Survivin Small Molecules Inhibitors: Recent Advances and Challenges
Abstract
:1. Introduction
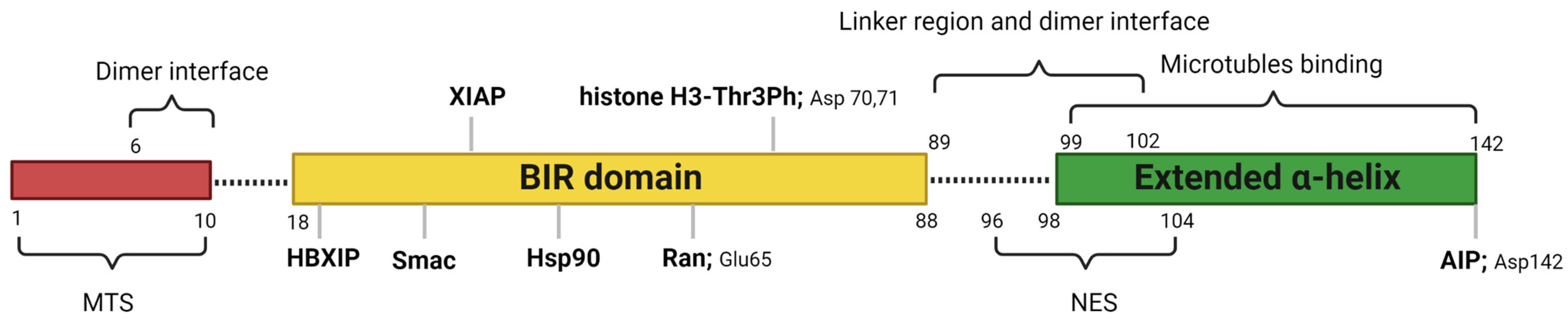
2. Structure and Cellular Functions of Survivin
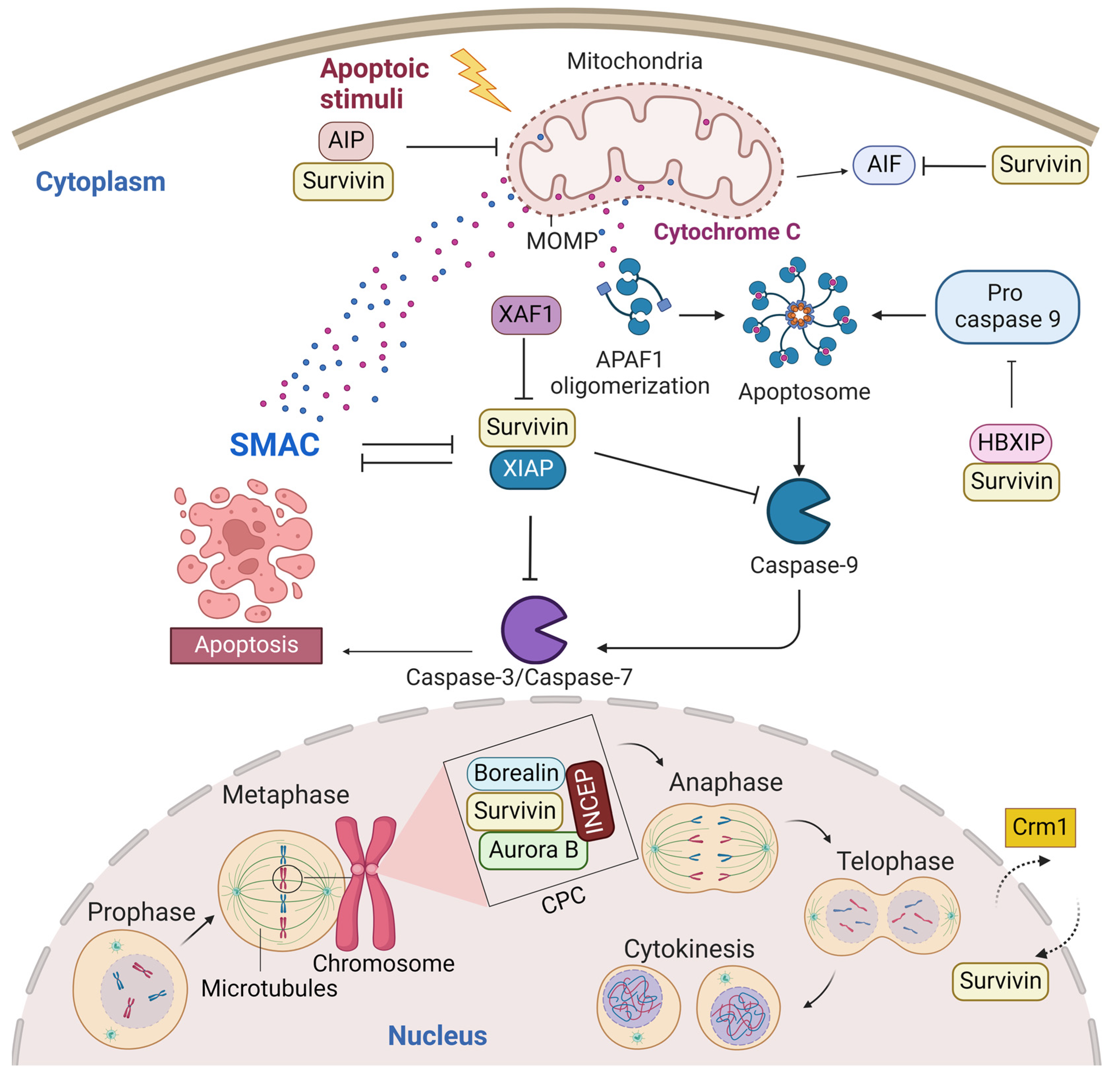
2.1. Cytoplasmic and Mitochondrial Survivin
2.2. Nuclear Survivin
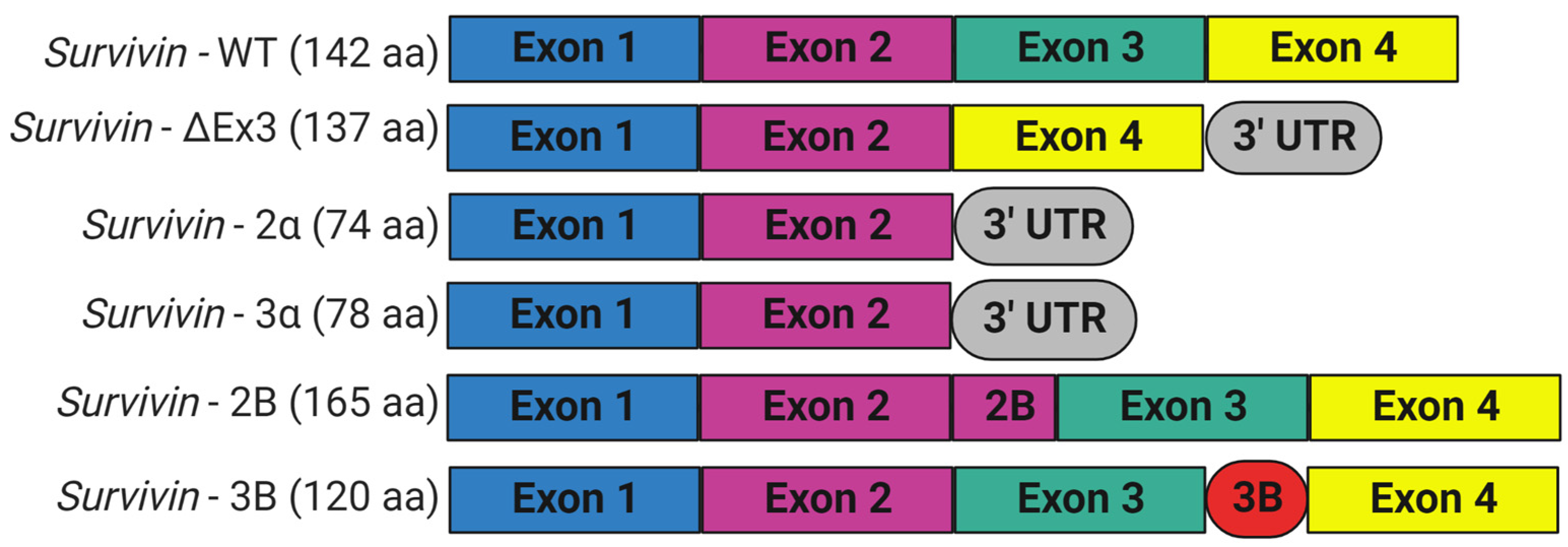
3. Expression and Isoforms of Survivin
4. Targeting Survivin for Cancer Therapy
4.1. Inhibitors That Decrease Survivin Gene Transcription
4.1.1. YM155 (1)
4.1.2. FL118 (2)
4.1.3. SF002-96-1 (3)
4.1.4. Terameprocol (Also Known as EM-1421, M4N, 4)
4.1.5. WM-127 (5)
4.1.6. GDP366 (6)
4.2. Inhibitors That Disrupt Survivin Homodimerization
4.2.1. Abbot 8 (7) and Its Analogs
4.2.2. S12 (10)
4.2.3. Indinavir (11) and Nelfinavir (12)
4.2.4. LQZ-7 (13) and Its Analogs
4.3. Inhibitors That Disrupt Survivin Interactions with its Partner Proteins
4.3.1. Shepherdin (16)
4.3.2. AICAR (17)
4.3.3. Deazaflavin Analog Compound 1 (18)
4.3.4. UC-112 (19) and Its Analogs
4.3.5. PZ-6-QN (26)
5. Final Remarks: Survivin in Cancer and the Efforts for Targeting Survivin So Far
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Crook, N.E.; Clem, R.J.; Miller, L.K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993, 67, 2168–2174. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Coulson, E.J.; Vaux, D.L. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem. Sci. 1998, 23, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Beilharz, T.; O’Connell, M.J.; Bugg, S.J.; van Driel, R.; Vaux, D.L.; Lithgow, T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA 1999, 96, 10170–10175. [Google Scholar] [CrossRef]
- Muchmore, S.W.; Chen, J.; Jakob, C.; Zakula, D.; Matayoshi, E.D.; Wu, W.; Zhang, H.; Li, F.; Ng, S.C.; Altieri, D.C. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell 2000, 6, 173–182. [Google Scholar] [CrossRef]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef]
- Verdecia, M.A.; Huang, H.; Dutil, E.; Kaiser, D.A.; Hunter, T.; Noel, J.P. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 2000, 7, 602–608. [Google Scholar] [CrossRef]
- Fortugno, P.; Wall, N.R.; Giodini, A.; O’Connor, D.S.; Plescia, J.; Padgett, K.M.; Tognin, S.; Marchisio, P.C.; Altieri, D.C. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J. Cell Sci. 2002, 115, 575–585. [Google Scholar] [CrossRef]
- Khan, S.; Aspe, J.R.; Asumen, M.G.; Almaguel, F.; Odumosu, O.; Acevedo-Martinez, S.; De Leon, M.; Langridge, W.H.; Wall, N.R. Extracellular, cell-permeable survivin inhibits apoptosis while promoting proliferative and metastatic potential. Br. J. Cancer 2009, 100, 1073–1086. [Google Scholar] [CrossRef]
- Altieri, D.C. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer 2003, 3, 46–54. [Google Scholar] [CrossRef]
- Humphry, N.J.; Wheatley, S.P. Survivin inhibits excessive autophagy in cancer cells but does so independently of its interaction with LC3. Biol. Open 2018, 7, bio037374. [Google Scholar] [CrossRef]
- Sanhueza, C.; Wehinger, S.; Castillo Bennett, J.; Valenzuela, M.; Owen, G.I.; Quest, A.F. The twisted survivin connection to angiogenesis. Mol. Cancer 2015, 14, 198. [Google Scholar] [CrossRef]
- Mull, A.N.; Klar, A.; Navara, C.S. Differential localization and high expression of SURVIVIN splice variants in human embryonic stem cells but not in differentiated cells implicate a role for SURVIVIN in pluripotency. Stem Cell Res. 2014, 12, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.K.; Kramer, O.H.; Knosel, T.; Engels, K.; Rodel, F.; Kovacs, A.F.; Dietmaier, W.; Klein-Hitpass, L.; Habtemichael, N.; Schweitzer, A.; et al. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 2007, 21, 207–216. [Google Scholar] [CrossRef]
- Li, F.; Yang, J.; Ramnath, N.; Javle, M.M.; Tan, D. Nuclear or cytoplasmic expression of survivin: What is the significance? Int. J. Cancer 2005, 114, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.A.; Span, S.W.; Ferreira, C.G.; Kruyt, F.A.; Giaccone, G. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein Survivin. Exp. Cell Res. 2002, 275, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.K.; Bier, C.; Habtemichael, N.; Stauber, R.H. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 2006, 7, 1259–1265. [Google Scholar] [CrossRef]
- Stauber, R.H.; Rabenhorst, U.; Rekik, A.; Engels, K.; Bier, C.; Knauer, S.K. Nucleocytoplasmic shuttling and the biological activity of mouse survivin are regulated by an active nuclear export signal. Traffic 2006, 7, 1461–1472. [Google Scholar] [CrossRef]
- Engelsma, D.; Rodriguez, J.A.; Fish, A.; Giaccone, G.; Fornerod, M. Homodimerization antagonizes nuclear export of survivin. Traffic 2007, 8, 1495–1502. [Google Scholar] [CrossRef]
- Dohi, T.; Okada, K.; Xia, F.; Wilford, C.E.; Samuel, T.; Welsh, K.; Marusawa, H.; Zou, H.; Armstrong, R.; Matsuzawa, S.; et al. An IAP-IAP complex inhibits apoptosis. J. Biol. Chem. 2004, 279, 34087–34090. [Google Scholar] [CrossRef] [Green Version]
- Liston, P.; Fong, W.G.; Kelly, N.L.; Toji, S.; Miyazaki, T.; Conte, D.; Tamai, K.; Craig, C.G.; McBurney, M.W.; Korneluk, R.G. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat. Cell. Biol. 2001, 3, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Arora, V.; Cheung, H.H.; Plenchette, S.; Micali, O.C.; Liston, P.; Korneluk, R.G. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J. Biol. Chem. 2007, 282, 26202–26209. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef]
- Song, Z.; Yao, X.; Wu, M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 2003, 278, 23130–23140. [Google Scholar] [CrossRef] [PubMed]
- Marusawa, H.; Matsuzawa, S.; Welsh, K.; Zou, H.; Armstrong, R.; Tamm, I.; Reed, J.C. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003, 22, 2729–2740. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Brouha, B.; Grossman, D. Rapid induction of mitochondrial events and caspase-independent apoptosis in Survivin-targeted melanoma cells. Oncogene 2004, 23, 39–48. [Google Scholar] [CrossRef]
- Liu, T.; Biddle, D.; Hanks, A.N.; Brouha, B.; Yan, H.; Lee, R.M.; Leachman, S.A.; Grossman, D. Activation of dual apoptotic pathways in human melanocytes and protection by survivin. J. Investig. Dermatol. 2006, 126, 2247–2256. [Google Scholar] [CrossRef]
- Dunajova, L.; Cash, E.; Markus, R.; Rochette, S.; Townley, A.R.; Wheatley, S.P. The N-terminus of survivin is a mitochondrial-targeting sequence and Src regulator. J. Cell Sci 2016, 129, 2707–2712. [Google Scholar] [CrossRef]
- Kang, B.H.; Altieri, D.C. Regulation of survivin stability by the aryl hydrocarbon receptor-interacting protein. J. Biol. Chem. 2006, 281, 24721–24727. [Google Scholar] [CrossRef]
- Fortugno, P.; Beltrami, E.; Plescia, J.; Fontana, J.; Pradhan, D.; Marchisio, P.C.; Sessa, W.C.; Altieri, D.C. Regulation of survivin function by Hsp90. Proc. Natl. Acad. Sci. USA 2003, 100, 13791–13796. [Google Scholar] [CrossRef] [Green Version]
- Colnaghi, R.; Connell, C.M.; Barrett, R.M.; Wheatley, S.P. Separating the anti-apoptotic and mitotic roles of survivin. J. Biol. Chem. 2006, 281, 33450–33456. [Google Scholar] [CrossRef] [PubMed]
- Connell, C.M.; Colnaghi, R.; Wheatley, S.P. Nuclear survivin has reduced stability and is not cytoprotective. J. Biol. Chem. 2008, 283, 3289–3296. [Google Scholar] [CrossRef]
- Krieg, S.; Roderburg, C.; Fung, S.; Luedde, T.; Knoefel, W.T.; Krieg, A. Nuclear survivin is a prognosticator in gastroenteropancreatic neuroendocrine neoplasms: A meta-analysis. J. Cancer Res. Clin. Oncol. 2022, 148, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Vay, C.; Babaei, S.; Safi, S.A.; Dizdar, L.; Rehders, A.; Haeberle, L.; Roderburg, C.; Loosen, S.H.; Esposito, I.; Knoefel, W.T.; et al. Clinicopathological and Prognostic Value of Survivin Expression in Surgically Resected Pancreatic Ductal Adenocarcinoma. Cancers 2022, 14, 3494. [Google Scholar] [CrossRef]
- Jeyaprakash, A.A.; Klein, U.R.; Lindner, D.; Ebert, J.; Nigg, E.A.; Conti, E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 2007, 131, 271–285. [Google Scholar] [CrossRef]
- Carmena, M.; Ruchaud, S.; Earnshaw, W.C. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell. Biol. 2009, 21, 796–805. [Google Scholar] [CrossRef]
- Jeyaprakash, A.A.; Basquin, C.; Jayachandran, U.; Conti, E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure 2011, 19, 1625–1634. [Google Scholar] [CrossRef]
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010, 330, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, S.P.; Carvalho, A.; Vagnarelli, P.; Earnshaw, W.C. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 2001, 11, 886–890. [Google Scholar] [CrossRef]
- Wheatley, S.P.; Barrett, R.M.; Andrews, P.D.; Medema, R.H.; Morley, S.J.; Swedlow, J.R.; Lens, S.M. Phosphorylation by aurora-B negatively regulates survivin function during mitosis. Cell Cycle 2007, 6, 1220–1230. [Google Scholar] [CrossRef]
- Lens, S.M.; Wolthuis, R.M.; Klompmaker, R.; Kauw, J.; Agami, R.; Brummelkamp, T.; Kops, G.; Medema, R.H. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003, 22, 2934–2947. [Google Scholar] [CrossRef]
- Carvalho, A.; Carmena, M.; Sambade, C.; Earnshaw, W.C.; Wheatley, S.P. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 2003, 116, 2987–2998. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Wong, L.; Pakusch, M.; Fowler, K.J.; Burrows, F.J.; Vaux, D.L.; Choo, K.H. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 2000, 10, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ambrosini, G.; Chu, E.Y.; Plescia, J.; Tognin, S.; Marchisio, P.C.; Altieri, D.C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 1998, 396, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Skoufias, D.A.; Mollinari, C.; Lacroix, F.B.; Margolis, R.L. Human survivin is a kinetochore-associated passenger protein. J. Cell Biol. 2000, 151, 1575–1582. [Google Scholar] [CrossRef]
- Rosa, J.; Canovas, P.; Islam, A.; Altieri, D.C.; Doxsey, S.J. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell 2006, 17, 1483–1493. [Google Scholar] [CrossRef]
- Xia, F.; Canovas, P.M.; Guadagno, T.M.; Altieri, D.C. A survivin-ran complex regulates spindle formation in tumor cells. Mol. Cell. Biol. 2008, 28, 5299–5311. [Google Scholar] [CrossRef]
- Garrett, S.; Auer, K.; Compton, D.A.; Kapoor, T.M. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 2002, 12, 2055–2059. [Google Scholar] [CrossRef]
- Gruss, O.J.; Wittmann, M.; Yokoyama, H.; Pepperkok, R.; Kufer, T.; Sillje, H.; Karsenti, E.; Mattaj, I.W.; Vernos, I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell. Biol. 2002, 4, 871–879. [Google Scholar] [CrossRef]
- Weis, K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell 2003, 112, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, S.; Pelus, L.M. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: Implication of survivin expression in normal hematopoiesis. Blood 2001, 98, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Foster, R.G.; Porter, S.B.; Pelus, L.M. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood 2002, 100, 2463–2471. [Google Scholar] [CrossRef] [PubMed]
- Martini, E.; Schneider, E.; Neufert, C.; Neurath, M.F.; Becker, C. Survivin is a guardian of the intestinal stem cell niche and its expression is regulated by TGF-beta. Cell Cycle 2016, 15, 2875–2881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tenev, T.; Martins, L.M.; Downward, J.; Lemoine, N.R. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J. Cell Sci. 2000, 113 Pt 23, 4363–4371. [Google Scholar] [CrossRef]
- Velculescu, V.E.; Madden, S.L.; Zhang, L.; Lash, A.E.; Yu, J.; Rago, C.; Lal, A.; Wang, C.J.; Beaudry, G.A.; Ciriello, K.M.; et al. Analysis of human transcriptomes. Nat. Genet. 1999, 23, 387–388. [Google Scholar] [CrossRef]
- Li, F.; Altieri, D.C. Transcriptional analysis of human survivin gene expression. Biochem. J. 1999, 344 Pt 2, 305–311. [Google Scholar] [CrossRef]
- Warrier, N.M.; Agarwal, P.; Kumar, P. Emerging Importance of Survivin in Stem Cells and Cancer: The Development of New Cancer Therapeutics. Stem Cell. Rev. Rep. 2020, 16, 828–852. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, J.; Li, F. Transcriptional and post-transcriptional controls of survivin in cancer cells: Novel approaches for cancer treatment. J. Exp. Clin. Cancer Res. 2006, 25, 391–402. [Google Scholar]
- Adamopoulos, P.G.; Tsiakanikas, P.; Adam, E.E.; Scorilas, A. Unraveling novel survivin mRNA transcripts in cancer cells using an in-house developed targeted high-throughput sequencing approach. Genomics 2021, 113, 573–581. [Google Scholar] [CrossRef]
- Sah, N.K.; Seniya, C. Survivin splice variants and their diagnostic significance. Tumour Biol. 2015, 36, 6623–6631. [Google Scholar] [CrossRef]
- Mahotka, C.; Wenzel, M.; Springer, E.; Gabbert, H.E.; Gerharz, C.D. Survivin-deltaEx3 and survivin-2B: Two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999, 59, 6097–6102. [Google Scholar] [PubMed]
- Badran, A.; Yoshida, A.; Ishikawa, K.; Goi, T.; Yamaguchi, A.; Ueda, T.; Inuzuka, M. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem. Biophys. Res. Commun. 2004, 314, 902–907. [Google Scholar] [CrossRef]
- Caldas, H.; Honsey, L.E.; Altura, R.A. Survivin 2alpha: A novel Survivin splice variant expressed in human malignancies. Mol. Cancer 2005, 4, 11. [Google Scholar] [CrossRef]
- Necochea-Campion, R.; Chen, C.S.; Mirshahidi, S.; Howard, F.D.; Wall, N.R. Clinico-pathologic relevance of Survivin splice variant expression in cancer. Cancer Lett. 2013, 339, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.K.; Bier, C.; Schlag, P.; Fritzmann, J.; Dietmaier, W.; Rodel, F.; Klein-Hitpass, L.; Kovacs, A.F.; Doring, C.; Hansmann, M.L.; et al. The survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex protein. Cell Cycle 2007, 6, 1502–1509. [Google Scholar] [CrossRef]
- Caldas, H.; Fangusaro, J.R.; Boue, D.R.; Holloway, M.P.; Altura, R.A. Dissecting the role of endothelial SURVIVIN DeltaEx3 in angiogenesis. Blood 2007, 109, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Faldt Beding, A.; Larsson, P.; Helou, K.; Einbeigi, Z.; Parris, T.Z. Pan-cancer analysis identifies BIRC5 as a prognostic biomarker. BMC Cancer 2022, 22, 322. [Google Scholar] [CrossRef]
- Sui, L.; Dong, Y.; Ohno, M.; Watanabe, Y.; Sugimoto, K.; Tokuda, M. Survivin expression and its correlation with cell proliferation and prognosis in epithelial ovarian tumors. Int. J. Oncol. 2002, 21, 315–320. [Google Scholar] [CrossRef]
- Ferrandina, G.; Legge, F.; Martinelli, E.; Ranelletti, F.O.; Zannoni, G.F.; Lauriola, L.; Gessi, M.; Gallotta, V.; Scambia, G. Survivin expression in ovarian cancer and its correlation with clinico-pathological, surgical and apoptosis-related parameters. Br. J. Cancer 2005, 92, 271–277. [Google Scholar] [CrossRef]
- Qi, G.; Tuncel, H.; Aoki, E.; Tanaka, S.; Oka, S.; Kaneko, I.; Okamoto, M.; Tatsuka, M.; Nakai, S.; Shimamoto, F. Intracellular localization of survivin determines biological behavior in colorectal cancer. Oncol. Rep. 2009, 22, 557–562. [Google Scholar] [CrossRef]
- Nestal de Moraes, G.; Delbue, D.; Silva, K.L.; Robaina, M.C.; Khongkow, P.; Gomes, A.R.; Zona, S.; Crocamo, S.; Mencalha, A.L.; Magalhaes, L.M.; et al. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell. Signal. 2015, 27, 2496–2505. [Google Scholar] [CrossRef] [PubMed]
- Shariat, S.F.; Lotan, Y.; Saboorian, H.; Khoddami, S.M.; Roehrborn, C.G.; Slawin, K.M.; Ashfaq, R. Survivin expression is associated with features of biologically aggressive prostate carcinoma. Cancer 2004, 100, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Gao, W.; Kang, Q.M.; Zhang, X.J.; Yang, S.G. Prognostic value of survivin in patients with gastric cancer: A systematic review with meta-analysis. PLoS ONE 2013, 8, e71930. [Google Scholar] [CrossRef] [PubMed]
- Adida, C.; Haioun, C.; Gaulard, P.; Lepage, E.; Morel, P.; Briere, J.; Dombret, H.; Reyes, F.; Diebold, J.; Gisselbrecht, C.; et al. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood 2000, 96, 1921–1925. [Google Scholar]
- Faccion, R.S.; Rezende, L.M.; Romano Sde, O.; Bigni Rde, S.; Mendes, G.L.; Maia, R.C. Centroblastic diffuse large B cell lymphoma displays distinct expression pattern and prognostic role of apoptosis resistance related proteins. Cancer Investig. 2012, 30, 404–414. [Google Scholar] [CrossRef] [PubMed]
- de Souza Reis, F.R.; de Faria, F.C.; Castro, C.P.; de Souza, P.S.; da Cunha Vasconcelos, F.; Bello, R.D.; da Silva, A.J.; Costa, P.R.; Maia, R.C. The therapeutical potential of a novel pterocarpanquinone LQB-118 to target inhibitor of apoptosis proteins in acute myeloid leukemia cells. Anti-cancer Agents Med. Chem. 2013, 13, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.; Uemura, A.; Harasawa, H.; Nagai, H.; Hirakata, Y.; Tomonaga, M.; Murata, K.; Sohda, H.; Nakagoe, T.; Shibasaki, S.; et al. Clinical relevance of survivin as a biomarker in neoplasms, especially in adult T-cell leukemias and acute leukemias. Int. J. Hematol. 2004, 80, 52–58. [Google Scholar] [CrossRef]
- Carter, B.Z.; Mak, D.H.; Schober, W.D.; Cabreira-Hansen, M.; Beran, M.; McQueen, T.; Chen, W.; Andreeff, M. Regulation of survivin expression through Bcr-Abl/MAPK cascade: Targeting survivin overcomes imatinib resistance and increases imatinib sensitivity in imatinib-responsive CML cells. Blood 2006, 107, 1555–1563. [Google Scholar] [CrossRef]
- Valent, P. Imatinib-resistant chronic myeloid leukemia (CML): Current concepts on pathogenesis and new emerging pharmacologic approaches. Biologics 2007, 1, 433–448. [Google Scholar]
- Grdina, D.J.; Murley, J.S.; Miller, R.C.; Mauceri, H.J.; Sutton, H.G.; Li, J.J.; Woloschak, G.E.; Weichselbaum, R.R. A survivin-associated adaptive response in radiation therapy. Cancer Res. 2013, 73, 4418–4428. [Google Scholar] [CrossRef]
- Virrey, J.J.; Guan, S.; Li, W.; Schonthal, A.H.; Chen, T.C.; Hofman, F.M. Increased survivin expression confers chemoresistance to tumor-associated endothelial cells. Am. J. Pathol. 2008, 173, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liang, L.; Yan, X.; Liu, N.; Gong, L.; Pan, S.; Lin, F.; Zhang, Q.; Zhao, H.; Zheng, F. Survivin status affects prognosis and chemosensitivity in epithelial ovarian cancer. Int. J. Gynecol. Cancer 2013, 23, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, B.; Fang, Y.; Liu, Y.; Wang, Y.; Li, J.; Zhou, W.; Wang, X. Overexpression of Class III beta-tubulin, Sox2, and nuclear Survivin is predictive of taxane resistance in patients with stage III ovarian epithelial cancer. BMC Cancer 2015, 15, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.W.; Xuan, Q.; Shu, Q.A.; Wu, S.S.; Chen, H.; Xiao, J.; Xiang, P.; Zhu, Y.P.; Wang, F.L.; Zhao, S.T. Correlation of tumor relapse and elevated expression of survivin and vascular endothelial growth factor in superficial bladder transitional cell carcinoma. Genet. Mol. Res. 2013, 12, 1045–1053. [Google Scholar] [CrossRef]
- Huang, W.; Mao, Y.; Zhan, Y.; Huang, J.; Wang, X.; Luo, P.; Li, L.I.; Mo, D.; Liu, Q.; Xu, H.; et al. Prognostic implications of survivin and lung resistance protein in advanced non-small cell lung cancer treated with platinum-based chemotherapy. Oncol. Lett. 2016, 11, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Salman, T.; Argon, A.; Kebat, T.; Vardar, E.; Erkan, N.; Alacacioglu, A. The prognostic significance of survivin expression in gallbladder carcinoma. APMIS 2016, 124, 633–638. [Google Scholar] [CrossRef]
- Yu, J.I.; Lee, H.; Park, H.C.; Choi, D.H.; Choi, Y.L.; Do, I.G.; Kim, H.C.; Lee, W.Y.; Yun, S.H.; Cho, Y.B.; et al. Prognostic significance of survivin in rectal cancer patients treated with surgery and postoperative concurrent chemo-radiation therapy. Oncotarget 2016, 7, 62676–62686. [Google Scholar] [CrossRef]
- Pu, Z.; Wang, Q.; Xie, H.; Wang, G.; Hao, H. Clinicalpathological and prognostic significance of survivin expression in renal cell carcinoma: A meta-analysis. Oncotarget 2017, 8, 19825–19833. [Google Scholar] [CrossRef]
- Ryan, B.; O’Donovan, N.; Browne, B.; O’Shea, C.; Crown, J.; Hill, A.D.; McDermott, E.; O’Higgins, N.; Duffy, M.J. Expression of survivin and its splice variants survivin-2B and survivin-DeltaEx3 in breast cancer. Br. J. Cancer 2005, 92, 120–124. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Meng, Y.; Chang, Y.; Xu, J.; Zhang, Q. Increased levels of LAPTM4B, VEGF and survivin are correlated with tumor progression and poor prognosis in breast cancer patients. Oncotarget 2017, 8, 41282–41293. [Google Scholar] [CrossRef]
- Ghanbari, P.; Mohseni, M.; Tabasinezhad, M.; Yousefi, B.; Saei, A.A.; Sharifi, S.; Rashidi, M.R.; Samadi, N. Inhibition of survivin restores the sensitivity of breast cancer cells to docetaxel and vinblastine. Appl. Biochem. Biotechnol. 2014, 174, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Li, S.D.; Huang, L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol. Pharm. 2006, 3, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, L. Tumor-targeted delivery of siRNA by non-viral vector: Safe and effective cancer therapy. Expert Opin. Drug Deliv. 2008, 5, 1301–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.D.; Huang, L. Surface-modified LPD nanoparticles for tumor targeting. Ann. N. Y. Acad. Sci. 2006, 1082, 1–8. [Google Scholar] [CrossRef]
- Chen, J.; Wu, W.; Tahir, S.K.; Kroeger, P.E.; Rosenberg, S.H.; Cowsert, L.M.; Bennett, F.; Krajewski, S.; Krajewska, M.; Welsh, K.; et al. Down-regulation of survivin by antisense oligonucleotides increases apoptosis, inhibits cytokinesis and anchorage-independent growth. Neoplasia 2000, 2, 235–241. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, T.; Liu, Z.; Tang, S.; Yue, M.; Feng, S.; Hu, M.; Xuan, L.; Chen, Y. Small interfering RNA targeting of the survivin gene inhibits human tumor cell growth in vitro. Exp. Ther. Med. 2017, 14, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhu, F.; Jiang, Y.; Sun, D.; Yang, B.; Yan, H. siRNA targeting survivin inhibits the growth and enhances the chemosensitivity of hepatocellular carcinoma cells. Oncol. Rep. 2013, 29, 1183–1188. [Google Scholar] [CrossRef]
- Zhao, G.; Wang, Q.; Gu, Q.; Qiang, W.; Wei, J.J.; Dong, P.; Watari, H.; Li, W.; Yue, J. Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget 2017, 8, 94666–94680. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, A.A.; Prasad, G.B.; Khan, N.; Tiwari, R.P.; Bisen, P.S. Growth inhibition and chemo-radiosensitization of head and neck squamous cell carcinoma (HNSCC) by survivin-siRNA lentivirus. Radiother. Oncol. 2016, 118, 359–368. [Google Scholar] [CrossRef]
- Lee, J.; Uy, B.R.; Liau, L.M. Brain Tumor Vaccines. Neurosurg. Clin. N. Am. 2021, 32, 225–234. [Google Scholar] [CrossRef]
- Li, F.; Aljahdali, I.; Ling, X. Cancer therapeutics using survivin BIRC5 as a target: What can we do after over two decades of study? J. Exp. Clin. Cancer Res. 2019, 38, 368. [Google Scholar] [CrossRef] [PubMed]
- Peery, R.C.; Liu, J.Y.; Zhang, J.T. Targeting survivin for therapeutic discovery: Past, present, and future promises. Drug Discov. Today 2017, 22, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, W. Recent Advances on Small-Molecule Survivin Inhibitors. Curr. Med. Chem. 2015, 22, 1136–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahara, T.; Kita, A.; Yamanaka, K.; Mori, M.; Amino, N.; Takeuchi, M.; Tominaga, F.; Hatakeyama, S.; Kinoyama, I.; Matsuhisa, A.; et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007, 67, 8014–8021. [Google Scholar] [CrossRef]
- Iwasa, T.; Okamoto, I.; Suzuki, M.; Nakahara, T.; Yamanaka, K.; Hatashita, E.; Yamada, Y.; Fukuoka, M.; Ono, K.; Nakagawa, K. Radiosensitizing effect of YM155, a novel small-molecule survivin suppressant, in non-small cell lung cancer cell lines. Clin. Cancer Res. 2008, 14, 6496–6504. [Google Scholar] [CrossRef]
- Tang, H.; Shao, H.; Yu, C.; Hou, J. Mcl-1 downregulation by YM155 contributes to its synergistic anti-tumor activities with ABT-263. Biochem. Pharmacol. 2011, 82, 1066–1072. [Google Scholar] [CrossRef]
- Cheng, Q.; Ling, X.; Haller, A.; Nakahara, T.; Yamanaka, K.; Kita, A.; Koutoku, H.; Takeuchi, M.; Brattain, M.G.; Li, F. Suppression of survivin promoter activity by YM155 involves disruption of Sp1-DNA interaction in the survivin core promoter. Int. J. Biochem. Mol. Biol. 2012, 3, 179–197. [Google Scholar]
- Yamauchi, T.; Nakamura, N.; Hiramoto, M.; Yuri, M.; Yokota, H.; Naitou, M.; Takeuchi, M.; Yamanaka, K.; Kita, A.; Nakahara, T.; et al. Sepantronium bromide (YM155) induces disruption of the ILF3/p54(nrb) complex, which is required for survivin expression. Biochem. Biophys. Res. Commun. 2012, 425, 711–716. [Google Scholar] [CrossRef]
- Hong, M.; Ren, M.Q.; Silva, J.; Paul, A.; Wilson, W.D.; Schroeder, C.; Weinberger, P.; Janik, J.; Hao, Z. YM155 inhibits topoisomerase function. Anti-cancer Drugs 2017, 28, 142–152. [Google Scholar] [CrossRef]
- Chen, J.; Pise-Masison, C.A.; Shih, J.H.; Morris, J.C.; Janik, J.E.; Conlon, K.C.; Keating, A.; Waldmann, T.A. Markedly additive antitumor activity with the combination of a selective survivin suppressant YM155 and alemtuzumab in adult T-cell leukemia. Blood 2013, 121, 2029–2037. [Google Scholar] [CrossRef]
- Kita, A.; Nakahara, T.; Yamanaka, K.; Nakano, K.; Nakata, M.; Mori, M.; Kaneko, N.; Koutoku, H.; Izumisawa, N.; Sasamata, M. Antitumor effects of YM155, a novel survivin suppressant, against human aggressive non-Hodgkin lymphoma. Leuk. Res. 2011, 35, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Kita, A.; Mitsuoka, K.; Kaneko, N.; Nakata, M.; Yamanaka, K.; Jitsuoka, M.; Miyoshi, S.; Noda, A.; Mori, M.; Nakahara, T.; et al. Sepantronium bromide (YM155) enhances response of human B-cell non-Hodgkin lymphoma to rituximab. J. Pharmacol. Exp. Ther. 2012, 343, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Nakata, M.; Kaneko, N.; Fushiki, H.; Kita, A.; Nakahara, T.; Koutoku, H.; Sasamata, M. YM155, a selective survivin suppressant, inhibits tumor spread and prolongs survival in a spontaneous metastatic model of human triple negative breast cancer. Int. J. Oncol. 2011, 39, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, T.; Kita, A.; Yamanaka, K.; Mori, M.; Amino, N.; Takeuchi, M.; Tominaga, F.; Kinoyama, I.; Matsuhisa, A.; Kudou, M.; et al. Broad spectrum and potent antitumor activities of YM155, a novel small-molecule survivin suppressant, in a wide variety of human cancer cell lines and xenograft models. Cancer Sci. 2011, 102, 614–621. [Google Scholar] [CrossRef]
- Yamanaka, K.; Nakahara, T.; Yamauchi, T.; Kita, A.; Takeuchi, M.; Kiyonaga, F.; Kaneko, N.; Sasamata, M. Antitumor activity of YM155, a selective small-molecule survivin suppressant, alone and in combination with docetaxel in human malignant melanoma models. Clin. Cancer Res. 2011, 17, 5423–5431. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Yadav, A.; Lang, J.C.; Cipolla, M.J.; Schmitt, A.C.; Arradaza, N.; Teknos, T.N.; Kumar, P. YM155 reverses cisplatin resistance in head and neck cancer by decreasing cytoplasmic survivin levels. Mol. Cancer Ther. 2012, 11, 1988–1998. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.; Stanzani, E.; Martinez-Soler, F.; Villanueva, A.; Vidal, A.; Condom, E.; Ponce, J.; Gil, J.; Tortosa, A.; Gimenez-Bonafe, P. YM155 sensitizes ovarian cancer cells to cisplatin inducing apoptosis and tumor regression. Gynecol. Oncol. 2014, 132, 211–220. [Google Scholar] [CrossRef]
- Yoon, D.H.; Shin, J.S.; Jin, D.H.; Hong, S.W.; Jung, K.A.; Kim, S.M.; Hong, Y.S.; Kim, K.P.; Lee, J.L.; Suh, C.; et al. The survivin suppressant YM155 potentiates chemosensitivity to gemcitabine in the human pancreatic cancer cell line MiaPaCa-2. Anti-cancer Res. 2012, 32, 1681–1688. [Google Scholar]
- Tolcher, A.W.; Mita, A.; Lewis, L.D.; Garrett, C.R.; Till, E.; Daud, A.I.; Patnaik, A.; Papadopoulos, K.; Takimoto, C.; Bartels, P.; et al. Phase I and pharmacokinetic study of YM155, a small-molecule inhibitor of survivin. J. Clin. Oncol. 2008, 26, 5198–5203. [Google Scholar] [CrossRef]
- Kelly, R.J.; Thomas, A.; Rajan, A.; Chun, G.; Lopez-Chavez, A.; Szabo, E.; Spencer, S.; Carter, C.A.; Guha, U.; Khozin, S.; et al. A phase I/II study of sepantronium bromide (YM155, survivin suppressor) with paclitaxel and carboplatin in patients with advanced non-small-cell lung cancer. Ann. Oncol. 2013, 24, 2601–2606. [Google Scholar] [CrossRef]
- Kudchadkar, R.; Ernst, S.; Chmielowski, B.; Redman, B.G.; Steinberg, J.; Keating, A.; Jie, F.; Chen, C.; Gonzalez, R.; Weber, J. A phase 2, multicenter, open-label study of sepantronium bromide (YM155) plus docetaxel in patients with stage III (unresectable) or stage IV melanoma. Cancer Med. 2015, 4, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Clemens, M.R.; Gladkov, O.A.; Gartner, E.; Vladimirov, V.; Crown, J.; Steinberg, J.; Jie, F.; Keating, A. Phase II, multicenter, open-label, randomized study of YM155 plus docetaxel as first-line treatment in patients with HER2-negative metastatic breast cancer. Breast Cancer Res. Treat. 2015, 149, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, K.P.; Lopez-Jimenez, J.; Smith, S.E.; Steinberg, J.; Keating, A.; Sasse, C.; Jie, F.; Thyss, A. A multicenter phase II study of sepantronium bromide (YM155) plus rituximab in patients with relapsed aggressive B-cell Non-Hodgkin lymphoma. Leuk. Lymphoma 2016, 57, 1848–1855. [Google Scholar] [CrossRef] [PubMed]
- Giaccone, G.; Zatloukal, P.; Roubec, J.; Floor, K.; Musil, J.; Kuta, M.; van Klaveren, R.J.; Chaudhary, S.; Gunther, A.; Shamsili, S. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J. Clin. Oncol. 2009, 27, 4481–4486. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Quinn, D.I.; Ferrari, A.; Ahmann, F.; Giaccone, G.; Drake, T.; Keating, A.; de Bono, J.S. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann. Oncol. 2012, 23, 968–973. [Google Scholar] [CrossRef]
- Iwai, M.; Minematsu, T.; Li, Q.; Iwatsubo, T.; Usui, T. Utility of P-glycoprotein and organic cation transporter 1 double-transfected LLC-PK1 cells for studying the interaction of YM155 monobromide, novel small-molecule survivin suppressant, with P-glycoprotein. Drug Metab. Dispos. 2011, 39, 2314–2320. [Google Scholar] [CrossRef] [PubMed]
- Lamers, F.; Schild, L.; Koster, J.; Versteeg, R.; Caron, H.N.; Molenaar, J.J. Targeted BIRC5 silencing using YM155 causes cell death in neuroblastoma cells with low ABCB1 expression. Eur. J. Cancer 2012, 48, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Cao, S.; Cheng, Q.; Keefe, J.T.; Rustum, Y.M.; Li, F. A novel small molecule FL118 that selectively inhibits survivin, Mcl-1, XIAP and cIAP2 in a p53-independent manner, shows superior antitumor activity. PLoS ONE 2012, 7, e45571. [Google Scholar] [CrossRef]
- Li, F.; Ling, X.; Harris, D.L.; Liao, J.; Wang, Y.; Westover, D.; Jiang, G.; Xu, B.; Boland, P.M.; Jin, C. Topoisomerase I (Top1): A major target of FL118 for its antitumor efficacy or mainly involved in its side effects of hematopoietic toxicity? Am. J. Cancer Res. 2017, 7, 370–382. [Google Scholar]
- Westover, D.; Ling, X.; Lam, H.; Welch, J.; Jin, C.; Gongora, C.; Del Rio, M.; Wani, M.; Li, F. FL118, a novel camptothecin derivative, is insensitive to ABCG2 expression and shows improved efficacy in comparison with irinotecan in colon and lung cancer models with ABCG2-induced resistance. Mol. Cancer 2015, 14, 92. [Google Scholar] [CrossRef]
- Ling, X.; Liu, X.; Zhong, K.; Smith, N.; Prey, J.; Li, F. FL118, a novel camptothecin analogue, overcomes irinotecan and topotecan resistance in human tumor xenograft models. Am. J. Transl. Res. 2015, 7, 1765–1781. [Google Scholar] [PubMed]
- Zhao, J.; Ling, X.; Cao, S.; Liu, X.; Wan, S.; Jiang, T.; Li, F. Antitumor activity of FL118, a survivin, Mcl-1, XIAP, and cIAP2 selective inhibitor, is highly dependent on its primary structure and steric configuration. Mol. Pharm. 2014, 11, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Wu, W.; Aljahdali, I.A.M.; Liao, J.; Santha, S.; Fountzilas, C.; Boland, P.M.; Li, F. FL118, acting as a ‘molecular glue degrader’, binds to dephosphorylates and degrades the oncoprotein DDX5 (p68) to control c-Myc, survivin and mutant Kras against colorectal and pancreatic cancer with high efficacy. Clin. Transl. Med. 2022, 12, e881. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Wu, W.; Fan, C.; Xu, C.; Liao, J.; Rich, L.J.; Huang, R.Y.; Repasky, E.A.; Wang, X.; Li, F. An ABCG2 non-substrate anticancer agent FL118 targets drug-resistant cancer stem-like cells and overcomes treatment resistance of human pancreatic cancer. J. Exp. Clin. Cancer Res. 2018, 37, 240. [Google Scholar] [CrossRef]
- Felix, S.; Sandjo, L.P.; Opatz, T.; Erkel, G. SF002-96-1, a new drimane sesquiterpene lactone from an Aspergillus species, inhibits survivin expression. Beilstein J. Org. Chem. 2013, 9, 2866–2876. [Google Scholar] [CrossRef]
- Smolewski, P. Terameprocol, a novel site-specific transcription inhibitor with anticancer activity. IDrugs 2008, 11, 204–214. [Google Scholar]
- Chang, C.C.; Heller, J.D.; Kuo, J.; Huang, R.C. Tetra-O-methyl nordihydroguaiaretic acid induces growth arrest and cellular apoptosis by inhibiting Cdc2 and survivin expression. Proc. Natl. Acad. Sci. USA 2004, 101, 13239–13244. [Google Scholar] [CrossRef] [Green Version]
- Park, R.; Chang, C.C.; Liang, Y.C.; Chung, Y.; Henry, R.A.; Lin, E.; Mold, D.E.; Huang, R.C. Systemic treatment with tetra-O-methyl nordihydroguaiaretic acid suppresses the growth of human xenograft tumors. Clin. Cancer Res. 2005, 11, 4601–4609. [Google Scholar] [CrossRef]
- Chao, A.; Lin, C.Y.; Wu, R.C.; Lee, Y.S.; Lee, L.Y.; Tsai, C.L.; Yang, L.Y.; Liu, H.; Chen, S.J.; Wang, T.H.; et al. The combination of everolimus and terameprocol exerts synergistic antiproliferative effects in endometrial cancer: Molecular role of insulin-like growth factor binding protein 2. J. Mol. Med. 2018, 96, 1251–1266. [Google Scholar] [CrossRef]
- Tibes, R.; McDonagh, K.T.; Lekakis, L.; Bogenberger, J.M.; Kim, S.; Frazer, N.; Mohrland, S.; Bassett, D.; Garcia, R.; Schroeder, K.; et al. Phase I study of the novel Cdc2/CDK1 and AKT inhibitor terameprocol in patients with advanced leukemias. Investig. New Drugs 2015, 33, 389–396. [Google Scholar] [CrossRef]
- Khanna, N.; Dalby, R.; Tan, M.; Arnold, S.; Stern, J.; Frazer, N. Phase I/II clinical safety studies of terameprocol vaginal ointment. Gynecol. Oncol. 2007, 107, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Grossman, S.A.; Ye, X.; Peereboom, D.; Rosenfeld, M.R.; Mikkelsen, T.; Supko, J.G.; Desideri, S.; Adult Brain Tumor, C. Phase I study of terameprocol in patients with recurrent high-grade glioma. Neuro Oncol. 2012, 14, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Que, R.; Liu, C.; Ji, W.; Sun, B.; Lin, X.; Zhang, Q.; Zhao, X.; Peng, Z.; Zhang, X.; et al. Survivin-targeted drug screening platform identifies a matrine derivative WM-127 as a potential therapeutics against hepatocellular carcinoma. Cancer Lett. 2018, 425, 54–64. [Google Scholar] [CrossRef]
- Shi, X.; Wang, D.; Ding, K.; Lu, Z.; Jin, Y.; Zhang, J.; Pan, J. GDP366, a novel small molecule dual inhibitor of survivin and Op18, induces cell growth inhibition, cellular senescence and mitotic catastrophe in human cancer cells. Cancer Biol. Ther. 2010, 9, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Carlos, J.; Lima, K.; Costa-Lotufo, L.V.; Leitao, A.; Machado-Neto, J.A. AD80, a multikinase inhibitor, exhibits antineoplastic effects in acute leukemia cellular models targeting the PI3K/STMN1 axis. Investig. New Drugs 2021, 39, 1139–1149. [Google Scholar] [CrossRef]
- Wendt, M.D.; Sun, C.; Kunzer, A.; Sauer, D.; Sarris, K.; Hoff, E.; Yu, L.; Nettesheim, D.G.; Chen, J.; Jin, S.; et al. Discovery of a novel small molecule binding site of human survivin. Bioorg. Med. Chem. Lett. 2007, 17, 3122–3129. [Google Scholar] [CrossRef]
- Chettiar, S.N.; Cooley, J.V.; Park, I.H.; Bhasin, D.; Chakravarti, A.; Li, P.K.; Li, C.; Jacob, N.K. Design, synthesis and biological studies of survivin dimerization modulators that prolong mitotic cycle. Bioorg. Med. Chem. Lett. 2013, 23, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Guvenc, H.; Pavlyukov, M.S.; Joshi, K.; Kurt, H.; Banasavadi-Siddegowda, Y.K.; Mao, P.; Hong, C.; Yamada, R.; Kwon, C.H.; Bhasin, D.; et al. Impairment of glioma stem cell survival and growth by a novel inhibitor for Survivin-Ran protein complex. Clin. Cancer Res. 2013, 19, 631–642. [Google Scholar] [CrossRef]
- Steigerwald, C.; Rasenberger, B.; Christmann, M.; Tomicic, M.T. Sensitization of colorectal cancer cells to irinotecan by the Survivin inhibitor LLP3 depends on XAF1 proficiency in the context of mutated p53. Arch. Toxicol. 2018, 92, 2645–2648. [Google Scholar] [CrossRef]
- Berezov, A.; Cai, Z.; Freudenberg, J.A.; Zhang, H.; Cheng, X.; Thompson, T.; Murali, R.; Greene, M.I.; Wang, Q. Disabling the mitotic spindle and tumor growth by targeting a cavity-induced allosteric site of survivin. Oncogene 2012, 31, 1938–1948. [Google Scholar] [CrossRef]
- Brun, S.N.; Markant, S.L.; Esparza, L.A.; Garcia, G.; Terry, D.; Huang, J.M.; Pavlyukov, M.S.; Li, X.N.; Grant, G.A.; Crawford, J.R.; et al. Survivin as a therapeutic target in Sonic hedgehog-driven medulloblastoma. Oncogene 2015, 34, 3770–3779. [Google Scholar] [CrossRef]
- Sarvagalla, S.; Cheung, C.H.; Tsai, J.-Y.; Hsieh, H.-P.; Coumar, M. Disruption of Protein-Protein Interactions: Hot spot detection, structure-based virtual screening and in vitro testing for anti-cancer drug target—Survivin. RSC Adv. 2016, 6, 31947–31959. [Google Scholar] [CrossRef]
- Gupta, V.; Samuleson, C.G.; Su, S.; Chen, T.C. Nelfinavir potentiation of imatinib cytotoxicity in meningioma cells via survivin inhibition. Neurosurg. Focus 2007, 23, E9. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Dong, Z.; Liu, J.; Peery, R.C.; Zhang, S.; Liu, J.Y.; Zhang, J.T. Effective Targeting of the Survivin Dimerization Interface with Small-Molecule Inhibitors. Cancer Res. 2016, 76, 453–462. [Google Scholar] [CrossRef]
- Peery, R.; Kyei-Baffour, K.; Dong, Z.; Liu, J.; de Andrade Horn, P.; Dai, M.; Liu, J.Y.; Zhang, J.T. Synthesis and Identification of a Novel Lead Targeting Survivin Dimerization for Proteasome-Dependent Degradation. J. Med. Chem. 2020, 63, 7243–7251. [Google Scholar] [CrossRef] [PubMed]
- Plescia, J.; Salz, W.; Xia, F.; Pennati, M.; Zaffaroni, N.; Daidone, M.G.; Meli, M.; Dohi, T.; Fortugno, P.; Nefedova, Y.; et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell 2005, 7, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Stella, S.; Tirro, E.; Conte, E.; Stagno, F.; Di Raimondo, F.; Manzella, L.; Vigneri, P. Suppression of survivin induced by a BCR-ABL/JAK2/STAT3 pathway sensitizes imatinib-resistant CML cells to different cytotoxic drugs. Mol. Cancer Ther. 2013, 12, 1085–1098. [Google Scholar] [CrossRef] [Green Version]
- Meli, M.; Pennati, M.; Curto, M.; Daidone, M.G.; Plescia, J.; Toba, S.; Altieri, D.C.; Zaffaroni, N.; Colombo, G. Small-molecule targeting of heat shock protein 90 chaperone function: Rational identification of a new anticancer lead. J. Med. Chem. 2006, 49, 7721–7730. [Google Scholar] [CrossRef]
- Oikawa, T.; Unno, Y.; Matsuno, K.; Sawada, J.; Ogo, N.; Tanaka, K.; Asai, A. Identification of a small-molecule inhibitor of the interaction between Survivin and Smac/DIABLO. Biochem. Biophys. Res. Commun. 2010, 393, 253–258. [Google Scholar] [CrossRef]
- Wang, J.; Li, W. Discovery of novel second mitochondria-derived activator of caspase mimetics as selective inhibitor of apoptosis protein inhibitors. J. Pharmacol. Exp. Ther. 2014, 349, 319–329. [Google Scholar] [CrossRef]
- Xiao, M.; Wang, J.; Lin, Z.; Lu, Y.; Li, Z.; White, S.W.; Miller, D.D.; Li, W. Design, Synthesis and Structure-Activity Relationship Studies of Novel Survivin Inhibitors with Potent Anti-Proliferative Properties. PLoS ONE 2015, 10, e0129807. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, B.; Mani, A.M.; Wu, Z.; Fan, Y.; Li, W.; Wu, Z.H. Survivin Inhibitors Mitigate Chemotherapeutic Resistance in Breast Cancer Cells by Suppressing Genotoxic Nuclear Factor-kappaB Activation. J. Pharmacol. Exp. Ther. 2018, 366, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, Q.; Wu, Z.; Tian, X.; Yan, H.; Wang, B.; Dong, P.; Watari, H.; Pfeffer, L.M.; Guo, Y.; et al. Ovarian Primary and Metastatic Tumors Suppressed by Survivin Knockout or a Novel Survivin Inhibitor. Mol. Cancer Ther. 2019, 18, 2233–2245. [Google Scholar] [CrossRef]
- Albadari, N.; Deng, S.; Chen, H.; Zhao, G.; Yue, J.; Zhang, S.; Miller, D.D.; Wu, Z.; Li, W. Synthesis and biological evaluation of selective survivin inhibitors derived from the MX-106 hydroxyquinoline scaffold. Eur. J. Med. Chem. 2021, 224, 113719. [Google Scholar] [CrossRef]
- Wang, Q.; Arnst, K.E.; Xue, Y.; Lei, Z.N.; Ma, D.; Chen, Z.S.; Miller, D.D.; Li, W. Synthesis and biological evaluation of indole-based UC-112 analogs as potent and selective survivin inhibitors. Eur. J. Med. Chem. 2018, 149, 211–224. [Google Scholar] [CrossRef]
- Park, S.H.; Shin, I.; Park, S.H.; Kim, N.D.; Shin, I. An Inhibitor of the Interaction of Survivin with Smac in Mitochondria Promotes Apoptosis. Chem. Asian J. 2019, 14, 4035–4041. [Google Scholar] [CrossRef] [PubMed]
- Houghton, P.J.; Germain, G.S.; Harwood, F.C.; Schuetz, J.D.; Stewart, C.F.; Buchdunger, E.; Traxler, P. Imatinib mesylate is a potent inhibitor of the ABCG2 (BCRP) transporter and reverses resistance to topotecan and SN-38 in vitro. Cancer Res. 2004, 64, 2333–2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shishido, Y.; Ueno, S.; Yamazaki, R.; Nagaoka, M.; Matsuzaki, T. ABCG2 inhibitor YHO-13351 sensitizes cancer stem/initiating-like side population cells to irinotecan. Anti-cancer Res. 2013, 33, 1379–1386. [Google Scholar]
- de Vries, N.A.; Zhao, J.; Kroon, E.; Buckle, T.; Beijnen, J.H.; van Tellingen, O. P-glycoprotein and breast cancer resistance protein: Two dominant transporters working together in limiting the brain penetration of topotecan. Clin. Cancer Res. 2007, 13, 6440–6449. [Google Scholar] [CrossRef]
- Tagen, M.; Zhuang, Y.; Zhang, F.; Harstead, K.E.; Shen, J.; Schaiquevich, P.; Fraga, C.H.; Panetta, J.C.; Waters, C.M.; Stewart, C.F. P-glycoprotein, but not multidrug resistance protein 4, plays a role in the systemic clearance of irinotecan and SN-38 in mice. Drug Metab. Lett. 2010, 4, 195–201. [Google Scholar] [CrossRef]
- Li, F.; Fountzilas, C.; Puzanov, I.; Attwood, K.M.; Morrison, C.; Ling, X. Multiple functions of the DEAD-box RNA helicase, DDX5 (p68), make DDX5 a superior oncogenic biomarker and target for targeted cancer therapy. Am. J. Cancer Res. 2021, 11, 5190–5213. [Google Scholar] [PubMed]
- Hwu, J.R.; Tseng, W.N.; Gnabre, J.; Giza, P.; Huang, R.C. Antiviral activities of methylated nordihydroguaiaretic acids. 1. Synthesis, structure identification, and inhibition of tat-regulated HIV transactivation. J. Med. Chem. 1998, 41, 2994–3000. [Google Scholar] [CrossRef] [PubMed]
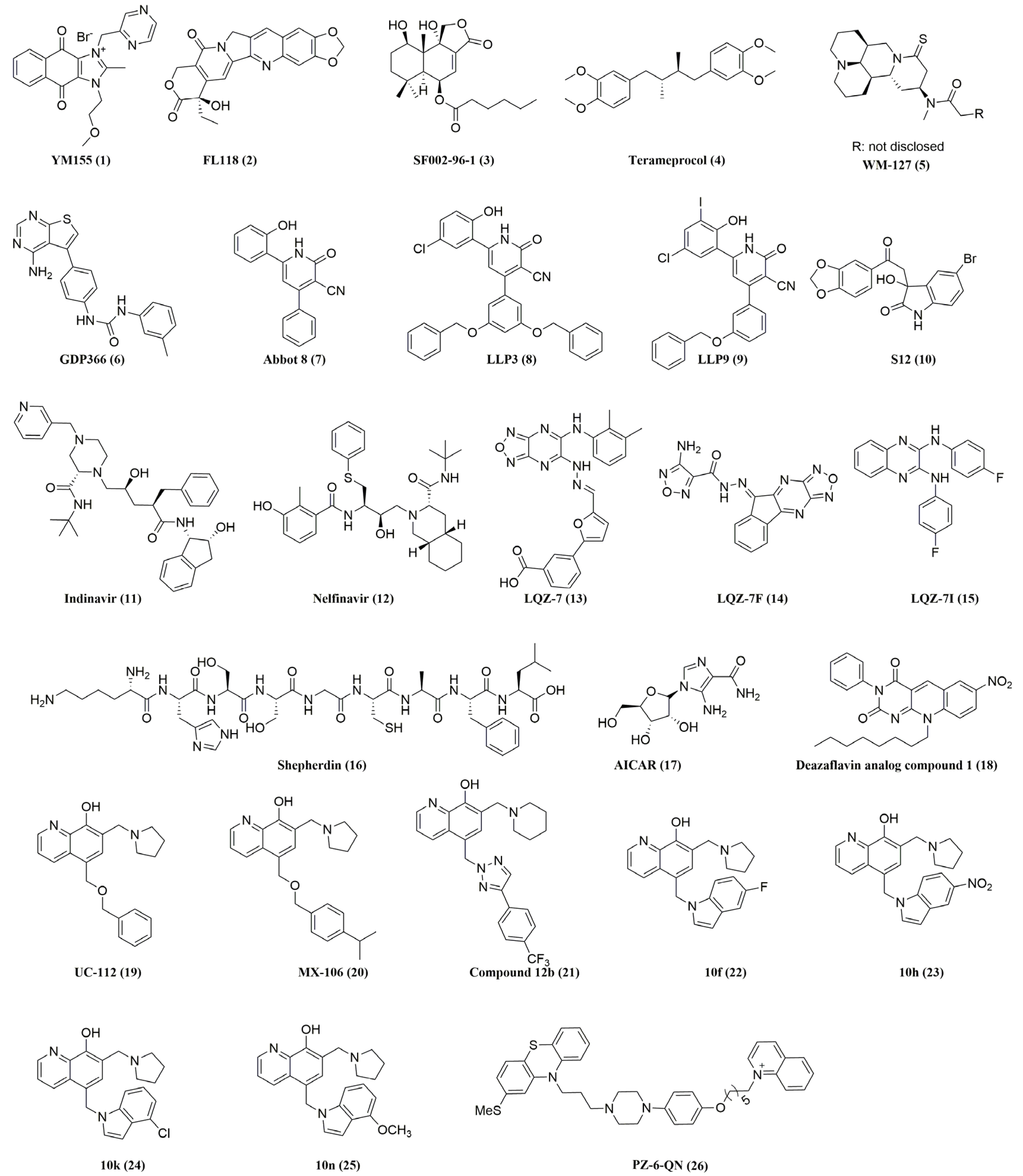
| Inhibitor | Mechanism of Action | Current Status | References |
|---|---|---|---|
| YM155 (1) | Inhibits survivin expression at both mRNA and protein levels; inhibits the survivin upstream transcription factors, Sp1 and ILF3, and their interactions with survivin promoter | Phase II; combination with docetaxel as first-line treatment for HER2-negative metastatic breast cancer; completed. Phase II; combination with rituximab for CD20-positive B cell non-Hodgkin’s lymphoma; completed Phase II; combination with docetaxel for stage III (unresectable) or stage IV melanoma; completed Phase II; alone for stage III (unresectable) or metastatic (stage IV) melanoma; completed. Phase II; combination with paclitaxel and carboplatin for advanced non-small cell lung carcinoma; completed Phase II; combination with docetaxel and prednisone for advanced hormone-refractory prostate cancer and other solid tumors; completed Phase II; alone for relapsed/refractory c-Myc rearranged high-grade B-cell lymphoma; recruiting | [104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127] |
| FL118 (2) | Inhibits survivin expression at both mRNA and protein levels | Preclinical | [128,129,130,131,132,133,134] |
| SF002-96-1 (3) | Inhibits survivin expression by inhibiting STAT3 and NF-κB | Preclinical | [135] |
| Terameprocol (4) | Inhibits survivin expression | Phase I; for intravenous administration in leukemia; completed Phase I; for intralesional injection in refractory malignant tumors of the head and neck; completed Phase I/II; for intravenous infusion administration in recurrent high-grade glioma; completed. Phase I; for oral administration in recurrent high-grade glioma; active Phase I/II; for intravaginal administration in cervical intraepithelial neoplasia induced by human papillomavirus; completed | [136,137,138,139,140,141,142] |
| WM-127 (5) | Inhibits survivin expression | Preclinical | [143] |
| GDP366 (6) | Inhibits survivin gene and protein expression | Preclinical | [144,145] |
| Abbot 8 (7), LLP3 (8), and LLP9 (9) | Disrupt survivin dimerization | Preclinical | [146,147,148,149] |
| S12 (10) | Disrupts survivin dimerization | Preclinical | [150,151] |
| Indinavir (11) and Nelfinavir (12) | Target survivin protein–protein interactions | Approved for HIV infection | [152,153] |
| LQZ-7 (13), LQZ-7F (14), and LQZ-7I (15) | Dissociate dimeric survivin and induce subsequent proteasome-dependent survivin degradation | Preclinical | [154,155] |
| Shepherdin (16) | Disrupts survivin interactions with Hsp90 and destabilizes survivin | Preclinical | [156,157] |
| AICAR (17) | Disrupts survivin interactions with Hsp90 and destabilizes survivin | Preclinical | [158] |
| Deazaflavin analog compound 1 (18) | Inhibits the interaction of survivin with Smac | Preclinical | [159] |
| UC-112 (19), MX-106 (20), Compound 12b (21), 10f (22), 10h (23), 10k (24) and 10n (25) | Increase the ubiquitin-mediated degradation of survivin | Preclinical | [160,161,162,163,164,165] |
| PZ-6-QN (26) | Disrupts the interaction of survivin with Smac in mitochondria | Preclinical | [166] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albadari, N.; Li, W. Survivin Small Molecules Inhibitors: Recent Advances and Challenges. Molecules 2023, 28, 1376. https://doi.org/10.3390/molecules28031376
Albadari N, Li W. Survivin Small Molecules Inhibitors: Recent Advances and Challenges. Molecules. 2023; 28(3):1376. https://doi.org/10.3390/molecules28031376
Chicago/Turabian StyleAlbadari, Najah, and Wei Li. 2023. "Survivin Small Molecules Inhibitors: Recent Advances and Challenges" Molecules 28, no. 3: 1376. https://doi.org/10.3390/molecules28031376
APA StyleAlbadari, N., & Li, W. (2023). Survivin Small Molecules Inhibitors: Recent Advances and Challenges. Molecules, 28(3), 1376. https://doi.org/10.3390/molecules28031376







