Emerging Roles of RNF168 in Tumor Progression
Abstract
:1. Introduction
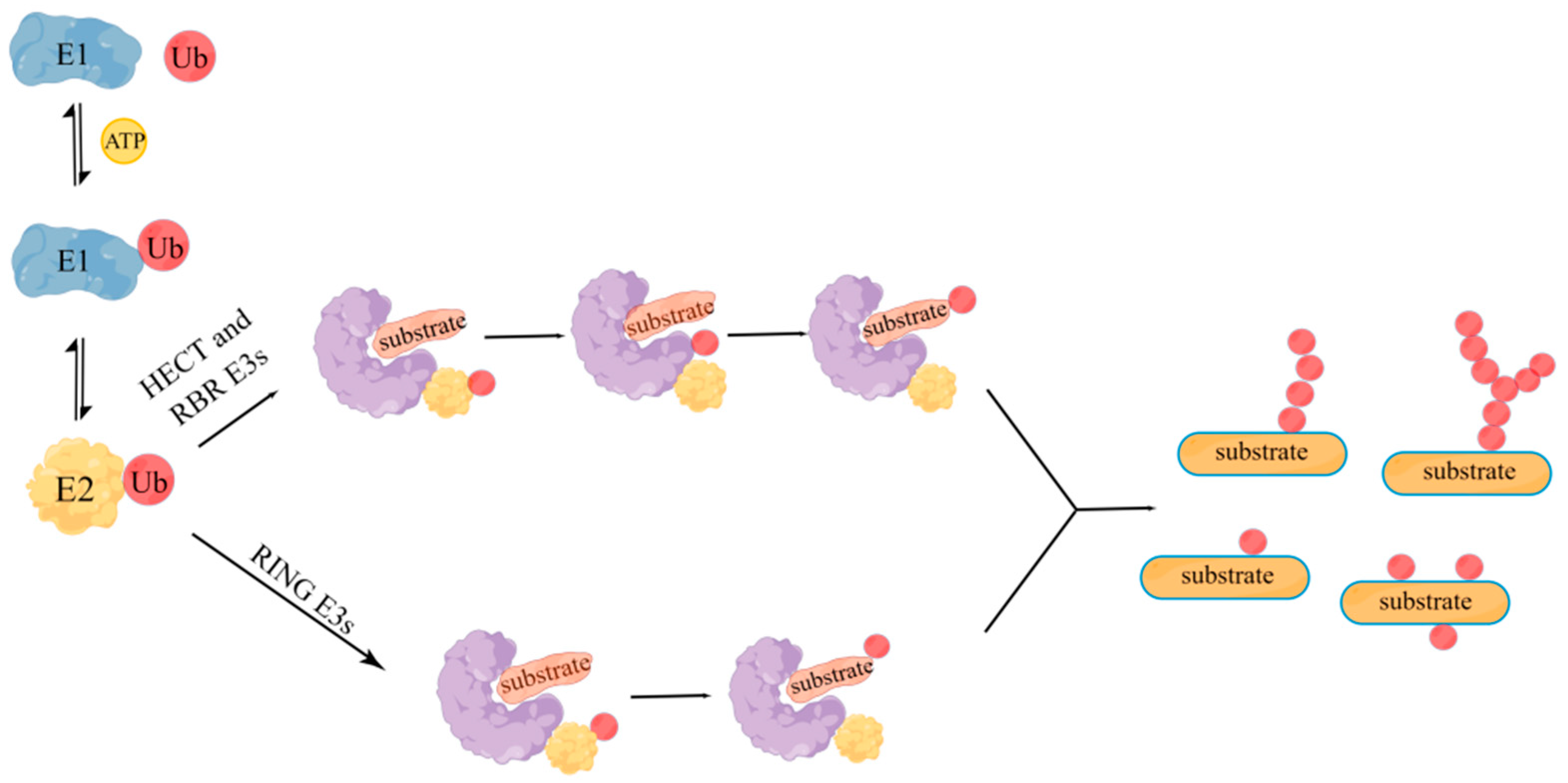
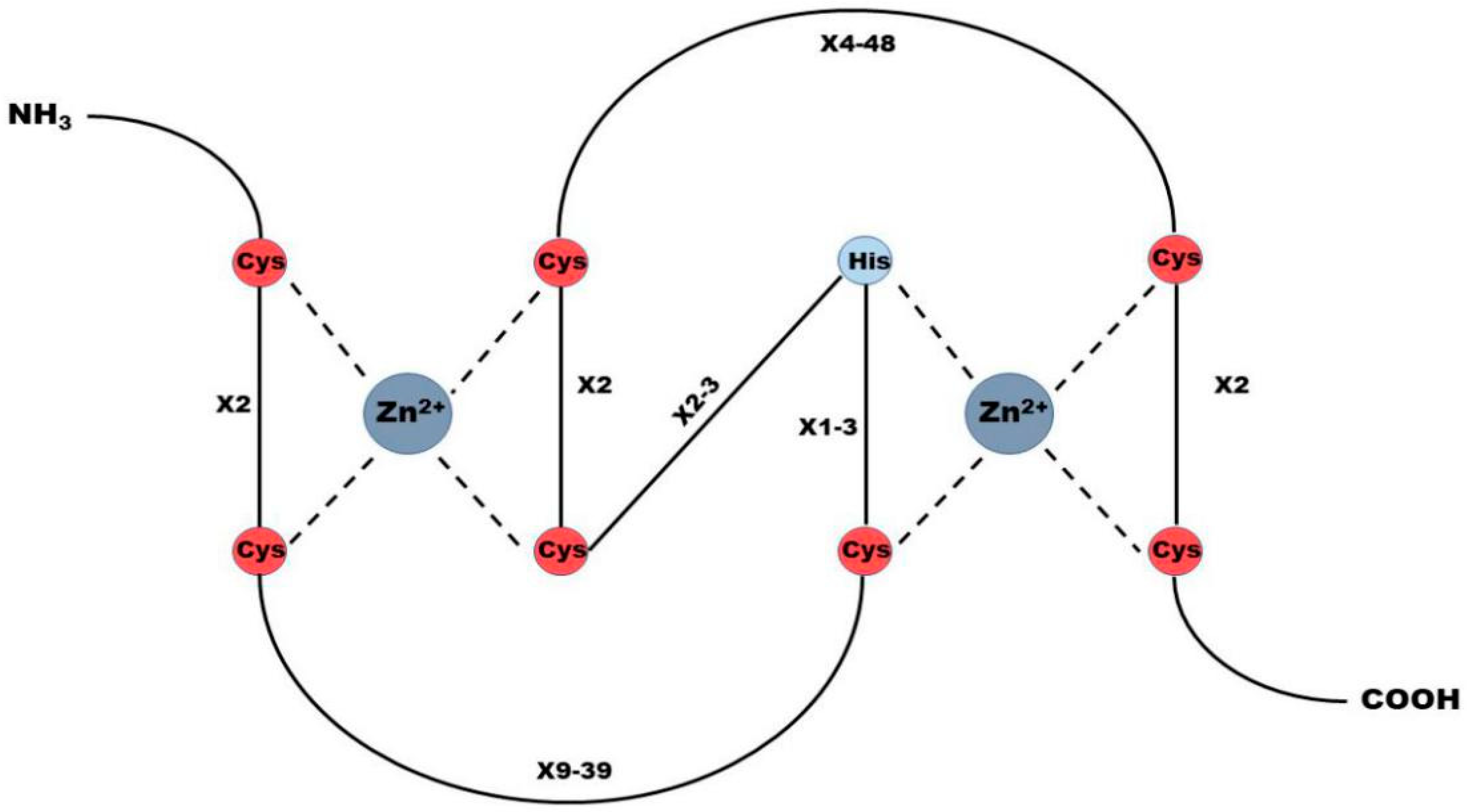
2. RING Finger Protein Family
3. Structure and Functions of RNF168
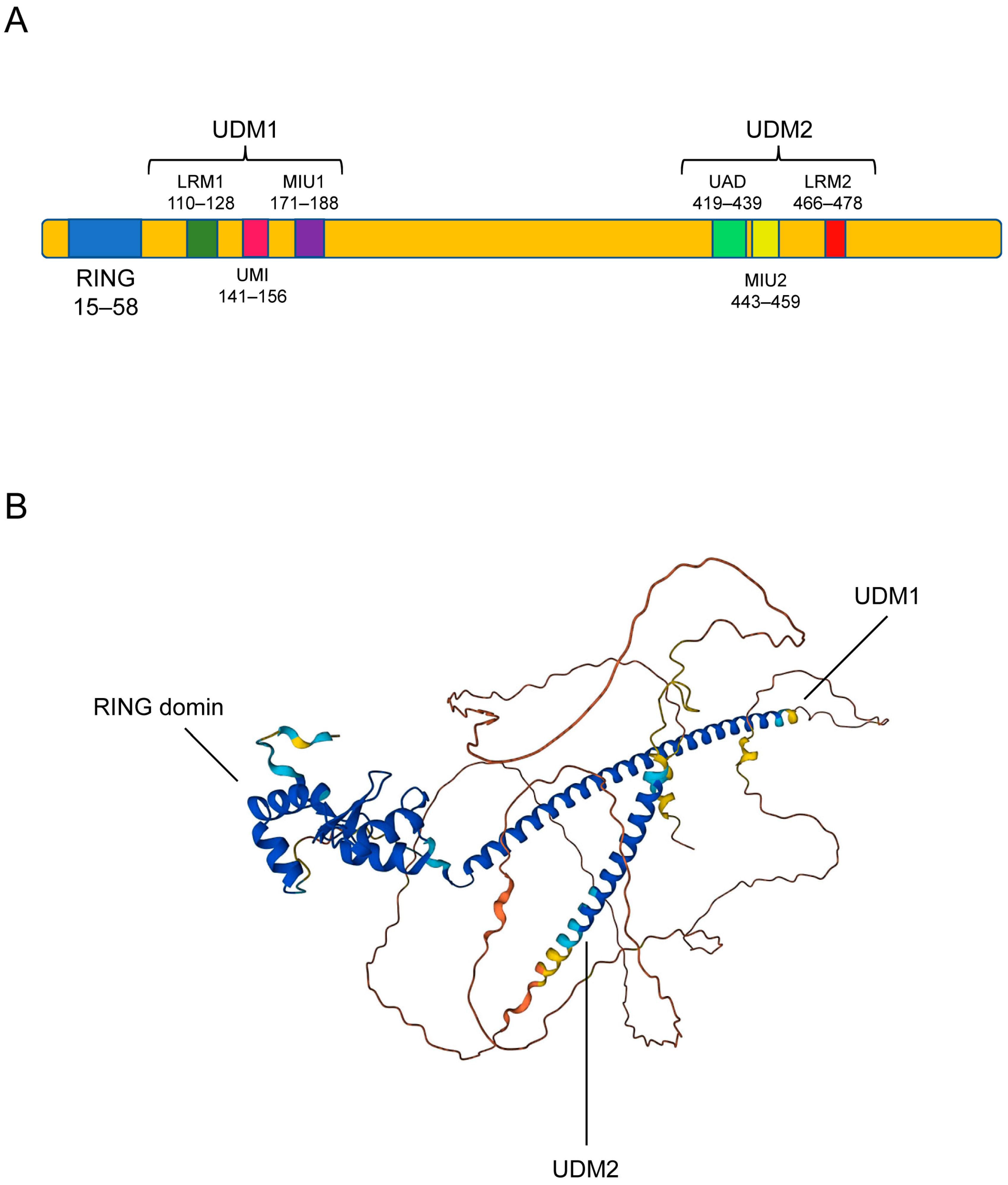
3.1. Structure of RNF168
3.2. Functions of RNF168
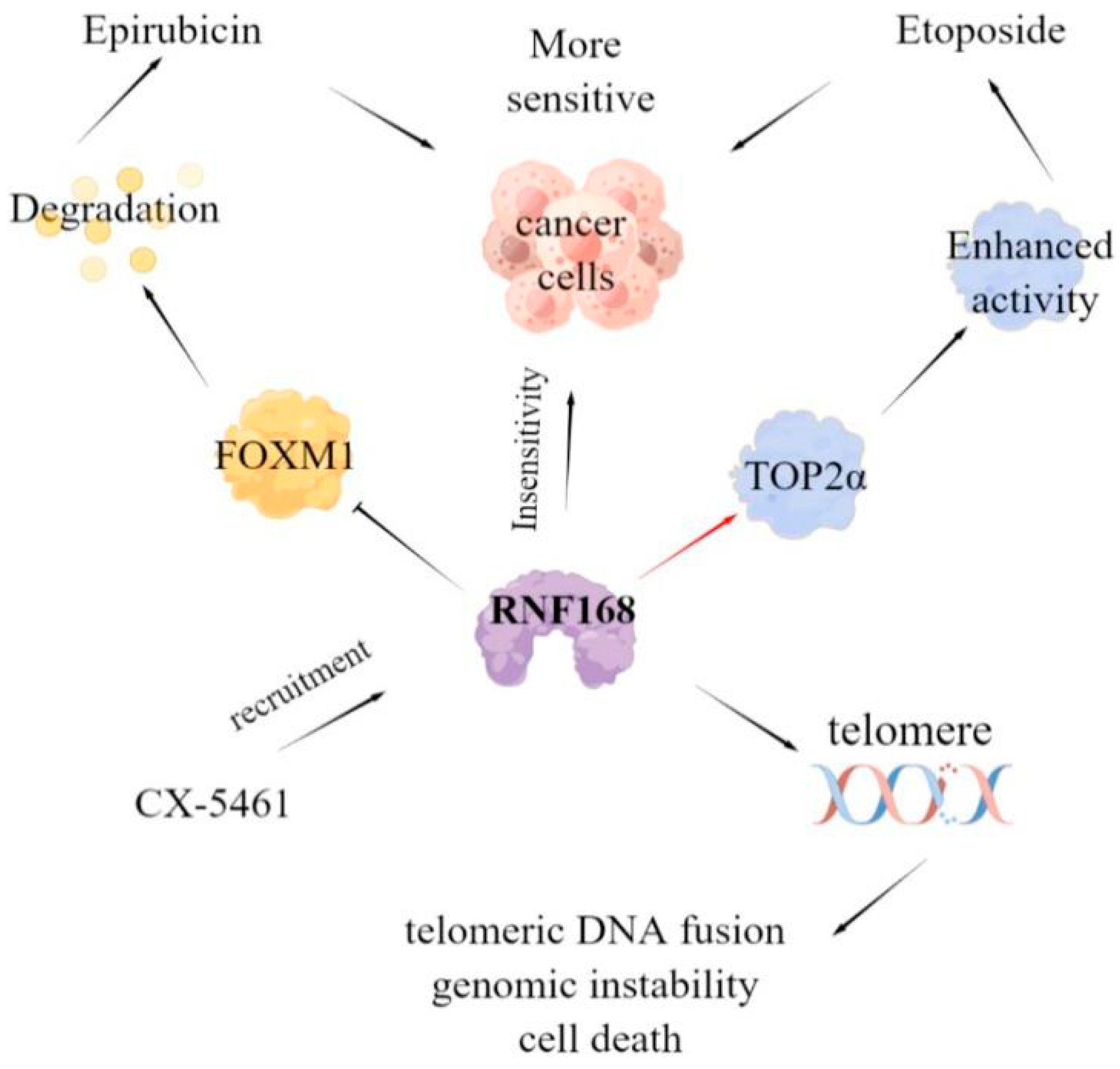
3.2.1. DNA Damage Response
3.2.2. Other Functions of RNF168
3.2.3. Regulation of RNF168 Function
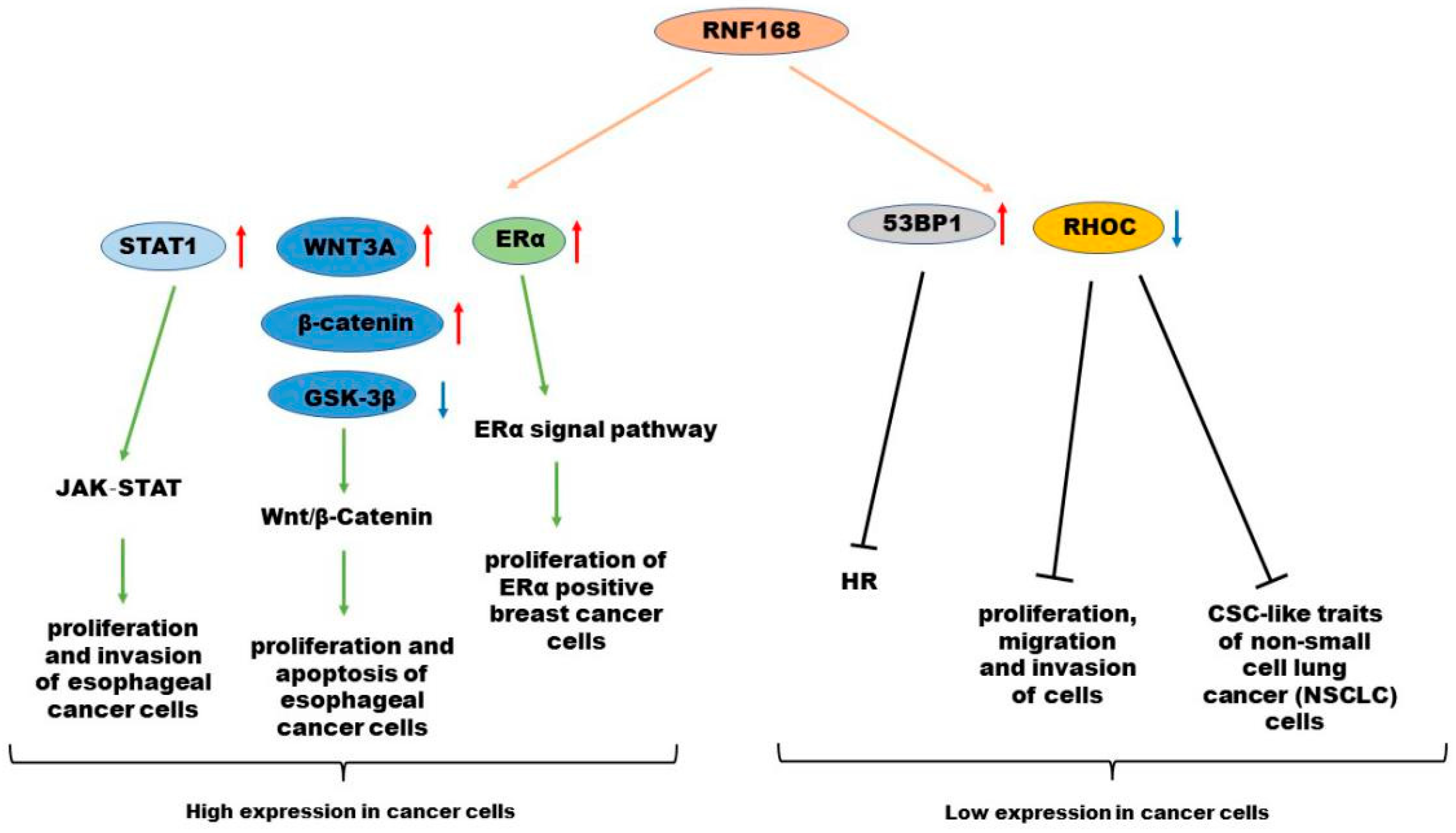
4. Roles of RNF168 in Various Cancer Types
4.1. High Expression of RNF168 in Cancers
4.1.1. Breast Cancer
4.1.2. Esophageal Cancer
4.2. Low Expression of RNF168 in Cancers
4.2.1. BRCA1-Mutant Cancer
4.2.2. Gastric Cancer
4.2.3. Lung Cancer
4.2.4. Glioblastoma
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RNF168 | RING finger protein 168 |
| PTM | Post-translational modification |
| PROTAC | Proteolysis-targeting Chimeras |
| HECT | Homologous to E6AP C-terminals |
| RBR | RING between RING |
| UDM | Ubiquitin-dependent double-strand break recruitment module |
| LRM | LR motif |
| UMI | UIM- and MIU-related UBD |
| MIU | Motif interacting with ubiquitin |
| UAD | Ubiquitin-associated domain |
| IRIF | Ionizing radiation-induced foci |
| DSB | DNA double-strand break |
| SSB | DNA single-strand break |
| ROS | Reactive oxygen species |
| DDR | DNA damage response |
| PIKK | Phosphatidylinositol-3 kinase-related kinases |
| HR | Homologous recombination (HR) |
| NHEJ | Non-homologous end connection |
| ATM | Ataxia-telangiectasia mutation |
| MDC1 | Mediator of DNA damage checkpoint 1 |
| L3MBTL2 | Lethal (3) malignant brain tumor like protein 2 |
| FOXM1 | Forkhead box M1 |
| TOP2α | Topoisomerase IIα |
| IR | Ionizing radiation |
| EBV | Epstein Barr virus |
| ERα | Estrogen receptor α |
| ESCC | Esophageal squamous cell carcinoma |
| GSK-3β | Glycogen synthase kinase 3β |
| RHOC | Ras homolog gene family member C |
| CSC | Cancer stem cell |
| NSCLC | Non-small cell lung cancer |
| MTAP | Methylthioadenosine phosphorylase |
| HD | Huntington’s disease |
| PD | Parkinson’s disease |
References
- Yang, L.; Chen, J.; Huang, X.; Zhang, E.; He, J.; Cai, Z. Novel Insights into E3 Ubiquitin Ligase in Cancer Chemoresistance. Am. J. Med. Sci. 2018, 355, 368–376. [Google Scholar] [CrossRef]
- Li, X.; Song, Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 2020, 13, 50. [Google Scholar] [CrossRef]
- He, S.; Gao, F.; Ma, J.; Ma, H.; Dong, G.; Sheng, C. Aptamer-PROTAC Conjugates (APCs) for Tumor-Specific Targeting in Breast Cancer. Angew. Chem. 2021, 133, 23487–23493. [Google Scholar] [CrossRef]
- Kelliher, J.L.; West, K.L.; Gong, Q.; Leung, J.W. Histone H2A variants alpha1-extension helix directs RNF168-mediated ubiquitination. Nat. Commun. 2020, 11, 2462. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.; Jin, D.; He, S.; Han, S.; Bai, Q. RNF168 is highly expressed in esophageal squamous cell carcinoma and contributes to the malignant behaviors in association with the Wnt/beta-catenin signaling pathway. Aging 2021, 13, 5403–5414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, J.; Wu, M.; Wu, H.; Arokiaraj, A.W.; Wang, C.; Zhang, W.; Tao, Y.; Huen, M.S.; Zang, J. Structural basis for role of ring finger protein RNF168 RING domain. Cell Cycle 2013, 12, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.A. Ubiquitination: Friend and foe in cancer. Int. J. Biochem. Cell Biol. 2018, 101, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Taguchi, K.; Tanaka, M. Ubiquitin, Autophagy and Neurodegenerative Diseases. Cells 2020, 9, 2022. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Y.; Zhou, L.; Yang, G.; Wang, Y.; Han, J.; Li, L.; Zhang, S. Role of RING-Type E3 Ubiquitin Ligases in Inflammatory Signalling and Inflammatory Bowel Disease. Mediat. Inflamm. 2020, 2020, 5310180. [Google Scholar] [CrossRef]
- Toma-Fukai, S.; Shimizu, T. Structural Diversity of Ubiquitin E3 Ligase. Molecules 2021, 26, 6682. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, L.F.; Munir, M.; Qiu, H.J. RING-Domain E3 Ligase-Mediated Host-Virus Interactions: Orchestrating Immune Responses by the Host and Antagonizing Immune Defense by Viruses. Front. Immunol. 2018, 9, 1083. [Google Scholar] [CrossRef] [PubMed]
- Gundogdu, M.; Walden, H. Structural basis of generic versus specific E2-RING E3 interactions in protein ubiquitination. Protein Sci. 2019, 28, 1758–1770. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Sun, Y.; Ahmed, R.I.; Ren, A.; Xie, M. Research Progress on Plant RING-Finger Proteins. Genes 2019, 10, 973. [Google Scholar] [CrossRef]
- Marsh, D.J.; Dickson, K.A. Writing Histone Monoubiquitination in Human Malignancy-The Role of RING Finger E3 Ubiquitin Ligases. Genes 2019, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Thorslund, T.; Ripplinger, A.; Hoffmann, S.; Wild, T.; Uckelmann, M.; Villumsen, B.; Narita, T.; Sixma, T.K.; Choudhary, C.; Bekker-Jensen, S.; et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 2015, 527, 389–393. [Google Scholar] [CrossRef]
- Kelliher, J.; Ghosal, G.; Leung, J. New answers to the old RIDDLE: RNF168 and the DNA damage response pathway. FEBS J. 2021, 289, 2467–2480. [Google Scholar] [CrossRef] [PubMed]
- Nakada, S. Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice. J. Radiat. Res. 2016, 57 (Suppl. 1), i33–i40. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Carusillo, A.; Mussolino, C. DNA Damage: From Threat to Treatment. Cells 2020, 9, 1665. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Krokidis, M.G.; Masi, A.; Terzidis, M.A. On the relevance of hydroxyl radical to purine DNA damage. Free. Radic. Res. 2021, 55, 384–404. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Aldred, M.A. DNA Damage and Repair in Pulmonary Arterial Hypertension. Genes 2020, 11, 1224. [Google Scholar] [CrossRef]
- Su, M.; Wang, H.; Wang, W.; Wang, Y.; Ouyang, L.; Pan, C.; Xia, L.; Cao, D.; Liao, Q. LncRNAs in DNA damage response and repair in cancer cells. Acta Biochim. Biophys. Sin. 2018, 50, 433–439. [Google Scholar] [CrossRef]
- Ma, S.; Rong, Z.; Liu, C.; Qin, X.; Zhang, X.; Chen, Q. DNA damage promotes microtubule dynamics through a DNA-PK-AKT axis for enhanced repair. J. Cell Biol. 2021, 220, e201911025. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.R.; Clifford, G.; Bonnet, C.; Groth, A.; Wilson, M.D.; Chapman, J.R. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature 2021, 596, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Burdak-Rothkamm, S.; Mansour, W.Y.; Rothkamm, K. DNA Damage Repair Deficiency in Prostate Cancer. Trends Cancer 2020, 6, 974–984. [Google Scholar] [CrossRef]
- Krais, J.J.; Johnson, N. Ectopic RNF168 expression promotes break-induced replication-like DNA synthesis at stalled replication forks. Nucleic Acids Res. 2020, 48, 4298–4308. [Google Scholar] [CrossRef]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair. (Amst) 2017, 56, 92–101. [Google Scholar] [CrossRef]
- Sharma, A.; Alswillah, T.; Singh, K.; Chatterjee, P.; Willard, B.; Venere, M.; Summers, M.K.; Almasan, A. USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy 2018, 14, 1976–1990. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Shi, R.; Zhu, X.; Zhou, J.; Peng, B.; Xu, X. RNF126 Quenches RNF168 Function in the DNA Damage Response. Genom. Proteom. Bioinform. 2018, 16, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hao, S.; Qiu, Z.; Wang, Y.; Zhao, Y.; Li, Y.; Gao, W.; Wu, Y.; Liu, C.; Xu, X.; et al. Cadmium disrupts the DNA damage response by destabilizing RNF168. Food Chem. Toxicol. 2019, 133, 110745. [Google Scholar] [CrossRef]
- Gatti, M.; Pinato, S.; Maspero, E.; Soffientini, P.; Polo, S.; Penengo, L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle 2012, 11, 2538–2544. [Google Scholar] [CrossRef]
- Nowsheen, S.; Aziz, K.; Aziz, A.; Deng, M.; Qin, B.; Luo, K.; Jeganathan, K.B.; Zhang, H.; Liu, T.; Yu, J.; et al. L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat. Cell Biol. 2018, 20, 455–464. [Google Scholar] [CrossRef]
- Mattiroli, F.; Vissers, J.H.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Horn, V.; Uckelmann, M.; Zhang, H.; Eerland, J.; Aarsman, I.; le Paige, U.B.; Davidovich, C.; Sixma, T.K.; van Ingen, H. Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat. Commun. 2019, 10, 1751. [Google Scholar] [CrossRef] [PubMed]
- Fradet-Turcotte, A.; Canny, M.D.; Escribano-Díaz, C.; Orthwein, A.; Leung, C.C.; Huang, H.; Landry, M.C.; Kitevski-LeBlanc, J.; Noordermeer, S.M.; Sicheri, F.; et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 2013, 499, 50–54. [Google Scholar] [CrossRef]
- Bohgaki, M. RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2013, 110, 20982–20987. [Google Scholar] [CrossRef]
- Lu, X.; Xu, M.; Zhu, Q.; Zhang, J.; Liu, G.; Bao, Y.; Gu, L.; Tian, Y.; Wen, H.; Zhu, W.G. RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage. FASEB J. 2021, 35, e21326. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Pinato, S.; Maiolica, A.; Rocchio, F.; Prato, M.G.; Aebersold, R.; Penengo, L. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015, 10, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Kongsema, M.; Zona, S.; Karunarathna, U.; Cabrera, E.; Man, E.P.S.; Yao, S.; Shibakawa, A.; Khoo, U.S.; Medema, R.H.; Freire, R.; et al. RNF168 cooperates with RNF8 to mediate FOXM1 ubiquitination and degradation in breast cancer epirubicin treatment. Oncogenesis 2016, 5, e252. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Sun, Y.; Huang, S.Y.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Guturi, K.; Bohgaki, M.; Bohgaki, T.; Srikumar, T.; Ng, D.; Kumareswaran, R.; El Ghamrasni, S.; Jeon, J.; Patel, P.; Eldin, M.S.; et al. RNF168 and USP10 regulate topoisomerase IIalpha function via opposing effects on its ubiquitylation. Nat. Commun. 2016, 7, 12638. [Google Scholar] [CrossRef] [PubMed]
- Masud, T.; Soong, C.; Xu, H.; Biele, J.; Bjornson, S.; McKinney, S.; Aparicio, S. Ubiquitin-mediated DNA damage response is synthetic lethal with G-quadruplex stabilizer CX-5461. Sci. Rep. 2021, 11, 9812. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.C.; Peng, B.; Ren, T.T.; Liu, S.P.; Du, J.R.; Chen, Z.H.; Zhang, T.T.; Gu, X.; Li, M.; Cao, S.L.; et al. A 1,2,3-Triazole Derivative of Quinazoline Exhibits Antitumor Activity by Tethering RNF168 to SQSTM1/P62. J. Med. Chem. 2022, 65, 15028–15047. [Google Scholar] [CrossRef]
- Tang, Y.; Mukherjee, J.; Pieper, R.O. MRE11 and UBR5 Co-Operate to Suppress RNF168-Mediated Fusion of Dysfunctional Telomeres. Front. Oncol. 2021, 11, 772233. [Google Scholar] [CrossRef]
- Yang, C.; Zang, W.; Tang, Z.; Ji, Y.; Xu, R.; Yang, Y.; Luo, A.; Hu, B.; Zhang, Z.; Liu, Z.; et al. A20/TNFAIP3 Regulates the DNA Damage Response and Mediates Tumor Cell Resistance to DNA-Damaging Therapy. Cancer Res. 2018, 78, 1069–1082. [Google Scholar] [CrossRef]
- de Poot, S.; Tian, G.; Finley, D. Meddling with Fate: The Proteasomal Deubiquitinating Enzymes. J. Mol. Biol. 2017, 429, 3525–3545. [Google Scholar] [CrossRef]
- Yu, M.; Liu, K.; Mao, Z.; Luo, J.; Gu, W.; Zhao, W. USP11 Is a Negative Regulator to gammaH2AX Ubiquitylation by RNF8/RNF168. J. Biol. Chem. 2016, 291, 959–967. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Hu, H.; Tong, X.; Li, L.; Liu, X.; Chen, M.; Yuan, H.; Xie, X.; Li, Q.; Zhang, Y.; et al. The mTOR-S6K pathway links growth signalling to DNA damage response by targeting RNF168. Nat. Cell Biol. 2018, 20, 320–331. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Zhang, W.; Alvarez, A.A.; Bai, Y.; Hu, B.; Cheng, S.Y.; Yang, K.; Li, Y.; Feng, H. Lipolytic inhibitor G0S2 modulates glioma stem-like cell radiation response. J. Exp. Clin. Cancer Res. 2019, 38, 147. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Hansen, L.J.; Singh, S.X.; Wang, F.; Sun, R.; Moure, C.J.; Roso, K.; Greer, P.K.; Yan, H.; He, Y. A PRMT5-RNF168-SMURF2 Axis Controls H2AX Proteostasis. Cell Rep. 2019, 28, 3199–3211.e5. [Google Scholar] [CrossRef]
- Salamun, S.G.; Sitz, J.; De La Cruz-Herrera, C.F.; Yockteng-Melgar, J.; Marcon, E.; Greenblatt, J.; Fradet-Turcotte, A.; Frappier, L. The Epstein-Barr Virus BMRF1 Protein Activates Transcription and Inhibits the DNA Damage Response by Binding NuRD. J. Virol. 2019, 93, e01070-19. [Google Scholar] [CrossRef] [PubMed]
- Sitz, J.; Blanchet, S.A.; Gameiro, S.F.; Biquand, E.; Morgan, T.M.; Galloy, M.; Dessapt, J.; Lavoie, E.G.; Blondeau, A.; Smith, B.C.; et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc. Natl. Acad. Sci. USA 2019, 116, 19552–19562. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, J.; Xu, J.; Yang, H.; Li, X.; Hou, Y.; Zhao, Y.; Xue, M.; Wang, B.; Yu, N.; et al. RNF168 facilitates oestrogen receptor a transcription and drives breast cancer proliferation. J. Cell. Mol. Med. 2018, 22, 4161–4170. [Google Scholar] [CrossRef]
- Rong, G.; Pan, Z.; Ding, M.; Wang, L. RNF168 suppresses the cancer stem cell-like traits of nonsmall cell lung cancer cells by mediating RhoC ubiquitination. Environ. Toxicol. 2022, 37, 603–611. [Google Scholar] [CrossRef]
- Xiao, M.; He, J.; Yin, L.; Chen, X.; Zu, X.; Shen, Y. Tumor-Associated Macrophages: Critical Players in Drug Resistance of Breast Cancer. Front. Immunol. 2021, 12, 799428. [Google Scholar] [CrossRef]
- Shams, A.; Binothman, N.; Boudreault, J.; Wang, N.; Shams, F.; Hamam, D.; Tian, J.; Moamer, A.; Dai, M.; Lebrun, J.J.; et al. Prolactin receptor-driven combined luminal and epithelial differentiation in breast cancer restricts plasticity, stemness, tumorigenesis and metastasis. Oncogenesis 2021, 10, 10. [Google Scholar] [CrossRef]
- Costa, R.; Han, H.S.; Gradishar, W.J. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: A review. Breast Cancer Res. Treat. 2018, 169, 397–406. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, W. Selective Estrogen Receptor Degraders (SERDs): A Promising Strategy for Estrogen Receptor Positive Endocrine-Resistant Breast Cancer. J. Med. Chem. 2020, 63, 15094–15114. [Google Scholar] [CrossRef]
- Niu, C. Risk factors for esophageal squamous cell carcinoma and its histological precursor lesions in China: A multicenter cross-sectional study. BMC Cancer 2021, 21, 1034. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Liu, F.; Xu, G. Predicting STAT1 as a prognostic marker in patients with solid cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920917558. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Xue, M.; Wang, W.; Xia, D.; Li, Y.; Zhou, X.; Pang, D.; Lu, K.; Hou, J.; Zhang, A.; et al. RNF168 facilitates proliferation and invasion of esophageal carcinoma, possibly via stabilizing STAT1. J. Cell. Mol. Med. 2019, 23, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Krais, J.J.; Wang, Y.; Bernhardy, A.J.; Clausen, E.; Miller, J.A.; Cai, K.Q.; Scott, C.L.; Johnson, N. RNF168-Mediated Ubiquitin Signaling Inhibits the Viability of BRCA1-Null Cancers. Cancer Res. 2020, 80, 2848–2860. [Google Scholar] [CrossRef] [PubMed]
- Jhanwar-Uniyal, M. BRCA1 in cancer, cell cycle and genomic stability. Front. Biosci. 2003, 8, s1107–s1117. [Google Scholar] [CrossRef]
- Zong, D.; Adam, S.; Wang, Y.; Sasanuma, H.; Callén, E.; Murga, M.; Day, A.; Kruhlak, M.J.; Wong, N.; Munro, M.; et al. BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation. Mol. Cell 2019, 73, 1267–1281.e7. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.S.; Abraham, K.J.; Guturi, K.K.N.; Halaby, M.J.; Khan, Z.; Palomero, L.; Ho, B.; Duan, S.; St-Germain, J.; Algouneh, A.; et al. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J. Clin. Investig. 2021, 131, e140105. [Google Scholar] [CrossRef]
- Abraham, H.G.; Ulintz, P.J.; Goo, L.; Yates, J.A.; Little, A.C.; Bao, L.; Wu, Z.; Merajver, S.D. RhoC Modulates Cell Junctions and Type I Interferon Response in Aggressive Breast Cancers. Front. Oncol. 2021, 11, 712041. [Google Scholar] [CrossRef]
- Thomas, P.; Pranatharthi, A.; Ross, C.; Srivastava, S. RhoC: A fascinating journey from a cytoskeletal organizer to a Cancer stem cell therapeutic target. J. Exp. Clin. Cancer Res. 2019, 38, 328. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, Y.C.; Sang, J.R.; Xu, W.R. RhoC protein stimulates migration of gastric cancer cells through interaction with scaffold protein IQGAP1. Mol. Med. Rep. 2011, 4, 697–703. [Google Scholar]
- Wu, Y.; Tao, Y.; Chen, Y.; Xu, W. RhoC regulates the proliferation of gastric cancer cells through interaction with IQGAP1. PLoS ONE 2012, 7, e48917. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, C.; Jiang, H.; Zhang, Z.; Xie, L.; He, X. MiR-493 suppresses the proliferation and invasion of gastric cancer cells by targeting RhoC. Iran. J. Basic Med. Sci. 2015, 18, 1027–1033. [Google Scholar]
- Xu, Y.; Feng, Y.; Sun, Z.; Li, Q. RNF168 promotes RHOC degradation by ubiquitination to restrain gastric cancer progression via decreasing HDAC1 expression. Biochem. Biophys. Res. Commun. 2021, 557, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hunt, C.P.; Sanders, L.H. DNA damage and repair in Parkinson’s disease: Recent advances and new opportunities. J. Neurosci. Res. 2021, 99, 180–189. [Google Scholar] [CrossRef]
- Maiuri, T.; Suart, C.E.; Hung, C.L.K.; Graham, K.J.; Bazan, C.A.B.; Truant, R.D.N.A. DNA Damage Repair in Huntington’s Disease and Other Neurodegenerative Diseases. Neurotherapeutics 2019, 16, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Filippini, S.E.; Vega, A. Breast cancer genes: Beyond BRCA1 and BRCA2. Front. Biosci. 2013, 18, 1358–1372. [Google Scholar]
- He, C.; Kawaguchi, K.; Toi, M. DNA damage repair functions and targeted treatment in breast cancer. Breast Cancer 2020, 27, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Farnood, P.R.; Pazhooh, R.D.; Asemi, Z.; Yousefi, B. DNA damage response and repair in pancreatic cancer development and therapy. DNA Repair. 2021, 103, 103116. [Google Scholar] [CrossRef] [PubMed]
- Reilly, N.M.; Novara, L.; Di Nicolantonio, F.; Bardelli, A. Exploiting DNA repair defects in colorectal cancer. Mol. Oncol. 2019, 13, 681–700. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Chroma, K.; Mistrik, M.; Moudry, P.; Gursky, J.; Liptay, M.; Strauss, R.; Skrott, Z.; Vrtel, R.; Bartkova, J.; Kramara, J.; et al. Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene 2017, 36, 2405–2422. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, T.; Qin, H.; Yuan, Z.; Zhang, Y.; Li, X.; Zheng, L. Emerging Roles of RNF168 in Tumor Progression. Molecules 2023, 28, 1417. https://doi.org/10.3390/molecules28031417
Xie T, Qin H, Yuan Z, Zhang Y, Li X, Zheng L. Emerging Roles of RNF168 in Tumor Progression. Molecules. 2023; 28(3):1417. https://doi.org/10.3390/molecules28031417
Chicago/Turabian StyleXie, Tianyuan, Hai Qin, Zhengdong Yuan, Yiwen Zhang, Xiaoman Li, and Lufeng Zheng. 2023. "Emerging Roles of RNF168 in Tumor Progression" Molecules 28, no. 3: 1417. https://doi.org/10.3390/molecules28031417
APA StyleXie, T., Qin, H., Yuan, Z., Zhang, Y., Li, X., & Zheng, L. (2023). Emerging Roles of RNF168 in Tumor Progression. Molecules, 28(3), 1417. https://doi.org/10.3390/molecules28031417






