimensional data matrix in CSV format was exported. The information in this three-dimensional matrix includes sample information, metabolite names, and mass-spectral-response intensities. Internal-standard peaks, as well as any known false positive-peaks including noise, column bleed, and derivatized-reagent peaks, were removed from the data matrix. Effect peaks were pooled. At the same time, the metabolites were searched and identified using the HMDB database (http://www.hmdb.ca/ (accessed on 3 January 2023)) and Metlin (https://metlin.scripps.edu/ (accessed on 3 January 2023)).
The data were further uploaded to the Majorbio cloud platform (https://cloud.majorbio.com (accessed on 3 January 2023)) for data analysis. At least 80% of the metabolite features detected in any sample were retained. After being filtered, the minimum metabolite-values were imputed for specific samples, in which the metabolite levels fell below the low limit of quantitation and each metabolic feature was normalized by sum. In order to reduce the errors caused by sample preparation and instrument instability, the response intensities of the samples’ mass-spectrum peaks were normalized using the sum-normalization method, and the normalized-data matrix was obtained. At the same time, the variables with relative standard deviation (RSD) >30% of the quality-control sample were removed, and log10 transformation was performed to obtain the final data matrix for subsequent analysis.
4.9. Statistics
All data are shown as the mean ± SEM. Experimental results were analyzed using one-way analysis of variance (ANOVA) with an additional Dunnett’s test in GraphPad Prism 8 software (San Diego, CA, USA). Results from the confocal laser scanning microscopy were analyzed using Image J and Contest 2 (Technical University of Denmark, Lyngby, Denmark). When p < 0.05 and p < 0.01, the difference was significant and extremely significant, respectively. * p < 0.05, ** p < 0.01.
5. Conclusions
Our work suggests that live L. paracasei ET-22 and its postbiotics and secretions display inhibition effects on the adhesion and formation of S. mutans biofilms. Postbiotics of L. paracasei ET-22 have great potential as a biological anticariogenic-agent that acts against S. mutans biofilms to prevent dental decay.
Author Contributions
Conceptualization, M.Z., Z.Z. and J.W.; methodology, Z.Z. and J.W.; software, Z.Z. and J.W.; validation, Z.S., J.F. and Z.Z.; investigation, W.Z.; resources, W.Z. and W.-H.L.; data curation, Z.Z., F.L. and J.W.; writing—original draft preparation, Z.Z.; writing—review and editing, J.W.; visualization, Z.Z., W.Z. and W.-H.L.; supervision, M.Z. and W.-L.H.; project administration, M.Z. and W.-L.H.; funding acquisition, M.Z., J.W. and W.-L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31601443, 32101938), China Postdoctoral Science Foundation (2022M723422).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available herein.
Acknowledgments
Thanks to all the technicians not shown in the author list, for their technical support to this study. Thanks to the Sanhe Fucheng Biotechnology Co. Ltd. for providing the L. paracasei ET-22 strain. At the same time, thank you for your support from the Inner Mongolia Dairy Technology Research Institute Co., Ltd. and Inner Mongolia Yili Industrial Group Co., Ltd.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Lin, Y.; Zhou, X.; Li, Y. Strategies for Streptococcus mutans biofilm dispersal through extracellular polymeric substances disruption. Mol. Oral Microbiol. 2022, 37, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, T.; Peng, W.; Zhu, Y. Effects of resveratrol on cariogenic virulence properties of Streptococcus mutans. BMC Microbiol. 2020, 20, 99. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Bidossi, A.; De Grandi, R.; Toscano, M.; Bottagisio, M.; De Vecchi, E.; Gelardi, M.; Drago, L. Probiotics Streptococcus salivarius 24SMB and Streptococcus oralis 89a interfere with biofilm formation of pathogens of the upper respiratory tract. BMC Infect. Dis. 2018, 18, 653. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Hwang, G.; Liu, Y.; Gao, L.; Kilpatrick-Liverman, L.; Santarpia, P.; Zhou, X.; Koo, H. L-arginine modifies the exopolysaccharide matrix and thwarts Streptococcus mutans outgrowth within mixed-species oral biofilms. J. Bacteriol. 2016, 198, 2651–2661. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.N.; Choi, H.M.; Jeon, J.G. Relationship between sucrose concentration and bacteria proportion in a multispecies biofilm. J. Oral Microbiol. 2021, 13, 1910443. [Google Scholar] [CrossRef]
- Alshahrani, A.M.; Gregory, R.L. In vitro Cariostatic effects of cinnamon water extract on nicotine-induced Streptococcus mutans biofilm. BMC Complement. Med. Ther. 2020, 20, 45. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, W.; Zhang, M.; Zhang, L.; Shen, Y.; Huang, S.; Li, M.; Qiu, W.; Pan, Y.; Zhou, L.; et al. The inhibitory effects of ficin on Streptococcus mutans biofilm formation. BioMed Res. Int. 2021, 2021, 6692328. [Google Scholar] [CrossRef]
- Bowen, W.H.; Koo, H. Biology of Streptococcus mutans-derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011, 45, 69–86. [Google Scholar] [CrossRef]
- Simón-Soro, A.; Mira, A. Solving the etiology of dental caries. Trends Microbiol. 2015, 23, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Söderling, E.M.; Marttinen, A.M.; Haukioja, A.L. Probiotic lactobacilli interfere with Streptococcus mutans biofilm formation in vitro. Curr. Microbiol. 2011, 62, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, C.; Chen, J.; Zhou, S.; Zhao, Y.; Xu, M.; Xu, H. Dual mode of anti-biofilm action of G3 against Streptococcus mutans. ACS Appl. Mater. Interfaces 2020, 12, 27866–27875. [Google Scholar] [CrossRef]
- Pedret, A.; Valls, R.M.; Calderón-Pérez, L.; Llauradó, E.; Companys, J.; Pla-Pagà, L.; Moragas, A.; Martín-Luján, F.; Ortega, Y.; Giralt, M.; et al. Effects of daily consumption of the probiotic Bifidobacterium animalis subsp. lactis CECT 8145 on anthropometric adiposity biomarkers in abdominally obese subjects: A randomized controlled trial. Int. J. Obes. 2019, 43, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Hannig, C.; Hannig, M. The oral cavity—A key system to understand substratum-dependent bioadhesion on solid surfaces in man. Clin. Oral Investig. 2009, 13, 123–139. [Google Scholar] [CrossRef]
- Maehata, H.; Arai, S.; Iwabuchi, N.; Abe, F. Immuno-modulation by heat-killed Lacticaseibacillus paracasei MCC1849 and its application to food products. Int. J. Immunopathol. Pharmacol. 2021, 35. [Google Scholar] [CrossRef]
- Gulube, Z.; Patel, M. Effect of Punica granatum on the virulence factors of cariogenic bacteria Streptococcus mutans. Microb. Pathog. 2016, 98, 45–49. [Google Scholar] [CrossRef]
- Torino, M.I.; Taranto, M.P.; Sesma, F.; De Valdez, G.F. Heterofermentative pattern and exopolysaccharide production by Lactobacillus helveticus ATCC 15807 in response to environmental pH. J. Appl. Microbiol. 2001, 91, 846–852. [Google Scholar] [CrossRef]
- Deng, Y.; Yang, Y.; Zhang, B.; Chen, H.; Lu, Y.; Ren, S.; Lei, L.; Hu, T. The vicK gene of Streptococcus mutans mediates its cariogenicity via exopolysaccharides metabolism. Int. J. Oral Sci. 2021, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.T.; Yates, D.; Ahn, S.J.; Burne, R.A. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol. 2010, 10, 111. [Google Scholar] [CrossRef] [PubMed]
- De Alvarenga, J.A.; De Barros, P.P.; De Camargo Ribeiro, F.; Rossoni, R.D.; Garcia, M.T.; Dos Santos Velloso, M.; Shukla, S.; Fuchs, B.B.; Shukla, A.; Mylonakis, E.; et al. Probiotic effects of Lactobacillus paracasei 28.4 to inhibit Streptococcus mutans in a gellan-based formulation. Probiotics Antimicrob. Proteins 2021, 13, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; He, X.; Chen, J.; Tang, B.; Zheng, T.; Jing, M.; Lin, Y.; Pan, Y.; Ma, Q.; Li, Y.; et al. Transcriptional profiling reveals the importance of RcrR in the regulation of multiple sugar transportation and biofilm formation in Streptococcus mutans. mSystems 2021, 6, e0078821. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, Q. Influence of surface energy of modified surfaces on bacterial adhesion. Biophys. Chem. 2005, 117, 39–45. [Google Scholar] [CrossRef]
- Teughels, W.; Van Assche, N.; Sliepen, I.; Quirynen, M. Effect of material characteristics and/or surface topography on biofilm development. Clin. Oral Implants Res. 2006, 17, 68–81. [Google Scholar] [CrossRef]
- Yang, J.; Deng, D.; Brandt, B.W.; Nazmi, K.; Wu, Y.; Crielaard, W.; Ligtenberg, A. Diversity of SpaP in genetic and salivary agglutinin mediated adherence among Streptococcus mutans strains. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Katsumata, T.; Nguyen-Tra Le, M.; Kawada-Matsuo, M.; Taniguchi, Y.; Ouhara, K.; Oogai, Y.; Nakata, M.; Mizuno, N.; Nishitani, Y.; Komatsuzawa, H. Comprehensive characterization of sortase A-dependent surface proteins in Streptococcus mutans. Microbiol. Immunol. 2022, 66, 145–156. [Google Scholar] [CrossRef]
- Ciandrini, E.; Campana, R.; Baffone, W. Live and heat-killed Lactobacillus spp. interfere with Streptococcus mutans and Streptococcus oralis during biofilm development on titanium surface. Arch. Oral Biol. 2017, 78, 48–57. [Google Scholar] [CrossRef]
- Kuhnert, W.L.; Quivey, R.G., Jr. Genetic and biochemical characterization of the F-ATPase operon from Streptococcus sanguis 10904. J. Bacteriol. 2003, 185, 1525–1533. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, B.; Weir, M.D.; Homayounfar, N.; Fay, G.G.; Martinho, F.; Lei, L.; Bai, Y.; Hu, T.; Xu, H. S. mutans gene-modification and antibacterial resin composite as dual strategy to suppress biofilm acid production and inhibit caries. J. Dent. 2020, 93, 103278. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; McLean, J.S.; Lux, R.; He, X.; Shi, W. The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of Streptococcus mutans. Sci. Rep. 2015, 5, 18015. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, Z.Z.; Xu, S.B.; Ma, M.; Wei, X. Farnesol inhibits development of caries by augmenting oxygen sensitivity and suppressing virulence-associated gene expression in Streptococcus mutans. J. Biomed. Res. 2017, 31, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, J.P.; Liao, S.; Yao, X.; Ahn, S.J.; Isoda, R.; Nguyen, A.H.; Wen, Z.T. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 2012, 78, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Al-Ansari, M.M.; Al-Dahmash, N.D.; Ranjitsingh, A. Synthesis of silver nanoparticles using gum arabic: Evaluation of its inhibitory action on Streptococcus mutans causing dental caries and endocarditis. J. Infect. Public Health 2021, 14, 324–330. [Google Scholar] [CrossRef]
- Lu, J.; Cheng, L.; Huang, Y.; Jiang, Y.; Chu, C.H.; Peng, X.; Li, M.; Xu, H.; Zhou, X.; Ren, B. Resumptive Streptococcus mutans persisters induced from dimethylaminododecyl methacrylate elevated the cariogenic virulence by up-regulating the Quorum-Sensing and VicRK pathway genes. Front. Microbiol. 2020, 10, 3102. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Jiang, W.; Wang, K.; Luo, J.; Li, W.; Zhou, X.; Zhang, L. Antimicrobial peptide GH12 suppresses cariogenic virulence factors of Streptococcus mutans. J. Oral Microbiol. 2018, 10, 1442089. [Google Scholar] [CrossRef]
- Wu, H.; Guang, C.; Zhang, W.; Mu, W. Recent development of phenyllactic acid: Physicochemical properties, biotechnological production strategies and applications. Crit. Rev. Biotechnol. 2021, 1–16. [Google Scholar] [CrossRef]
- Liu, F.; Sun, Z.; Wang, F.; Liu, Y.; Zhu, Y.; Du, L.; Wang, D.; Xu, W. Inhibition of biofilm formation and exopolysaccharide synthesis of Enterococcus faecalis by phenyllactic acid. Food Microbiol. 2020, 86, 103344. [Google Scholar] [CrossRef]
- Shakya, S.; Danshiitsoodol, N.; Noda, M.; Inoue, Y.; Sugiyama, M. 3-Phenyllactic acid generated in medicinal plant extracts fermented with plant-derived lactic acid bacteria inhibits the biofilm synthesis of Aggregatibacter actinomycetemcomitans. Front. Microbiol. 2022, 13, 991144. [Google Scholar] [CrossRef]
- Jiang, X.; Jiang, C.; Yu, T.; Jiang, X.; Kang, R.; Ren, S.; Chen, H.; Zhang, Y.; Li, Y.; Meng, H.; et al. Phenyllactic acid application to control Listeria monocytogenes biofilms and its growth in milk and spiced beef. Int. J. Food Microbiol. 2022, 381, 109910. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, D.; Sun, J.; Sun, Z.; Liu, F.; Du, L.; Wang, D. Synergistic antibiofilm effects of ultrasound and phenyllactic acid against Staphylococcus aureus and Salmonella enteritidis. Foods 2021, 10, 2171. [Google Scholar] [CrossRef] [PubMed]
- Shariff, M.; Chatterjee, M.; Morris, S.D.; Paul, V.; Vasudevan, A.K.; Mohan, C.G.; Paul-Prasanth, B.; Biswas, R. Enhanced inhibition of Pseudomonas aeruginosa virulence factor production and biofilm development by sublethal concentrations of eugenol and phenyllactic acid. Lett. Appl. Microbiol. 2022, 75, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Pertusati, F.; Pileggi, E.; Richards, J.; Wootton, M.; Van Leemputte, T.; Persoons, L.; De Coster, D.; Villanueva, X.; Daelemans, D.; Steenackers, H.; et al. Drug repurposing: Phosphate prodrugs of anticancer and antiviral FDA-approved nucleosides as novel antimicrobials. J. Antimicrob. Chemother. 2020, 75, 2864–2878. [Google Scholar] [CrossRef] [PubMed]
- Vermilyea, D.M.; Ottenberg, G.K.; Davey, M.E. Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381. npj Biofilms Microbiomes 2019, 5, 7. [Google Scholar] [CrossRef]
- Xiong, Z.; Zhu, X.; Geng, J.; Xu, Y.; Wu, R.; Li, C.; Fan, D.; Qin, X.; Du, Y.; Tian, Y.; et al. Intestinal Tuft-2 cells exert antimicrobial immunity via sensing bacterial metabolite N-undecanoylglycine. Immunity 2022, 55, 686–700. [Google Scholar] [CrossRef]
- Wasfi, R.; El-Rahman, O.A.A.; Zafer, M.M.; Ashour, H.M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell Mol. Med. 2018, 22, 1972–1983. [Google Scholar] [CrossRef]
- Choi, J.H.; Moon, C.M.; Shin, T.S.; Kim, E.K.; McDowell, A.; Jo, M.K.; Joo, Y.H.; Kim, S.E.; Jung, H.K.; Shim, K.N.; et al. Lactobacillus paracasei-derived extracellular vesicles attenuate the intestinal inflammatory response by augmenting the endoplasmic reticulum stress pathway. Exp. Mol. Med. 2020, 52, 423–437. [Google Scholar] [CrossRef]
- Ayebo, A.D.; Angelo, I.A.; Shahani, K.M. Effect of ingesting Lactobacillus acidophilus milk upon fecal flora and enzyme activity in humans. Milchwissenschaft 1980, 35, 730–733. [Google Scholar] [CrossRef]
- Jiang, Q.; Stamatova, I.; Kainulainen, V.; Korpela, R.; Meurman, J.H. Interactions between Lactobacillus rhamnosus GG and oral micro-organisms in an in vitro biofilm model. BMC Microbiol. 2016, 16, 149. [Google Scholar] [CrossRef]
- Bijle, M.N.; Neelakantan, P.; Ekambaram, M.; Lo, E.; Yiu, C. Effect of a novel synbiotic on Streptococcus mutans. Sci. Rep. 2020, 10, 7951. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yin, L.; Ramage, G.; Li, B.; Tao, Y.; Zhi, Q.; Lin, H.; Zhou, Y. Assessing the impact of curcumin on dual-species biofilms formed by Streptococcus mutans and Candida albicans. Microbiologyopen 2019, 8, e937. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Huang, Y.; Chen, M.; Hu, C.; Chen, Y. Functional synergy of antimicrobial peptides and chlorhexidine acetate against gram-negative/gram-positive bacteria and a fungus in vitro and in vivo. Infect. Drug Resist. 2019, 12, 3227–3239. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D. Real-time quantitative PCR. Methods 2001, 4, 383–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, Y.; Zuo, Y.; Tang, C.; Zhou, F.; Cui, X.; Wang, L. Effects of Rhein-8-O-β-D-glucopyranoside on the biofilm formation of Streptococcus mutans. Curr. Microbiol. 2021, 78, 323–328. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).