Nucleotide Imbalance, Provoked by Downregulation of Aspartate Transcarbamoylase Impairs Cold Acclimation in Arabidopsis
Abstract
:1. Introduction
2. Results
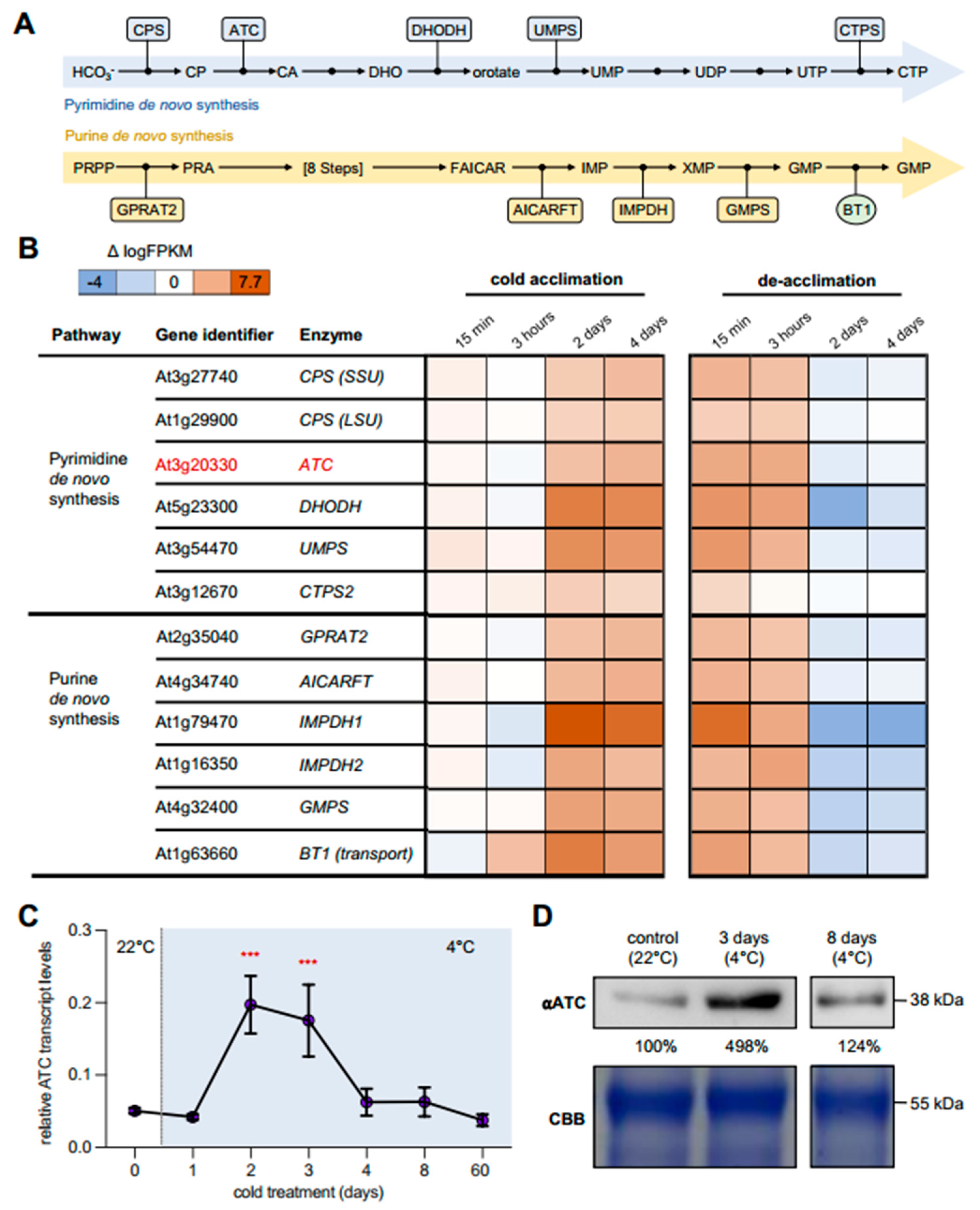
2.1. Cold Leads to Increased Expression of Genes Involved in Nucleotide De Novo Synthesis
2.2. ATC Abundancy Is Strongly Increased Up on Cold Acclimation and Adaptation
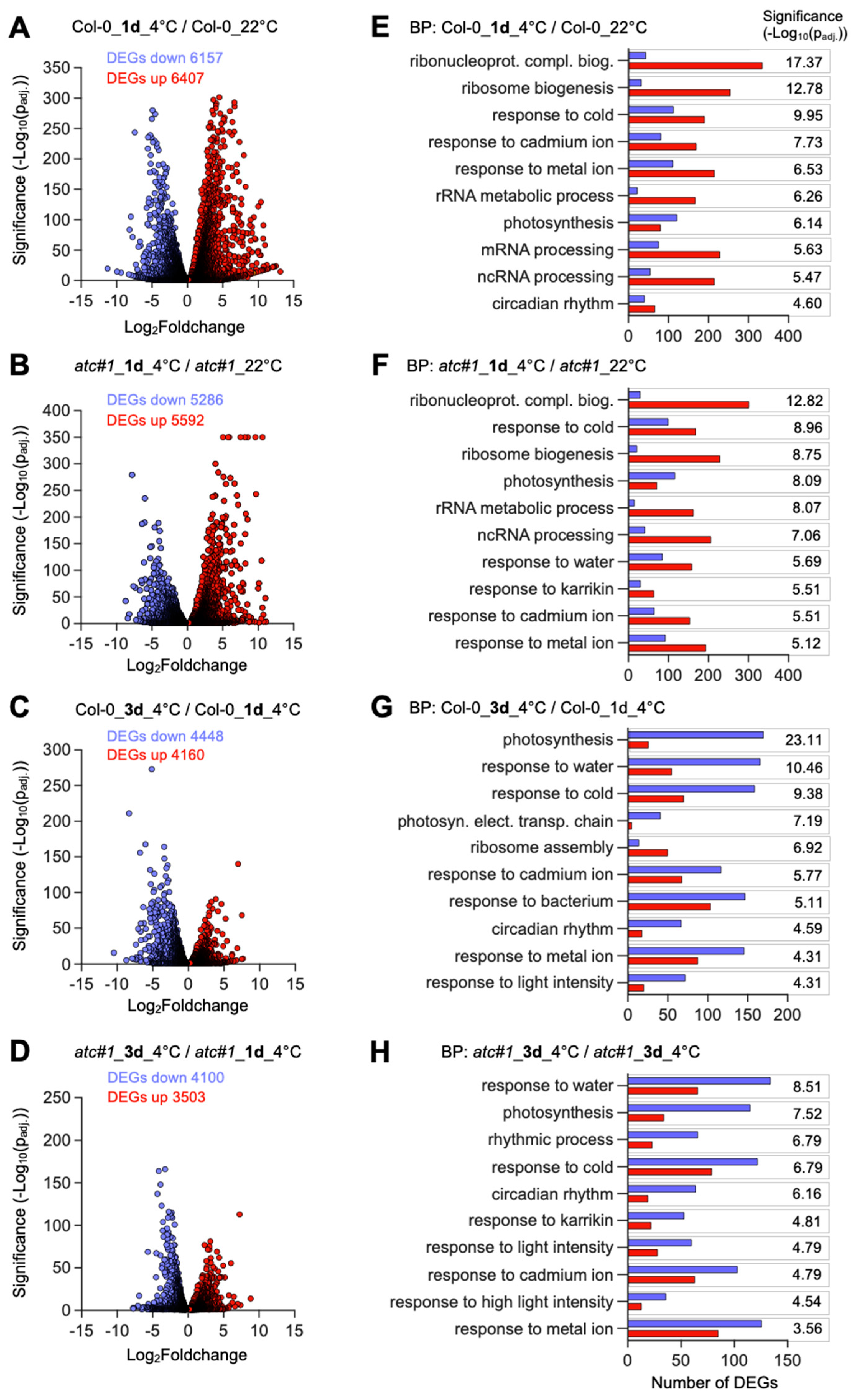
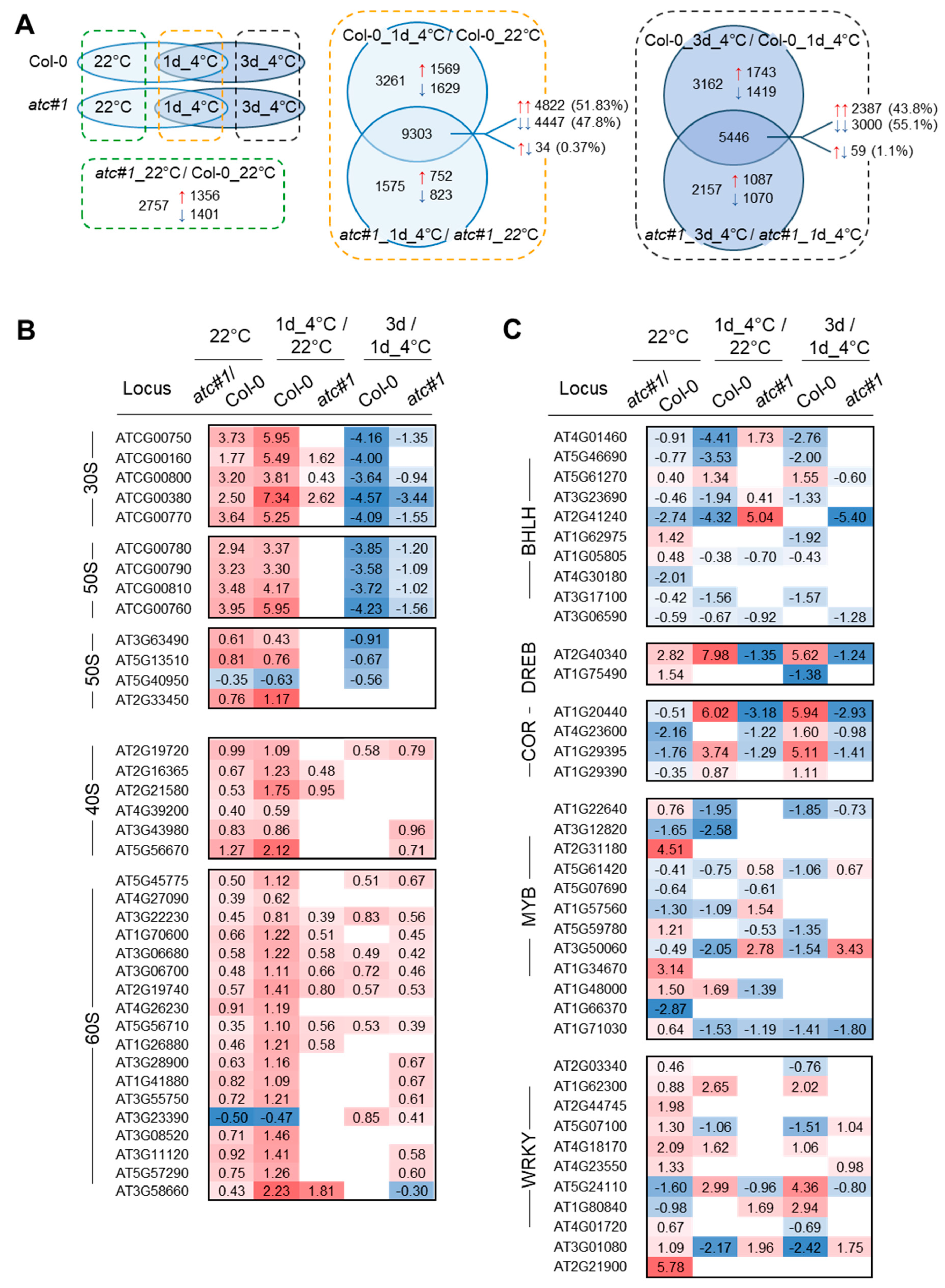
2.3. Cold Acclimation Imposes Global Adaptations of the Transcriptome
2.4. Transcriptional Response in Functional Groups “Ribosomal Protein” and “Transcription Factor”
2.5. Transcriptional Response in Functional Groups “Nucleotide Metabolism”, Intracellular Transport” and “Central Carbohydrate Metabolism and Respiration”
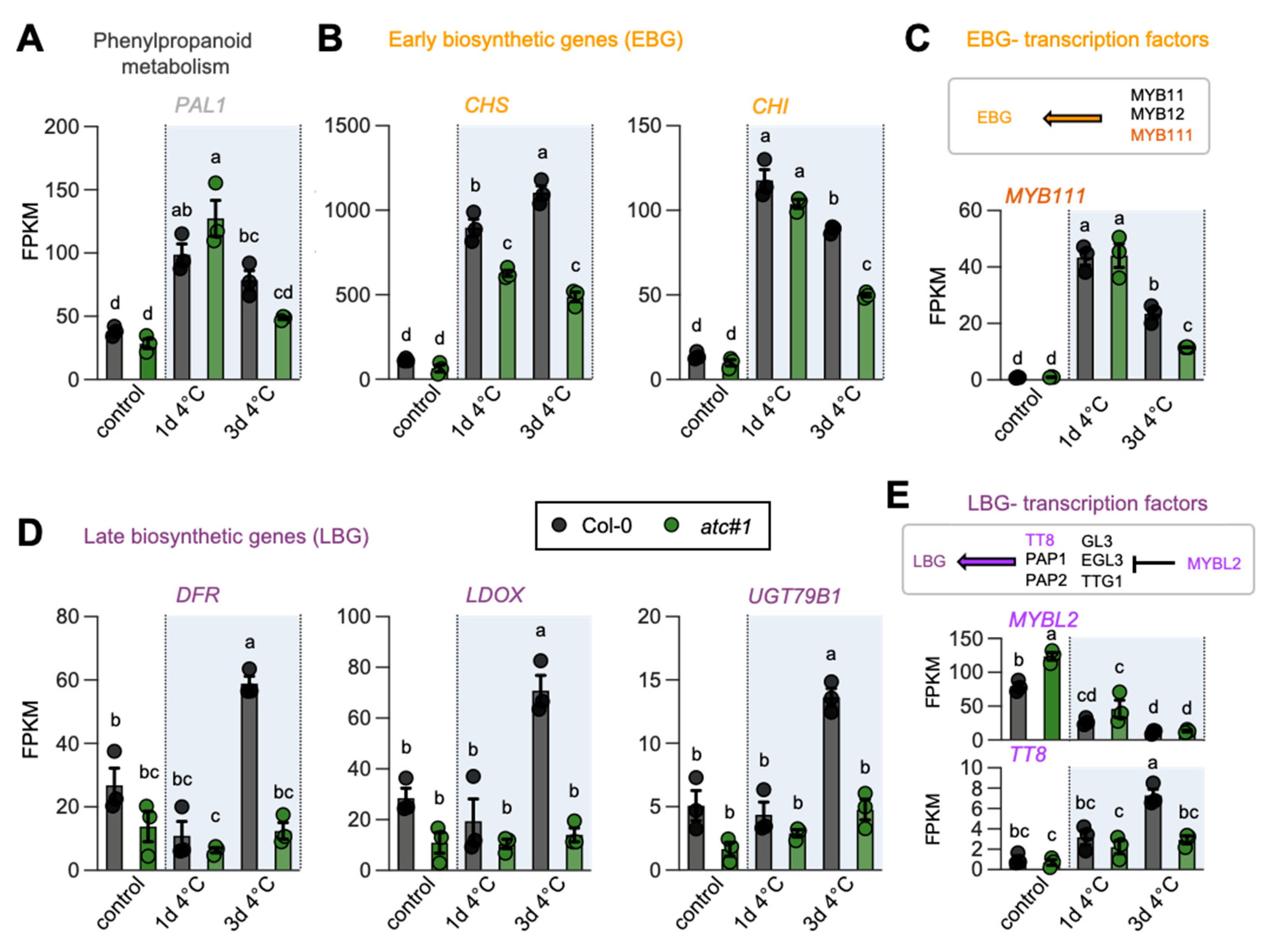
2.6. ATC Knockdown Showed Diminished Activation of Flavonoid Biosynthesis in the Cold
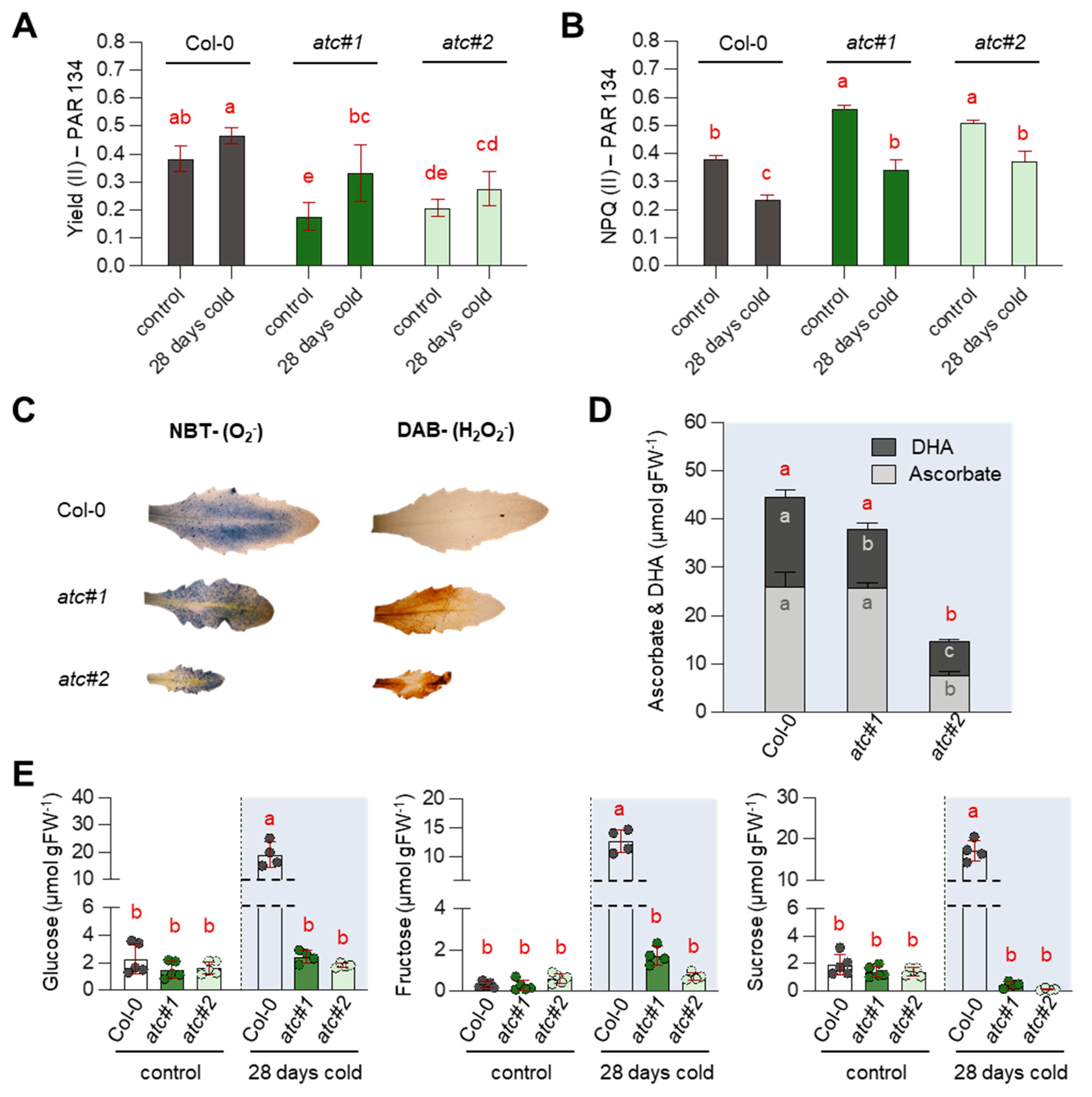
2.7. Alterations in Photosynthetic Rate Led to ROS Accumulation & Reduced Sugar Accumulation in atc#1 and 2
3. Discussion
4. Materials and Methods
4.1. Plant Growth
4.2. RNA Extraction
4.3. cDNA Synthesis and Gene Expression Analyses
4.4. RNA Seq and Transcriptome Analyses
4.5. Protein Extraction
4.6. Immunoblotting and Western Blot Detection
4.7. Chlorophyll and Anthocyanin Extraction and Quantification
4.8. Pulse-Amplitude-Modulation (PAM) Fluorometry Measurements
4.9. NBT (O2−) and DAB (H2O2) Staining
4.10. Whole-Leaf Ascorbate Determination
4.11. Sugar Extraction
4.12. Quantification of Aminoacids
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guy, C. Molecular responses of plants to cold shock and cold acclimation. J. Mol. Microbiol. Biotechnol. 1999, 1, 231–242. [Google Scholar] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Kopka, J.; Sung, D.Y.; Zhao, W.; Popp, M.; Porat, R.; Guy, C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007, 50, 967–981. [Google Scholar] [CrossRef]
- Kleine, T.; Nägele, T.; Neuhaus, H.E.; Schmitz-Linneweber, C.; Fernie, A.R.; Geigenberger, P.; Grimm, B.; Kaufmann, K.; Klipp, E.; Meurer, J.; et al. Acclimation in plants—The Green Hub consortium. Plant J. 2021, 106, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Schwenkert, S.; Fernie, A.R.; Geigenberger, P.; Leister, D.; Möhlmann, T.; Naranjo, B.; Neuhaus, H.E. Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci 2022, 27, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Huner, N.P.; Oquist, G.; Hurry, V.M.; Krol, M.; Falk, S.; Griffith, M. Photosynthesis, photoinhibition and low temperature acclimation in cold tolerant plants. Photosynth. Res. 1993, 37, 19–39. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Kleine, T.; Schneider, K.; Mühlhaus, T.; Lehmann, M.; Leister, D. Translational Components Contribute to Acclimation Responses to High Light, Heat, and Cold in Arabidopsis. iScience 2020, 23, 101331. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef]
- Araguirang, G.E.; Richter, A.S. Activation of anthocyanin biosynthesis in high light—What is the initial signal? New Phytol. 2022, 236, 2037–2043. [Google Scholar] [CrossRef]
- Hörmiller, I.I.; Nägele, T.; Augustin, H.; Stutz, S.; Weckwerth, W.; Heyer, A.G. Subcellular reprogramming of metabolism during cold acclimation in Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 602–610. [Google Scholar] [CrossRef]
- Wanner, L.A.; Junttila, O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 1999, 120, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Mahmood, K.; Rothstein, S.J. ROS Induces Anthocyanin Production Via Late Biosynthetic Genes and Anthocyanin Deficiency Confers the Hypersensitivity to ROS-Generating Stresses in Arabidopsis. Plant Cell Physiol. 2017, 58, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Zirngibl, M.E.; Araguirang, G.E.; Kitashova, A.; Jahnke, K.; Rolka, T.; Kuhn, C.; Nägele, T.; Richter, A.S. Triose phosphate export from chloroplasts and cellular sugar content regulate anthocyanin biosynthesis during high light acclimation. Plant Commun. 2023, 4, 100423. [Google Scholar] [CrossRef] [PubMed]
- Van Buskirk, H.A.; Thomashow, M.F. Arabidopsis transcription factors regulating cold acclimation. Physiol. Plant. 2006, 126, 72–80. [Google Scholar] [CrossRef]
- Bittner, A.; Hause, B.; Baier, M. Cold-priming causes dampening of oxylipin biosynthesis and signalling during the early cold- and light-triggering response of Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 7163–7179. [Google Scholar] [CrossRef]
- Bittner, A.; van Buer, J.; Baier, M. Cold priming uncouples light- and cold-regulation of gene expression in Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 281. [Google Scholar] [CrossRef] [PubMed]
- Busche, M.; Scarpin, M.R.; Hnasko, R.; Brunkard, J.O. TOR coordinates nucleotide availability with ribosome biogenesis in plants. Plant Cell 2021, 33, 1615–1632. [Google Scholar] [CrossRef]
- Bellin, L.; Del Cano-Ochoa, F.; Velazquez-Campoy, A.; Möhlmann, T.; Ramon-Maiques, S. Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase. Nat. Commun. 2021, 12, 947. [Google Scholar] [CrossRef]
- Bellin, L.; Melzer, M.; Hilo, A.; Amaya, D.L.G.; Keller, I.; Meurer, J.; Möhlmann, T. Pyrimidine nucleotide availability is essential for efficient photosynthesis, ROS scavenging, and organelle development. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chen, X.; Kim, S.H.; Rhee, S.; Witte, C.P. A plastid nucleoside kinase is involved in inosine salvage and control of purine nucleotide biosynthesis. Plant Cell 2023, 35, 510–528. [Google Scholar] [CrossRef] [PubMed]
- Kurt, F.; Filiz, E. Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100 and bHLH101 genes in Arabidopsis, tomato, rice, soybean and maize: Insights into iron (Fe) homeostasis. Biometals 2018, 31, 489–504. [Google Scholar] [CrossRef]
- Van Dingenen, J.; Antoniou, C.; Filippou, P.; Pollier, J.; Gonzalez, N.; Dhondt, S.; Goossens, A.; Fotopoulos, V.; Inze, D. Strobilurins as growth-promoting compounds: How Stroby regulates Arabidopsis leaf growth. Plant Cell Environ. 2017, 40, 1748–1760. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.X.; Liu, D.; Pan, Y.; Gong, W.; Ma, L.G.; Luo, J.C.; Deng, X.W.; Zhu, Y.X. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 2005, 59, 853–868. [Google Scholar] [CrossRef]
- Ohkubo, T.; Kameyama, A.; Kamiya, K.; Kondo, M.; Hara, M. F-segments of Arabidopsis dehydrins show cryoprotective activities for lactate dehydrogenase depending on the hydrophobic residues. Phytochemistry 2020, 173, 112300. [Google Scholar] [CrossRef] [PubMed]
- Okawa, K.; Nakayama, K.; Kakizaki, T.; Yamashita, T.; Inaba, T. Identification and characterization of Cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Environ. 2008, 31, 1470–1483. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, H. Coordinated Shoot and Root Responses to Light Signaling in Arabidopsis. Plant Commun. 2020, 1, 100026. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Zanor, M.I.; Müller-Röber, B.; Valle, E.M. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 2013, 83, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Linka, N.; Weber, A.P. Intracellular metabolite transporters in plants. Mol. Plant 2010, 3, 21–53. [Google Scholar] [CrossRef]
- Kunz, H.H.; Häusler, R.E.; Fettke, J.; Herbst, K.; Niewiadomski, P.; Gierth, M.; Bell, K.; Steup, M.; Flügge, U.I.; Schneider, A. The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol. 2010, 1, 115–128. [Google Scholar] [CrossRef]
- Selinski, J.; Scheibe, R. Malate valves: Old shuttles with new perspectives. Plant Biol. 2019, 21, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Scheibe, R.; Day, D.A.; Whelan, J. Alternative Oxidase Is Positive for Plant Performance. Trends Plant Sci. 2018, 23, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Jethva, J.; Lichtenauer, S.; Schmidt-Schippers, R.; Steffen-Heins, A.; Poschet, G.; Wirtz, M.; van Dongen, J.T.; Eirich, J.; Finkemeier, I.; Bilger, W.; et al. Mitochondrial alternative NADH dehydrogenases NDA1 and NDA2 promote survival of reoxygenation stress in Arabidopsis by safeguarding photosynthesis and limiting ROS generation. New Phytol. 2022. [Google Scholar] [CrossRef]
- Chen, C.T.; Slocum, R.D. Expression and functional analysis of aspartate transcarbamoylase and role of de novo pyrimidine synthesis in regulation of growth and development in Arabidopsis. Plant Physiol. Biochem. 2008, 46, 150–159. [Google Scholar] [CrossRef]
- Giermann, N.; Schröder, M.; Ritter, T.; Zrenner, R. Molecular analysis of de novo pyrimidine synthesis in solanaceous species. Plant Mol. Biol. 2002, 50, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Nägele, T.; Heyer, A.G. Approximating subcellular organisation of carbohydrate metabolism during cold acclimation in different natural accessions of Arabidopsis thaliana. New Phytol. 2013, 198, 777–787. [Google Scholar] [CrossRef]
- van Buer, J.; Prescher, A.; Baier, M. Cold-priming of chloroplast ROS signalling is developmentally regulated and is locally controlled at the thylakoid membrane. Sci. Rep. 2019, 9, 3022. [Google Scholar] [CrossRef]
- Vu, D.P.; Martins Rodrigues, C.; Jung, B.; Meissner, G.; Klemens, P.A.W.; Holtgräwe, D.; Fürtauer, L.; Nägele, T.; Nieberl, P.; Pommerrenig, B.; et al. Vacuolar sucrose homeostasis is critical for plant development, seed properties, and night-time survival in Arabidopsis. J. Exp. Bot. 2020, 71, 4930–4943. [Google Scholar] [CrossRef] [PubMed]
- Pommerrenig, B.; Ludewig, F.; Cvetkovic, J.; Trentmann, O.; Klemens, P.A.W.; Neuhaus, H.E. In Concert: Orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant Cell Physiol. 2018, 59, 1290–1299. [Google Scholar] [CrossRef]
- Noguchi, K.; Yoshida, K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 2008, 8, 87–99. [Google Scholar] [CrossRef]
- Dyson, B.C.; Miller, M.A.; Feil, R.; Rattray, N.; Bowsher, C.G.; Goodacre, R.; Lunn, J.E.; Johnson, G.N. FUM2, a cytosolic fumarase, is essential for acclimation to low temperature in Arabidopsis thaliana. Plant Physiol. 2016, 172, 118–127. [Google Scholar] [CrossRef]
- Hodges, M. Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp. Bot 2002, 53, 905–916. [Google Scholar] [CrossRef]
- Maurino, V.G.; Engqvist, M.K. 2-Hydroxy Acids in Plant Metabolism. Arab. Book 2015, 13, e0182. [Google Scholar] [CrossRef]
- Werner, A.K.; Witte, C.P. 2011. The biochemistry of nitrogen mobilization: Purine ring catabolism. Trends Plant Sci. 2002, 16, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar]
- Schreiber, U.; Quayle, P.; Schmidt, S.; Escher, B.I.; Müller, J.F. Methodology and evaluation of a highly sensitive algae toxicity test based on multiwell chlorophyll fluorescence imaging. Biosensens. Bioelectron. 2007, 22, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Fryer, M.J.; Oxborough, K.; Mullineaux, P.M.; Baker, N.R. Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 2002, 53, 1249–1254. [Google Scholar]
- Gillespie, K.M.; Ainsworth, E.A. Measurement of reduced, oxidized and total ascorbate content in plants. Nat. Prot. 2007, 2, 871–874. [Google Scholar] [CrossRef]
- Stitt, M.; Lilley, R.M.; Gerhardt, R.; Heldt, H.W. Metabolite levels in specific cells and subcellular compartments of plant leaves. Meth. Enzymol. 1989, 174, 518–552. [Google Scholar]

| 22 °C | 1 d 4 °C/22 °C | 3 d/1 d 4 °C | |||||
|---|---|---|---|---|---|---|---|
| Locus | Name | atc#1/Col-0 | Col-0 | atc#1 | Col-0 | atc#1 | Function |
| AT4G18440 | ADSL | −1.27 | −0.98 | −0.46 | −1.09 | −0.50 | de novo |
| AT2G37690 | AIR carb. | 0.69 | 1.90 | 1.36 | 0.88 | 0.43 | de novo |
| AT2G37250 | AMK1 | 1.37 | 1.56 | −0.43 | de novo | ||
| AT5G35170 | AMK5 | −0.72 | −1.14 | −1.16 | −0.50 | de novo | |
| AT3G01820 | AMK7 | 2.97 | 3.16 | −0.80 | de novo | ||
| AT4G34740 | ATase2 | 2.34 | 2.34 | −0.63 | −0.58 | de novo | |
| AT5G23300 | DHODH | 1.62 | 1.09 | de novo | |||
| AT1G30820 | CTPS1 | 1.84 | 2.29 | −2.82 | −2.31 | de novo | |
| AT3G12670 | CTPS2 | 1.95 | 2.03 | −0.99 | −0.82 | de novo | |
| AT2G16370 | DHFR-TS | −1.45 | −1.22 | de novo | |||
| AT3G06200 | GMK | 1.52 | 1.05 | de novo | |||
| AT1G63660 | GMPS | 0.73 | 1.53 | 0.88 | de novo | ||
| AT1G79470 | IMPDH1 | 0.76 | 1.80 | 0.96 | de novo | ||
| AT1G16350 | IMPDH2 | 0.51 | −0.73 | −0.92 | 1.14 | 0.92 | de novo |
| AT2G21790 | RNR1 | 0.65 | 1.03 | 0.52 | 0.41 | de novo | |
| AT3G60180 | UMK1 | −1.28 | −1.39 | 0.95 | de novo | ||
| AT4G20070 | AAH | 2.57 | 2.26 | −1.60 | −1.34 | catabolism | |
| AT5G43600 | UAH | −1.57 | −1.42 | 0.53 | 0.61 | catabolism | |
| AT4G04955 | ALN | −2.43 | 2.20 | 1.00 | catabolism | ||
| AT1G05620 | NSH2 | −1.39 | −0.75 | 1.25 | 0.75 | catabolism | |
| AT3G17810 | PYD1 | −0.53 | 0.44 | 1.00 | −1.50 | −1.18 | catabolism |
| AT5G12200 | PYD2 | −0.41 | −1.19 | −0.90 | catabolism | ||
| AT5G64370 | PYD3 | −0.42 | −1.17 | −0.69 | catabolism | ||
| AT2G26230 | UOX | −0.37 | 1.01 | 1.25 | −0.53 | catabolism | |
| AT1G80050 | APT2 | −1.58 | −1.89 | 1.02 | salvage | ||
| AT4G22570 | APT3 | −0.74 | −2.90 | −2.66 | 1.29 | 1.75 | salvage |
| AT5G11160 | APT5 | 1.13 | 1.54 | −0.59 | salvage | ||
| AT1G72040 | dNK | 0.63 | 1.14 | 0.65 | salvage | ||
| AT1G71750 | HGPRT | 1.06 | 1.00 | salvage | |||
| AT5G40870 | UCK1 | −0.85 | −1.04 | 0.56 | salvage | ||
| AT5G13490 | AAC2 | 0.97 | 2.09 | transporter | |||
| AT5G61810 | APC1 | 2.23 | 1.88 | −1.55 | −1.34 | transporter | |
| AT4G32400 | BT1 | 2.01 | 1.58 | −0.70 | −0.72 | transporter | |
| AT4G05120 | ENT3 | −1.30 | −0.69 | 0.80 | transporter | ||
| AT4G05110 | ENT6 | −1.06 | transporter | ||||
| AT5G03555 | PLUTO | 2.08 | 2.39 | −1.28 | −1.29 | transporter | |
| AT5G56450 | pmANT1 | −1.00 | −0.77 | transporter | |||
| Locus | Name | 22 °C | 1 d 4 °C/22 °C | 3 d/1 d 4 °C | Subcell. Localization | Substrate | ||
|---|---|---|---|---|---|---|---|---|
| atc#1/Col-0 | Col-0 | atc#1 | Col-0 | atc#1 | ||||
| AT1G14140 | UCP3 | −1.16 | −0.93 | 0.62 | Mt | protons | ||
| AT5G58970 | UCP2 | −0.86 | −1.14 | −1.10 | 1.17 | 1.71 | Mt | protons |
| AT5G09470 | DIC3 | 3.33 | 4.26 | Mt | dicarboxylate | |||
| AT4G24570 | DIC2 | −0.89 | 3.23 | 4.04 | Mt | dicarboxylate | ||
| AT2G22500 | DIC1 | −0.85 | 2.15 | 2.97 | Mt | dicarboxylate | ||
| AT3G48850 | PIC2 | 1.41 | 4.35 | −1.48 | −2.67 | Mt | phosphate | |
| AT2G17270 | MPT1 | 2.32 | 2.42 | −1.04 | −1.00 | Mt | phosphate | |
| AT5G01340 | SFC | 3.15 | 3.46 | −1.16 | Mt | succinate/fumarate | ||
| AT5G27520 | PNC2 | 1.97 | 2.05 | −1.48 | −1.49 | Mt, P | adenine nucleotide | |
| AT5G66380 | FOLT1 | −0.76 | −1.05 | 0.65 | PL | folate | ||
| AT5G16150 | pGlcT | −0.44 | −0.87 | −1.03 | −1.20 | −0.34 | PL | hexose |
| AT1G68570 | NPF3.1 | 2.69 | 2.35 | −0.66 | −0.72 | PL | nitrite | |
| AT4G32400 | BT1-like | 2.01 | 1.58 | −0.70 | −0.72 | PL | nucleotide | |
| AT1G61800 | GPT2 | 5.90 | 7.61 | 0.39 | −1.65 | PL | Glc6P, TP | |
| AT1G58030 | CAT2 | −1.40 | −1.32 | 0.62 | TP | amino acid | ||
| AT5G40890 | CLCa | −2.34 | −1.50 | 0.56 | TP | Cl, nitrate | ||
| AT3G16240 | TIP2.1 | −3.42 | −3.74 | −0.48 | 0.97 | TP | H2O | |
| AT3G26520 | TIP1.2 | −2.08 | −1.85 | −0.49 | 0.39 | TP | H2O | |
| AT3G18440 | ALMT9 | 1.40 | 0.92 | −0.71 | −0.86 | TP | malate | |
| AT5G47560 | TDT | 1.82 | 0.96 | −2.45 | −1.47 | TP | malate | |
| AT1G20840 | TST1 | −0.84 | −1.50 | −1.10 | 0.89 | 0.66 | TP | monosaccharide |
| AT4G35300 | TST2 | 2.94 | 2.61 | −1.07 | −1.22 | TP | mono-, disaccharide | |
| Locus | Name | 22 °C | 1 d 4 °C/22 °C | 3 d/1 d 4 °C | Localization | Function | ||
|---|---|---|---|---|---|---|---|---|
| atc#1/Col-0 | Col-0 | atc#1 | Col-0 | atc#1 | ||||
| AT5G50950 | FUM2 | −1.49 | −2.38 | −2.14 | −0.74 | Mt, Ct | fumarase | |
| AT1G07180 | NDA1 | 0.59 | 1.07 | −0.73 | Mt, P | dehydrogenase | ||
| AT3G22370 | AOX1A | 0.70 | 3.00 | 2.47 | −1.74 | −1.97 | Mt | reductase |
| AT3G55410 | E1-OGDH1 | −0.33 | 0.99 | 1.37 | Mt | dehydrogenase | ||
| AT5G55070 | E2-OGDH2 | −0.62 | 1.06 | 1.63 | −0.43 | Mt | dehydrogenase | |
| AT4G35260 | IDH1 | −0.51 | 0.80 | 1.04 | Mt | dehydrogenase | ||
| AT3G27380 | SDH2-1 | −0.43 | 1.68 | 2.22 | −1.14 | −1.59 | Mt | dehydrogenase |
| AT3G03250 | UGP2 | −0.57 | 1.19 | 0.95 | −0.40 | −0.52 | Ct, PM | pyrophosphorylase |
| AT5G51830 | FRK1 | −0.53 | 2.46 | 3.27 | −1.22 | −1.93 | Ct | kinase |
| AT5G56630 | PFK7 | −0.35 | 2.15 | 1.87 | −1.26 | −1.44 | Ct | kinase |
| AT5G56350 | PK | −0.46 | 1.59 | 2.04 | −0.76 | Ct | kinase | |
| AT4G26520 | FBA7 | 0.40 | −1.85 | −1.62 | 0.61 | Ct | aldolase | |
| AT3G52930 | FBA8 | −0.38 | 1.48 | 1.26 | −0.47 | −0.49 | all | aldolase |
| AT1G36380 | Cyt C red | −0.77 | −1.10 | −0.61 | ? | unknown | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellin, L.; Garza Amaya, D.L.; Scherer, V.; Pruß, T.; John, A.; Richter, A.; Möhlmann, T. Nucleotide Imbalance, Provoked by Downregulation of Aspartate Transcarbamoylase Impairs Cold Acclimation in Arabidopsis. Molecules 2023, 28, 1585. https://doi.org/10.3390/molecules28041585
Bellin L, Garza Amaya DL, Scherer V, Pruß T, John A, Richter A, Möhlmann T. Nucleotide Imbalance, Provoked by Downregulation of Aspartate Transcarbamoylase Impairs Cold Acclimation in Arabidopsis. Molecules. 2023; 28(4):1585. https://doi.org/10.3390/molecules28041585
Chicago/Turabian StyleBellin, Leo, Diana Laura Garza Amaya, Vanessa Scherer, Tobias Pruß, Annalisa John, Andreas Richter, and Torsten Möhlmann. 2023. "Nucleotide Imbalance, Provoked by Downregulation of Aspartate Transcarbamoylase Impairs Cold Acclimation in Arabidopsis" Molecules 28, no. 4: 1585. https://doi.org/10.3390/molecules28041585
APA StyleBellin, L., Garza Amaya, D. L., Scherer, V., Pruß, T., John, A., Richter, A., & Möhlmann, T. (2023). Nucleotide Imbalance, Provoked by Downregulation of Aspartate Transcarbamoylase Impairs Cold Acclimation in Arabidopsis. Molecules, 28(4), 1585. https://doi.org/10.3390/molecules28041585






