Compilation of the Antimicrobial Compounds Produced by Burkholderia Sensu Stricto
Abstract
:1. Introduction
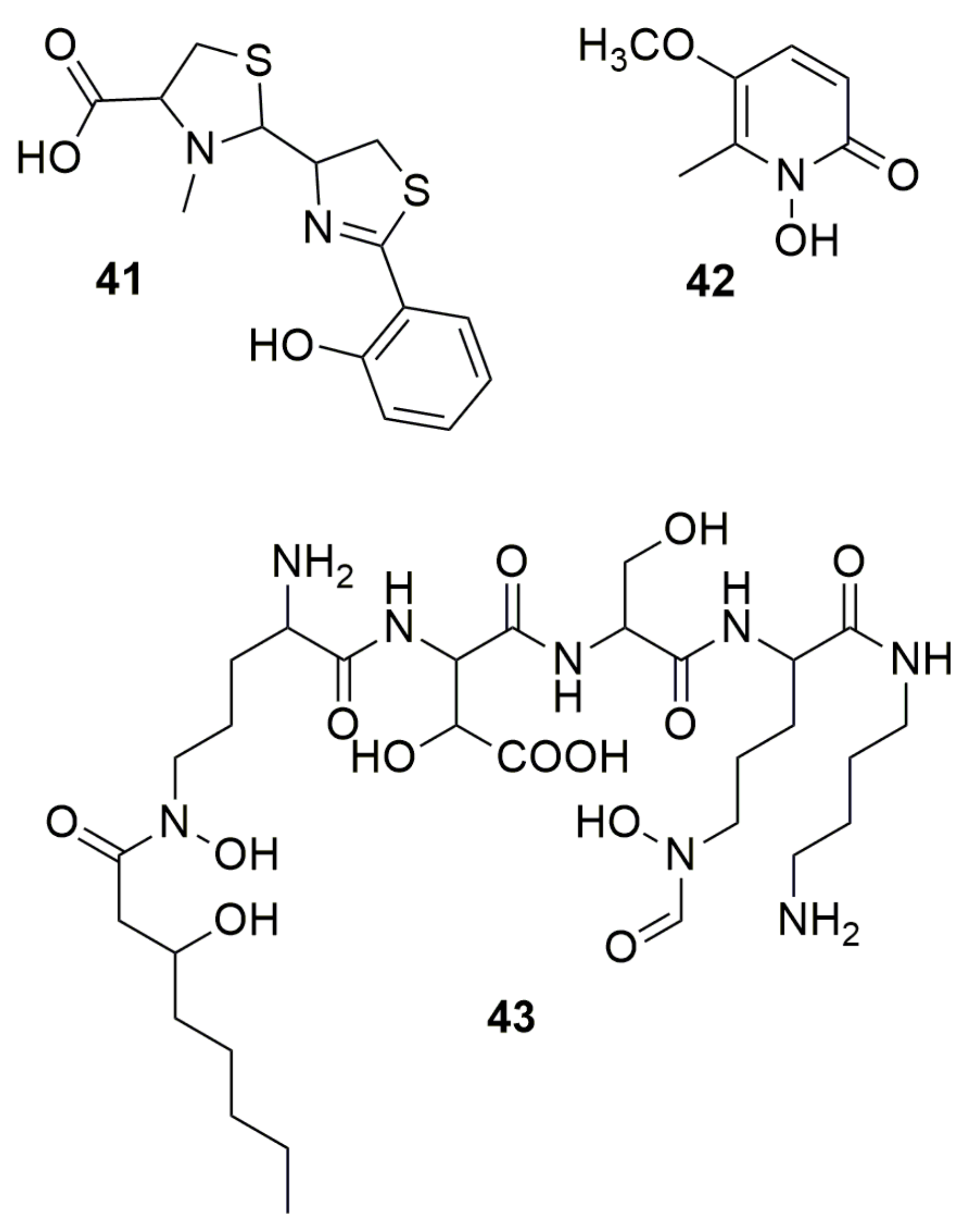
2. N-Containing Heterocycles
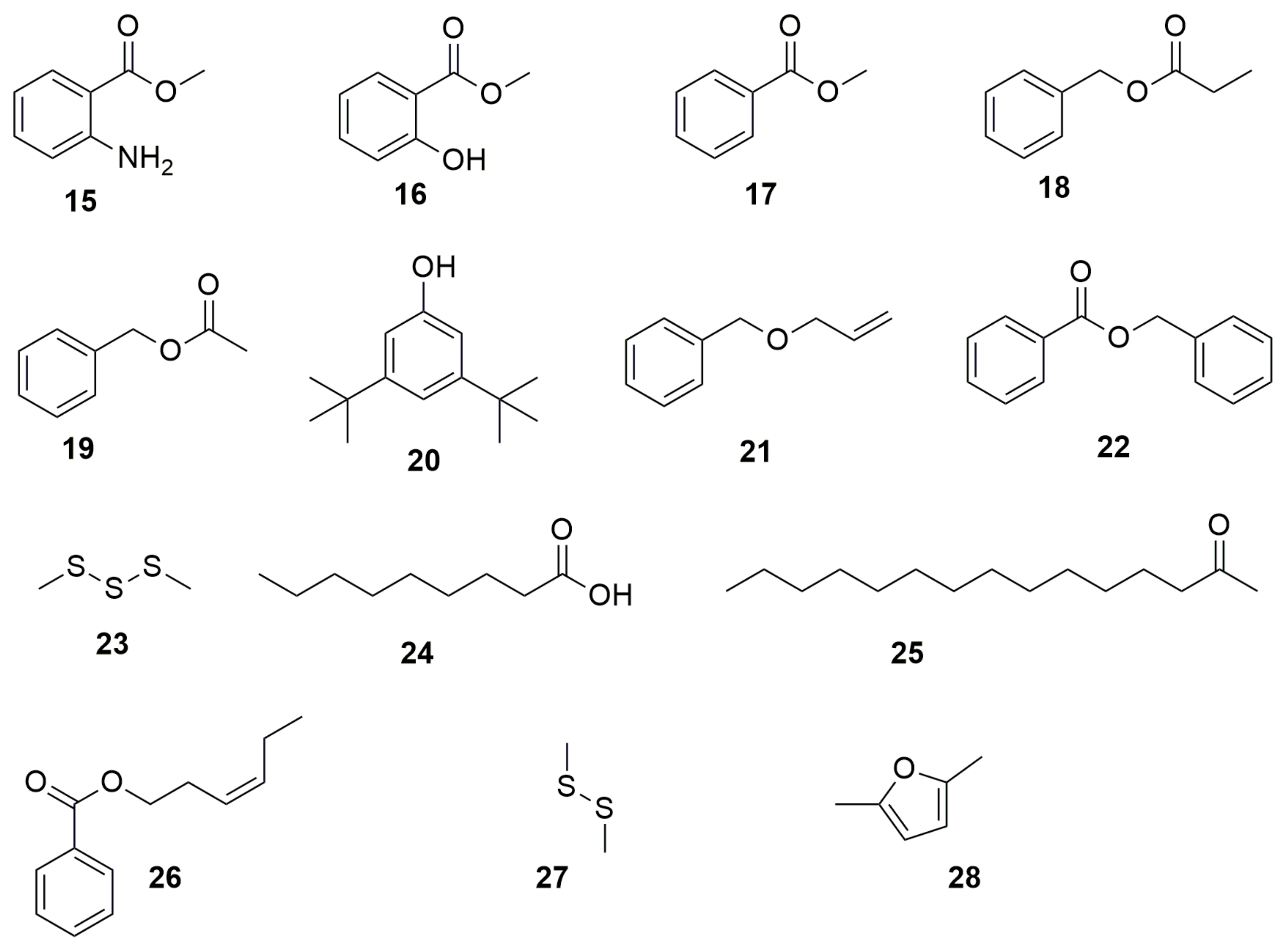
3. Volatile Organic Compounds
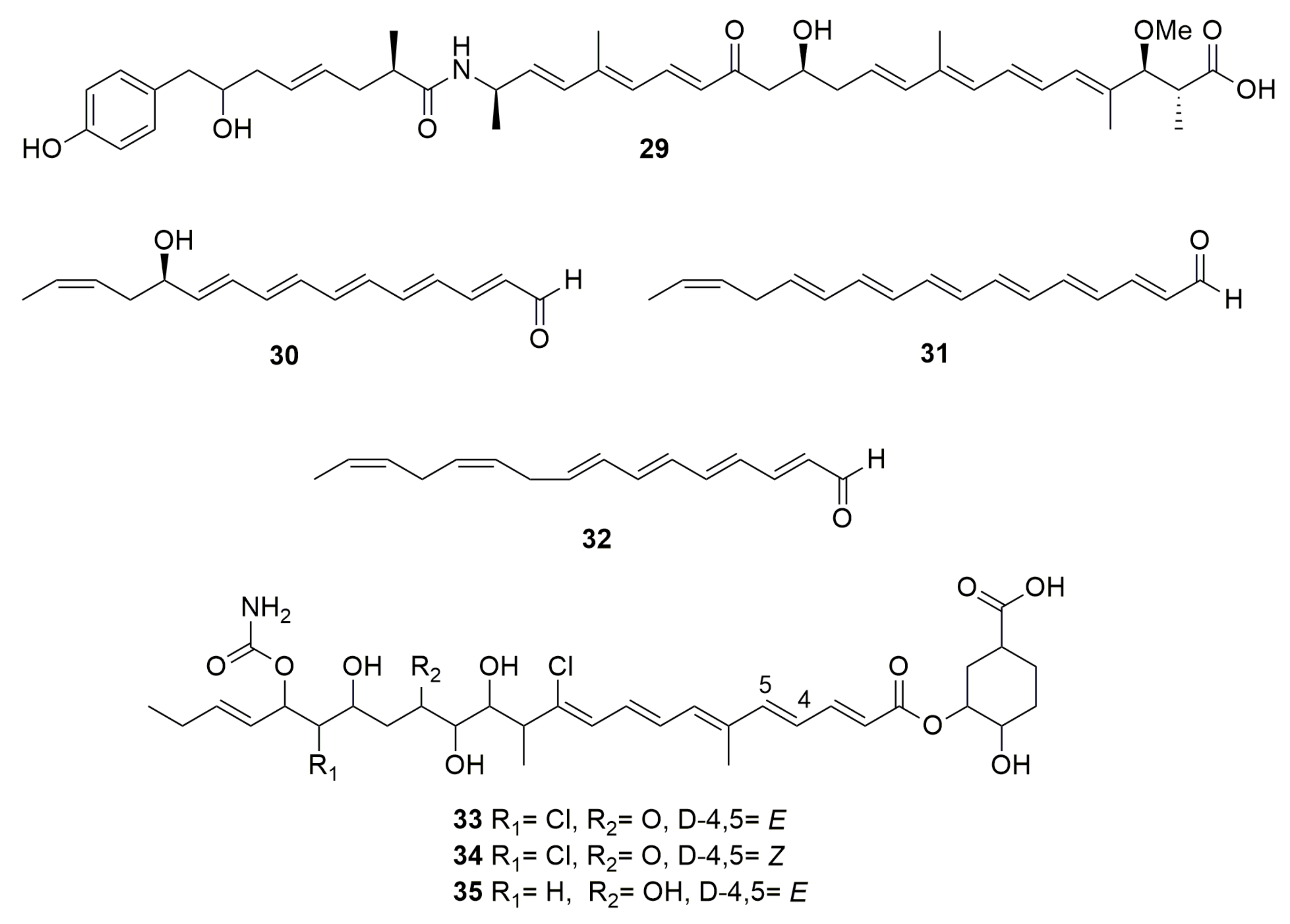
4. Polyenes
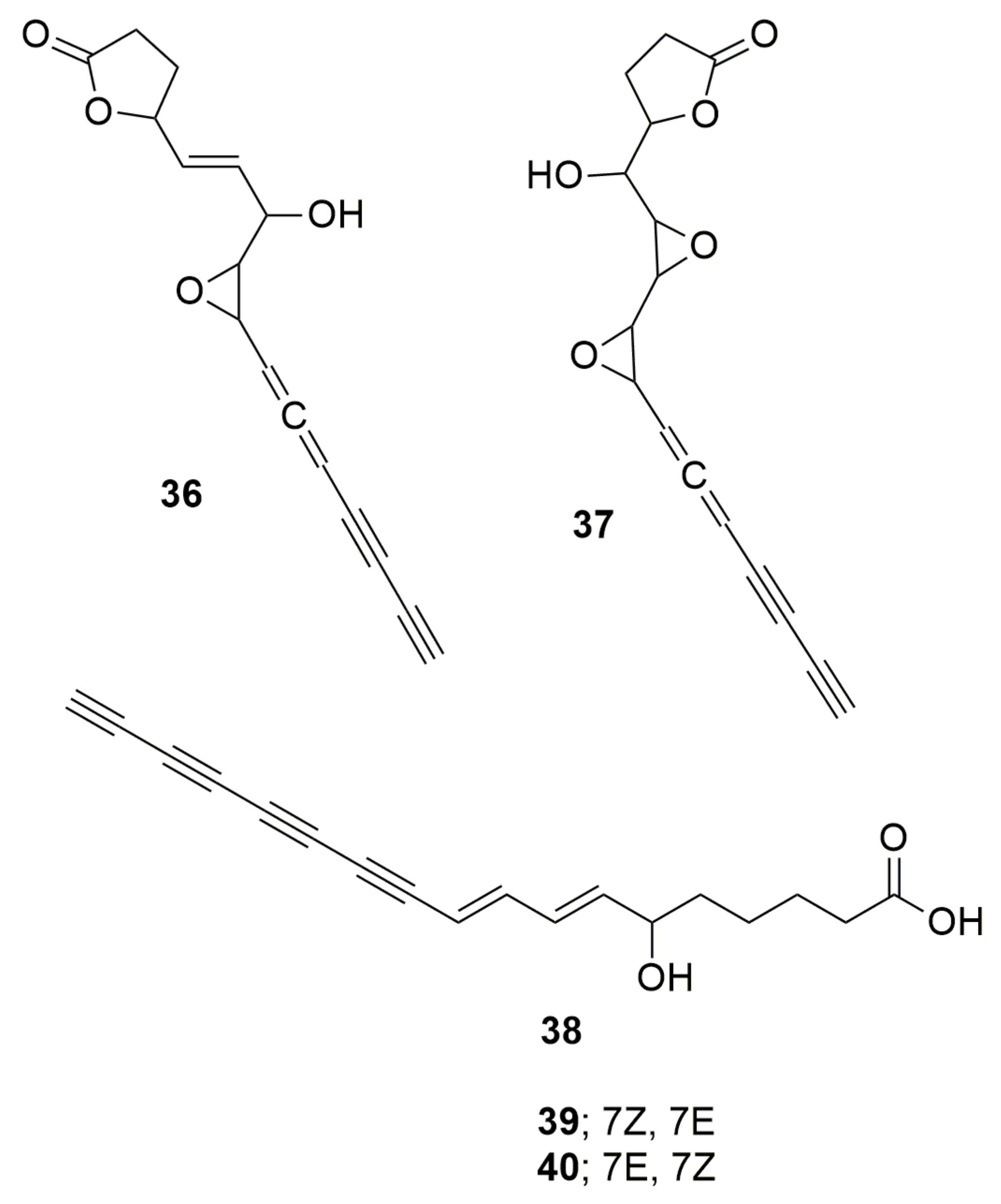
5. Polyynes
6. Siderophores
7. Macrolides
8. Bacteriocins
9. Quinolones
10. Other NPR-PK Compounds
11. Other Antimicrobial Compounds
12. Compounds with Dual Effect
13. Metabolism as Control
14. Data Mining
15. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven Burkholderia species to the genus Paraburkholderia and proposal of Caballeronia gen. nov., a new genus to accommodate twelve species of Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Santos, L.; Castro, D.B.A.; Ferreira-Tonin, M.; Correa, D.B.A.; Weir, B.S.; Park, D.; Mariscal-Ottoboni, L.M.; Rodrigues-Neto, J.; Lanza-Destefano, S.A. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Anton. Leeuw. Int. J. Gen. 2017, 110, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Estrada-de los Santos, P.; Palmer, M.; Chavez-Ramírez, B.; Beukes, C.; Steenkamp, E.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, C.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov. and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.H.; Lv, Y.Y.; Gao, Z.G.; Qiu, L.H. Pararobbsia silviterrae gen. nov., sp. nov., isolated from forest soil and reclassification of Burkholderia alpina as Pararobbsia alpina comb. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 1412–1420. [Google Scholar] [CrossRef]
- Khakhum, N.; Tapia, D.; Torres, A.G. Burkholderia mallei and glanders. In Defense Against Biological Attacks; Springer: Manhattan, NY, USA, 2019; pp. 161–183. [Google Scholar] [CrossRef]
- Gassiep, I.; Armostrong, M.; Norton, R. Human melioidosis. Clin. Microbiol. Rev. 2020, 33, e00006-19. [Google Scholar] [CrossRef]
- Hall, C.M.; Baker, A.L.; Sahl, J.W.; Mayo, M.; Scholz, H.C.; Kaestli, M.; Schupp, J.; Martz, M.; Settles, E.W.; Busch, J.D.; et al. Expanding the Burkholderia pseudomallei complex with the addition of two novel species: Burkholderia mayonis sp. nov. and Burkholderia savanae sp. nov. Appl. Environ. Microbiol. 2022, 11, e0158321. [Google Scholar] [CrossRef]
- Pereira-Andrade, J.; de Souza, H.G.; Carvalho-Ferreira, L.; Cnockaert, M.; De Canck, E.; Wieme, A.D.; Peeters, C.; Gross, E.; De Souza, J.T.; Santos-Marbach, P.A.; et al. Burkholderia perseverans sp. nov., a bacterium isolated from the Restinga ecosystem, a producer of volatile and diffusible compounds that inhibit plant pathogens. Braz. J. Microbiol 2021, 52, 2145–2152. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef]
- Morales-Ruiz, L.M.; Rodriguez-Cisneros, M.; Kerber-Diaz, J.C.; Rojas-Rojas, F.U.; Ibarra, J.A.; Estrada-de los Santos, P. Burkholderia orbicola sp. nov., a novel species within the Burkholderia cepacia complex. Arch. Microbiol. 2022, 204, 178. [Google Scholar] [CrossRef]
- Rose, H.; Baldwin, A.; Dowson, C.G.; Mahenthiralingam, E. Biocide susceptibility of the Burkholderia cepacia complex. J. Ant. Chemother. 2009, 63, 502–510. [Google Scholar] [CrossRef]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef]
- Parke, J.L.; Gurian-Sherman, D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Ann. Rev. Phytopathol. 2001, 39, 225–258. [Google Scholar] [CrossRef]
- Chain, P.S.G.; Denef, V.J.; Konstantinidis, K.T.; Vergez, L.M.; Agullo, L.; Reyes, V.L.; Hauser, L.; Cordova, M.; Gomez, L.; Gonzalez, M.; et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 2006, 103, 15280–15287. [Google Scholar] [CrossRef]
- O’Sullivan, L.A.; Weightman, A.J.; Jones, T.H.; Marchbank, A.M.; Tiedje, J.M.; Mahenthiralingam, E. Identifying the genetic basis of ecologically and biotechnologically useful functions of the bacterium Burkholderia vietnamiensis. Environ. Microbiol. 2007, 9, 1017–1034. [Google Scholar] [CrossRef]
- Bach, E.; Pereira-Passaglia, L.M.; Jiao, J.; Gross, H. Burkholderia in the genomic era: From taxonomy to the discovery of new antimicrobial secondary metabolites. Rev. Microbiol. 2022, 48, 121–160. [Google Scholar] [CrossRef]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandmme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef]
- Valenca, C.A.S.; Barbosa, A.A.T.; Souto, E.B.; Caramao, E.B.; Jain, S. Volatile nitrogenous compounds from bacteria: Source of novel bioactive compounds. Chem. Biodivers. 2021, 18, e2100549. [Google Scholar] [CrossRef]
- Prasad, J.; Pandey, P.; Anand, R.; Raghuwanshi, R. Drought Exposed Burkholderia seminalis JRBHU6 exhibits antimicrobial potential through pyrazine-1,4-dione derivatives targeting multiple bacterial and fungal proteins. Front. Microbiol. 2021, 12, 633036. [Google Scholar] [CrossRef]
- Petri, G.L.; Spano, V.; Spatola, R.; Holl, R.; Raimondi, M.V.; Barraja, P.; Montalbano, A. Bioactive pyrrole-based compound with target selectivity. Eur. J. Med. Chem. 2020, 208, 112783. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.E.; Teh, K.L. Antibacterial iminopyrrolidines from Burkholderia plantarii, a bacterial pathogen of rice. Org. Biomol. Chem. 2005, 3, 3540. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.E.; Greenwood, D.R.; Sarajioni, V. An antibacterial pyrazole derivative from Burkholderia glumae, a bacterial pathogen of rice. Phytochemistry 2008, 69, 2704–2707. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.; Chaudhari, A.; Prabha, R.; Shukla, R.; Singh, D.P. Microbial pyrrolnitrin: Natural metabolite with immense practical utility. Biomolecules 2019, 9, 443. [Google Scholar] [CrossRef] [PubMed]
- Arima, K.; Imanaka, H.; Kousaka, M.; Fukuta, A.; Tamura, G. Pyrrolnitrin, a new antibiotic substance produced by Pseudomonas. Agric. Biol. Chem. 1964, 28, 575–576. [Google Scholar] [CrossRef]
- Kilani, J.; Fillinger, S. Phenylpyrroles: 30 years, two molecules and (nearly) no resistance. Front. Microbiol. 2016, 7, 2014. [Google Scholar] [CrossRef]
- El-Banna, N.; Winkelmann, G. Pyrrolnitrin from Burkholderia cepacia: Antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 1998, 85, 69–78. [Google Scholar] [CrossRef]
- Sultan, M.Z.; Park, K.; Lee, S.Y.; Park, J.K.; Varughese, T.; Moon, S.S. Novel oxidized derivatives of antifungal pyrrolnitrin from the bacterium Burkholderia cepacia K87. J. Antibiot. 2008, 61, 420–425. [Google Scholar] [CrossRef]
- Jung, B.K.; Hong, S.J.; Park, G.S.; Kim, M.C.; Shin, J.H. Isolation of Burkholderia cepacia JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl. Biol. Chem. 2018, 61, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Webster, G.; Jones, C.; Mullins, A.J.; Mahenthiralingam, E. A rapid screening method for the detection of specialized metabolites from bacteria: Induction and suppression of metabolites from Burkholderia species. J. Microbiol. Methods 2020, 178, 106057. [Google Scholar] [CrossRef]
- Yan, J.; Liu, W.; Cai, J.; Wang, Y.; Li, D.; Hua, H.; Cao, H. Advances in phenazines over the past decade: Review of their pharmacological activities, mechanisms of action, biosynthetic pathways, and synthetic strategies. Mar. Drugs 2021, 19, 610. [Google Scholar] [CrossRef]
- Cartwright, D.K.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 1995, 43, 211–216. [Google Scholar] [CrossRef]
- Hendry, S.; Steinke, S.; Wittstein, K.; Stadler, M.; Harmrolfs, K.; Adewunmi, Y.; Sahukhal, G.; Elasri, M.; Thomashow, L.; Weller, D.; et al. Functional analysis of phenazines biosynthesis genes in Burkholderia spp. Appl. Environ. Microbiol. 2021, 87, e02348-20. [Google Scholar] [CrossRef]
- Han, J.W.; Kim, J.D.; Lee, J.M.; Ham, J.H.; Lee, D.; Kim, B.S. Structural elucidation and antimicrobial activity of new phencomycin derivatives isolated from Burkholderia glumae strain 411gr-6. J. Antibiot. 2014, 67, 721–723. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, M.; Du, J.; Huang, T.; Liu, J.; Dong, T.; Chen, Y. Isolation of Burkholderia sp. HQB-1, a promising biocontrol bacterium to protect banana against Fusarium wilt through phenazine-1-carboxylic acid secretion. Front. Microbiol. 2020, 11, 605152. [Google Scholar] [CrossRef]
- Chen, J.H.; Xiang, W.; Cao, K.X.; Lu, X.; Yao, S.C.; Hung, D.; Huang, R.S.; Li, L.B. Characterization of volatile organic compounds emitted from endophytic Burkholderia cenocepacia ETR-B22 by SPME-GC-MS and their inhibitory activity against various plant fungal pathogens. Molecules 2020, 25, 3765. [Google Scholar] [CrossRef]
- Xu, T.; Shi, L.; Zhang, Y.; Wang, K.; Yang, Z.; Ke, S. Synthesis and biological evaluation of marine alkaloid-oriented β-carboline analogues. Eur. J. Med. Chem. 2019, 25, 3765. [Google Scholar] [CrossRef]
- Huang, D.; Zhang, Z.; Li, Y.; Liu, F.; Huang, W.; Min, Y.; Wang, K.; Yang, J.; Cao, C.; Gong, Y.; et al. Carboline derivatives based on natural pityriacitrin as potential antifungal agents. Phytochemi. Lett. 2022, 48, 100–105. [Google Scholar] [CrossRef]
- Lin, Y.T.; Lee, C.C.; Leu, W.M.; Wu, J.J.; Huang, Y.C.; Meng, M. Fungicidal activity of volatile organic compounds emitted by Burkholderia gladioli strain BBB-01. Molecules 2021, 26, 745. [Google Scholar] [CrossRef]
- Hunter, W.J.; Manter, D.K. Antimicrobial properties of an oxidizer produced by Burkholderia cenocepacia P525. Curr. Microbiol. 2014, 68, 610–614. [Google Scholar] [CrossRef]
- Wu, Y.; Seyedsayamdost, M.R. The polyene natural product thailandamide A inhibits fatty acid biosynthesis in Gram-positive and Gram-negative bacteria. Biochemistry 2018, 57, 4247–4251. [Google Scholar] [CrossRef] [PubMed]
- Park, J.D.; Moon, K.; Miller, C.; Rose, J.; Xu, F.; Ebmeier, C.C.; Jacobsen, J.R.; Mao, D.; Old, W.M.; DeShazer, D.; et al. Thailandenes, cryptic polyene natural products isolated from Burkholderia thailandensis using phenotype-guided transposon mutagenesis. ACS Chem. Biol. 2020, 15, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Song, L.; Sass, A.; White, J.; Wilmot, C.; Marchbank, A.; Boaisha, O.; Paine, J.; Knight, D.; Challis, G.L. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem. Biol. 2011, 18, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Parmeggiani, A.; Krab, I.M.; Watanabe, T.; Nielsen, R.C.; Dahlberg, C.; Nyborg, J.; Nissen, P. Enacyloxin IIa pinpoints a binding pocket of elongation factor Tu for development of novel antibiotics. J. Biol. Chem. 2006, 281, 2893–2900. [Google Scholar] [CrossRef]
- Ross, C.; Opel, V.; Scherlach, K.; Hertweck, C. Biosynthesis of antifungal and antibacterial polyketides by Burkholderia gladioli in coculture with Rhizopus microsporus. Mycoses 2014, 57, 48–55. [Google Scholar] [CrossRef]
- Parker, W.L.; Rathnum, M.L.; Seiner, V.; Trejo, W.H.; Principe, P.A.; Sykes, R.B. Cepacin A and cepacin B, two new antibiotics produced by Pseudomonas cepacia. J. Antibiot. 1984, 37, 431–440. [Google Scholar] [CrossRef]
- Mullins, A.J.; Murray, J.A.H.; Bull, M.J.; Jenner, M.; Jones, C.; Webster, G.; Green, A.E.; Neill, D.R.; Connor, T.R.; Parkhill, J.; et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat. Microbiol. 2019, 4, 996–1005. [Google Scholar] [CrossRef]
- Kusumi, T.; Ohtani, I.; Nishiyama, K.; Kakisawa, H. Caryoynencins, potent antibiotics from a plant pathogen. Tetrahedron 1987, 28, 3981–3984. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Park, H.J.; Ishizuka, S.; Omata, K.; Hirama, M. Chemistry and antimicrobial activity of caryoynencin analogs. J. Med. Chem. 1995, 38, 5015–5022. [Google Scholar] [CrossRef]
- Florez, L.V.; Scherlach, K.; Gaube, P.; Ross, C.; Sitte, E.; Hermes, C.; Rodrigues, A.; Hertweck, C.; Kaltenpoth, M. Antibiotic-producing symbionts dynamically transition between plant pathogenicity and insect-defensive mutualism. Nat. Commun. 2017, 8, 15172. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637. [Google Scholar] [CrossRef]
- Finking, R.; Marahiel, M.A. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- Hur, G.H.; Vickery, C.R.; Burkart, M.D. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep. 2012, 29, 1074–1098. [Google Scholar] [CrossRef]
- Jaremko, M.J.; Davis, T.D.; Corpuz, J.C.; Burkart, M.D. Type II non-ribosomal peptide synthetase proteins: Structure, mechanism, and protein–protein interactions. Nat. Prod. Rep. 2020, 37, 355–379. [Google Scholar] [CrossRef]
- Adler, C.; Corbalan, N.S.; Seyedsayamdost, M.R.; Pomares, M.F.; de Cristobal, R.E.; Clardy, J.; Kolter, R.; Vincent, P.A. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS ONE 2012, 7, e46754. [Google Scholar] [CrossRef]
- Ong, K.S.; Aw, Y.K.; Lee, L.H.; Yule, C.M.; Cheow, Y.L.; Lee, S.M. Burkholderia paludis sp. nov., an antibiotic-siderophore producing novel Burkholderia cepacia complex species, isolated from Malaysian tropical peat swamp soil. Front. Microbiol. 2016, 7, 2046. [Google Scholar] [CrossRef]
- Ong, K.S.; Cheow, Y.L.; Lee, S.M. The role of reactive oxygen species in the antimicrobial activity of pyochelin. J. Adv. Res. 2017, 8, 393–398. [Google Scholar] [CrossRef]
- da Araujo, F.D.S.; Araujo, W.L.; Eberlin, M.N. Potential of Burkholderia seminalis TC3.4.2R3 as biocontrol agent against Fusarium oxysporum evaluated by mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 2017, 28, 901–907. [Google Scholar] [CrossRef]
- Meyer, J.M.; Hohnadel, D.; Halle, F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J. Gen. Microbiol. 1989, 135, 1479–1487. [Google Scholar] [CrossRef] [Green Version]
- Itoh, J.; Miyadoh, S.; Takahasi, S.; Amano, S.; Ezaki, N.; Yamada, Y. Studies on antibiotics BN-227 and BN-227-F, new antibiotics. I. Taxonomy, isolation and characterization. J. Antibiot. 1979, 32, 1089–1095. [Google Scholar] [CrossRef]
- Itoh, J.; Amano, S.; Ogawa, Y.; Kodama, Y.; Ezaki, N.; Yamada, Y. Studies on antibiotics BN-227 and BN-227-F, new antibiotics. II. Chemical structure of antibiotics BN-227 and BN-227-F. J. Antibiot. 1980, 33, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Darling, P.; Chan, M.; Cox, A.D.; Sokol, P. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infec. Immun. 1998, 66, 874–877. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.S. Iron acquisition mechanisms of the Burkholderia cepacia complex. BioMetals 2007, 20, 431–452. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Foxfire, A.; Xu, J.; Baird, S.M.; Jia, J.; Delgado, K.H.; Shin, R.; Smith, L.; Lu, S.E. The siderophore product ornibactin is required for the bactericidal activity of Burkholderia contaminans MS14. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Rojas-Rojas, F.U.; Salazar-Gomez, A.; Vargas-Diaz, M.E.; Vasquez-Murrieta, M.S.; Hirsch, A.M.; De Mot, R.; Ghequire, M.G.K.; Ibarra, J.A.; Estrada-de los Santos, P. Broad-spectrum antimicrobial activity by Burkholderia cenocepacia TAtl-371, a strain isolated from the tomato rhizosphere. Microbiology 2018, 164, 1072–1086. [Google Scholar] [CrossRef]
- Lenz, K.D.; Klosterman, K.E.; Mukundan, H.; Kubicek-Sutherland, J.Z. Macrolides: From toxins to therapeutics. Toxins 2021, 13, 347. [Google Scholar] [CrossRef]
- Song, L.; Jenner, M.; Masschelein, J.; Jones, C.; Bull, M.J.; Harris, S.R.; Harkoorn, R.C.; Vocat, A.; Romero-Canelon, I.; Coupland, P.; et al. Discovery and biosynthesis of gladiolin: A Burkholderia gladioli antibiotic with promising activity against Mycobacterium tuberculosis. J. Am. Chem. Soc. 2017, 139, 7974–7981. [Google Scholar] [CrossRef]
- Riley, M.A. Bacteriocins, biology, ecology, and evolution. In Encyclopedia of Microbiology; Schaechter, M., Ed.; Elsevier: Oxford, UK, 2009; pp. 32–44. [Google Scholar]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: Resistance is futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef]
- Yao, G.W.; Duarte, I.; Le, T.T.; Carmody, L.; LiPuma, J.J.; Young, R.; Gonzalez, C.F. A broad-host-range tailocin from Burkholderia cenocepacia. Appl. Environ. Microbiol. 2017, 83, e03414-16. [Google Scholar] [CrossRef] [Green Version]
- Principe, A.; Fernandez, M.; Torasso, M.; Godino, A.; Fischer, S. Effectiveness of tailocins produced by Pseudomonas fluorescens SF4c in controlling the bacterial-spot disease in tomatoes caused by Xanthomonas vesicatoria. Microbiol. Res. 2018, 212–213, 94–102. [Google Scholar] [CrossRef]
- Ghequire, M.G.K.; De Canck, E.; Wattiau, P.; Van Winge, I.; Loris, R.; Coenye, T.; De Mot, R. Antibacterial activity of a lectin-like Burkholderia cenocepacia protein. MicrobiologyOpen 2013, 2, 566–575. [Google Scholar] [CrossRef]
- Ghequire, M.G.K.; De Mot, R. Distinct colicin M-like bacteriocin-immunity pairs in Burkholderia. Sci. Rep. 2015, 5, 17368. [Google Scholar] [CrossRef]
- Marshall, K.; Shakya, S.; Greenhill, A.R.; Padilla, G.; Baker, A.; Warner, J.M. Antibiosis of Burkholderia ubonensis against Burkholderia pseudomallei, the causative agent for melioidosis. J. Trop. Med. Public Health 2010, 41, 904–912. [Google Scholar]
- Knappe, T.A.; Linne, U.; Zirah, S.; Rebuffat, S.; Xie, X.; Marahiel, M.A. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. Chem. Biol. 2008, 16, 1290–1298. [Google Scholar] [CrossRef]
- Rebufat, S.; Blond, A.; Destoumieux-Garzon, D.; Goulard, C.; Peduzzi, J. Microcin J25, from the macrocyclic to the lasso structure: Implications for biosynthetic, evolutionary, and biotechnological perspectives. Curr. Protein Pept. Sci. 2004, 5, 383–391. [Google Scholar] [CrossRef]
- Knappe, T.A.; Linne, U.; Robbel, L.; Marahiel, M.A. Insights into the biosynthesis and stability of the lasso peptide capistruin. Chem. Biol. 2009, 16, 1290–1298. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Parry, M.E.; Zong, C.; Jaramillo-Cartagena, A.; Darst, S.A.; Connell, N.D.; Rusoo, R.; Link, A.J. Discovery of ubonodin, an antimicrobial lasso peptide active against members of the Burkholderia cepacia complex. ChemBioChem. 2020, 21, 1335–1340. [Google Scholar] [CrossRef]
- Millanao, A.; Mora, A.; Villagra, N.; Bucarey, S.; Hidalgo, A. Biological effects of quinolones: A family of broad-spectrum antimicrobial agents. Molecules 2021, 26, 7153. [Google Scholar] [CrossRef]
- Heeb, S.; Fletcher, M.P.; Chhabra, S.R.; Diggle, S.P.; Williams, P.; Camara, M. Quinolones: From antibiotics to autoinducers. FEMS Microbiol. Rev. 2011, 35, 247–274. [Google Scholar] [CrossRef]
- Wu, Y.; Seyedsayamdost, M.R. Synergy and target promiscuity drive structural divergence in bacterial alkylquinolone biosynthesis. Cell Chem. Biol. 2017, 24, 1437–1444. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hoffmann, J.P.; Chou, C.W.; Honer zu Bentrup, K.; Fuselier, J.A.; Bitoun, J.P.; Wimley, W.C.; Morici, L.A. Burkholderia thailandensis outer membrane vesicles exert antimicrobial activity against drug-resistant and competitor microbial species. J. Microbiol. 2020, 58, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Yoshihisa, H.; Sato, Z.; Hirayama, F.; Konno, K.; Shirahama, H.; Suzui, T. Production of antibiotics by Pseudomonas cepacia as an agent for biological control of soilborne plant pathogens. Soil Biol. Biochem. 1989, 21, 723–728. [Google Scholar] [CrossRef]
- Saalim, M.; Villegas-Moreno, J.; Clark, B.R. Bacterial alkyl-4-quinolones: Discovery, structural diversity and biological properties. Molecules 2020, 25, 5689. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Yamashita, T.; Furihata, K.; Nagai, K.; Suzuki, K.I.; Hayakawa, Y.; Shin-ya, K. Burkholone, a new cytotoxic antibiotic against IGF-I dependent cells from Burkholderia sp. J. Antibiot. 2007, 60, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Baserga, R.; Hongo, A.; Rubini, M.; Prisco, M.; Valentinis, B. The IGF-I receptor in cell growth, transformation and apoptosis. Biochim. Biophys. Acta 1997, 1332, F105–F126. [Google Scholar] [CrossRef]
- Niehs, S.P.; Kumpfmuller, J.; Dose, B.; Little, R.F.; Ishida, K.; Florez, L.V.; Kaltenpoth, M.; Hertweck, C. Insect-associated bacteria assemble the antifungal butanolide gladiofungin by non-canonical polyketide chain termination. Angew. Chem. Int. Ed. Engl. 2020, 59, 23122–23126. [Google Scholar] [CrossRef]
- Nakou, I.T.; Jenner, M.; Dashti, Y.; Romero-Canelón, I.; Masschelein, J.; Mahenthiralingam, E.; Challis, G.L. Genomics-driven discovery of a novel glutarimide antibiotic from Burkholderia gladioli reveals an unusual polyketide synthase chain release mechanism. Angew. Chem. Int. Ed. Engl. 2020, 59, 23145–23153. [Google Scholar] [CrossRef]
- Oka, M.; Yaginuma, K.; Numata, K.; Konishi, M.; Oki, T.; Kawaguchi, H. Glidobactins A, B and C, new antitumor antibiotics. II. Structure elucidation. J. Antibiot. 1988, 41, 1338–1350. [Google Scholar] [CrossRef]
- Oka, M.; Nishiyama, Y.; Ohta, S.; Kamei, H.; Konishi, M.; Miyaki, T.; Oki, T.; Kawaguchi, H. Glidobactins A, B and C, new antitumor antibiotics. I. Production, isolation, chemical properties and biological activity. J. Antibiot. 1988, 41, 1331–1337. [Google Scholar] [CrossRef]
- Shoji, J.; Hinoo, H.; Kato, T.; Hattori, T.; Hirooka, K.; Tawara, K.; Shiratori, O.; Terui, Y. Isolation of cepafungins I, II and III from Pseudomonas species. J. Antibiot. 1990, 43, 783–787. [Google Scholar] [CrossRef] [Green Version]
- Terui, Y.; Nishikawa, J.; Hinoo, H.; Kato, T.; Shoji, J. Structures of cepafungins I, II and III. J. Antibiot. 1990, 43, 788–795. [Google Scholar] [CrossRef]
- Schellenberg, B.; Bigles, L.; Dudler, R. Identification of genes involved in the biosynthesis of the cytotoxic compound glidobactin from a soil bacterium. Environ. Microbiol. 2007, 9, 1640–1650. [Google Scholar] [CrossRef]
- Biggins, J.B.; Kang, H.S.; Ternei, M.A.; DeShazer, D.; Brady, S.F. The chemical arsenal of Burkholderia pseudomallei is essential for pathogenicity. J. Am. Chem. Soc. 2014, 136, 9484–9490. [Google Scholar] [CrossRef]
- Lu, S.E.; Novak, J.; Austin, F.W.; Gu, G.; Ellis, D.; Kirk, M.; Wilson-Stanford, S.; Tonelli, M.; Smith, L. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 2009, 48, 8312–8321. [Google Scholar] [CrossRef]
- Ellis, D.; Gosai, J.; Emrick, C.; Heintz, R.; Romans, L.; Gordon, D.; Lu, S.E.; Austin, F.; Smith, L. Occidiofungin’s chemical stability and in vitro potency against Candida species. Antimicrob. Agents Chemother. 2012, 56, 765–769. [Google Scholar] [CrossRef]
- Ravichandran, A.; Geng, M.; Hull, K.G.; Romo, D.; Lu, S.E.; Albee, A.; Nutter, C.; Gordon, D.M.; Ghannoum, M.A.; Lockless, S.W.; et al. Occidiofungin, and actin binding antifungal with in vivo efficacy in a vulvovaginal candidiasis infection. bioRxiv 2018, 368720. [Google Scholar] [CrossRef]
- Emrick, D.; Ravichandran, A.; Gosai, J.; Lu, S.; Gordon, D.M.; Smith, L. The antifungal occidiofungin triggers an apoptotic mechanism of cell death in yeast. J. Nat. Prod. 2013, 76, 829–838. [Google Scholar] [CrossRef]
- Ma, J.; Guo, F.; Jin, Z.; Geng, M.; Ju, M.; Ravichandran, A.; Orugunty, R.; Smith, L.; Zhu, G.; Zhang, H. Novel antiparasitic activity of the antifungal lead occidiofungin. Antimicrob. Agents Chemother. 2020, 64, e00244-20. [Google Scholar] [CrossRef]
- Wang, X.Q.; Liu, A.X.; Guerrero, A.; Liu, J.; Yu, X.Q.; Deng, P.; Ma, L.; Baird, S.M.; Smith, L.; Lu, S.E. Occidiofungin is an important component responsible for the antifungal activity of Burkholderia pyrrocinia strain Lyc2. J. Appl. Microbiol. 2016, 120, 607–618. [Google Scholar] [CrossRef]
- Hing, S.L.; Ravichandran, A.; Escano, J.; Cooley, J.; Autin, F.; Lu, S.E.; Pruett, S.; Smith, L. Toxicological evaluation of occidiofungin against mice and human cancer cell lines. Sci. Res. 2014, 5, 1085–1093. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.; Suh, J.W.; Kim, S.; Hyun, B.; Kim, C.; Lee, C. Cepacidine A, a novel antifungal antibiotic produced by Pseudomonas cepacia. II. Physico-chemical properties and structure elucidation. J. Antibiot. 1994, 47, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, S.; Hyun, B.; Suh, J.W.; Yon, C.; Kim, C.; Lim, Y.; Kim, C. Cepacidine A, a novel antifungal antibiotic produced by Pseudomonas cepacia. I. Taxonomy, production, isolation and biological activity. J. Antibiot. 1994, 47, 1402–1405. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Suh, J.W.; Cho, Y.H. Immunosuppressive activity of cepacidine A, a novel antifungal antibiotic produced by Pseudomonas cepacia. J. Microbiol. Biotechnol. 1999, 9, 672–674. [Google Scholar]
- Lee, C.H.; Kempf, H.J.; Lim, Y.; Cho, Y.H. Biocontrol activity of Pseudomonas cepacia AF2001 and anthelmintic activity of its novel metabolite, cepacidine A. J. Microbiol. Biotechnol. 2000, 10, 568–571. [Google Scholar]
- Kang, Y.; Carlson, R.; Tharpe, W.; Schell, M.A. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 1998, 64, 3939–3947. [Google Scholar] [CrossRef]
- Dose, B.; Niehs, S.P.; Scherlach, K.; Florez, L.V.; Kaltenpoth, M.; Hertweck, C. Unexpected bacterial origin of the antibiotic icosalide: Two-tailed depsipeptide assembly in multifarious Burkholderia symbionts. ACS Chem. Biol. 2018, 13, 2414–2420. [Google Scholar] [CrossRef]
- Jenner, M.; Jian, X.; Dashti, Y.; Masschelein, J.; Hobson, C.; Roberts, D.; Jones, C.; Harris, S.; Parkhill, J.; Raja, H.A.; et al. An unusual Burkholderia gladioli double chain-initiating nonribosomal peptide synthetase assembles “fungal” icosalide antibiotics. Chem. Sci. 2019, 10, 5489–5494. [Google Scholar] [CrossRef]
- Boros, C.; Smith, C.J.; Vasina, Y.; Che, Y.; Dix, A.B.; Darveaux, B.; Pearce, C. Isolation and identification of the icosalides—Cyclic peptolides with selective antibiotic and cytotoxic activities. J. Antibiot. 2006, 59, 486–494. [Google Scholar] [CrossRef]
- Chandler, J.R.; Truong, T.T.; Silva, P.M.; Seyedsayamdost, M.R.; Carr, G.; Radey, M.; Jacobs, M.A.; Sims, E.H.; Clardy, J.; Greenberg, E.P. Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. mBio 2012, 3, e00499-12. [Google Scholar] [CrossRef]
- Bisacchi, G.S.; Hockstein, D.R.; Koster, W.H.; Parker, W.L.; Rathnum, M.L.; Unger, S.E. Xylocandin: A new complex of antifungal peptides. II. Structural studies and chemical modifications. J. Antibiot. 1987, 40, 1520–1529. [Google Scholar] [CrossRef]
- Meyers, E.; Bissachi, G.S.; Dean, L.; Liu, W.C.; Minassian, B.; Slusarchyk, D.S.; Sykes, R.B.; Tanaka, S.K.; Trejo, W. Xylocandin: A new complex of antifungal peptides. I. Taxonomy, isolation and biological activity. J. Antibiot. 1987, 40, 1515–1519. [Google Scholar] [CrossRef]
- Jenul, C.; Sieber, S.; Daeppen, C.; Mathew, A.; Lardi, M.; Pessi, G.; Hoepfner, D.; Neuburger, M.; Linden, A.; Gademann, K.; et al. Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Biggins, J.B.; Liu, X.; Feng, Z.; Brady, S.F. Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J. Am. Chem. Soc. 2011, 133, 1638–1641. [Google Scholar] [CrossRef]
- Florez, L.V.; Scherlach, K.; Miller, I.J.; Rodrigues, A.; Kwan, J.C.; Hertweck, C.; Kaltenpoth, M. An antifungal polyketide associated with horizontally acquired genes supports symbiont-mediated defense in Lagria villosa beetles. Nat. Commun. 2018, 9, 2478. [Google Scholar] [CrossRef]
- Imada, A.; Kitano, K.; Kintaka, K.; Muroi, M.; Asai, M. Sulfazecin and isosulfazecin, novel β-lactam antibiotics of bacterial origin. Nature 1981, 289, 590–591. [Google Scholar] [CrossRef]
- Loveridge, E.J.; Jones, C.; Bull, M.J.; Moody, S.C.; Kahl, M.W.; Khan, Z.; Neilson, L.; Tomeva, M.; Adams, S.E.; Wood, A.C.; et al. Reclassification of the specialized metabolite producer Pseudomonas mesoacidophila ATCC 31433 as a member of the Burkholderia cepacia complex. J. Bacteriol. 2017, 199, e00125-17. [Google Scholar] [CrossRef]
- Imada, A.; Kintaka, K.; Nakao, M.; Shinagawa, S. Bulgecin, a bacterial metabolite which in concert with β-lactam antibiotics causes bulge formation. J. Antibiot. 1982, 35, 1400–1403. [Google Scholar] [CrossRef]
- Quan, C.S.; Zheng, W.; Liu, Q.; Ohta, Y.; Fan, S.D. Isolation and characterization of a novel Burkholderia cepacia with strong antifungal activity against Rhizoctornia solani. Appl. Environ. Biotechnol. 2006, 72, 1276–1284. [Google Scholar] [CrossRef]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef]
- Ye, L.; Cornelis, P.; Guillemyn, K.; Ballet, S.; Christophersen, C.; Hammerich, O. Structure revision of N-mercapto-4-formylcarbostyril produced by Pseudomonas fluorescens G308 to 2-(2-hydroxyphenyl)thiazole-4-carbaldehyde (aeruginaldehyde). Nat. Prod. Commun. 2014, 9, 789–794. [Google Scholar] [CrossRef]
- Trottmann, F.; Franke, J.; Ishida, K.; Garcia-Altares, M.; Hertweck, C. A pair of bacterial siderophores releases and traps an intercellular signal molecule: An unusual case of natural nitrone bioconjugation. Angew. Chem. Int. Ed. 2019, 58, 200–2004. [Google Scholar] [CrossRef] [PubMed]
- Eustaquio, A.S.; Janso, J.E.; Ratnayake, A.S.; O’Donnell, C.J.; Koehn, F.E. Spliceostatin hemiketal biosynthesis in Burkholderia spp. is catalyzed by an iron/-ketoglutarate-dependent dioxygenase. Proc. Natl. Acad. Sci. USA 2014, 111, E3376–E3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Ratnayake, A.S.; Janso, J.E.; Min, H.; Yang, H.Y.; Loganzo, F.; Shor, B.; O’Donnell, J.C.; Koehn, F.E. Cytotoxic spliceostatins from Burkholderia sp. and their semisynthetic analogues. J. Nat. Prod. 2014, 77, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Prasad, C. Bioactive cyclic dipeptides. Peptides 1995, 16, 151–164. [Google Scholar] [CrossRef]
- Wang, J.H.; Quan, C.S.; Qi, X.H.; Li, X.; Fan, S.D. Determination of diketopiperazines of Burkholderia cepacia CF-66 by gas chromatography–mass spectrometry. Anal. Bioanal. Chem. 2010, 396, 1773–1779. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Chiarelli, L.R.; Makarov, V.; Brackman, G.; Israyilova, A.; Azzalin, A.; Forneris, F.; Riabova, O.; Savina, S.; Coenye, T.; et al. Discovery of new diketopiperazines inhibiting Burkholderia cenocepacia quorum sensing in vitro and in vivo. Sci. Rep. 2016, 6, 32487. [Google Scholar] [CrossRef]
- Rojas-Rojas, F.U.; Sanchez-Lopez, D.; Tapia-Garcia, E.Y.; Arroyo-Herrera, I.; Maymon, M.; Humm, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; et al. Draft genome of Burkholderia cenocepacia TAtl-371, a strain from the Burkholderia cepacia complex retains antagonism in different carbon and nitrogen sources. Curr. Microbiol. 2019, 76, 566–574. [Google Scholar] [CrossRef]
- Jiao, Y.; Yoshihara, T.; Ishikuri, S.; Uchino, H.; Ichihara, A. Structural identification of cepaciamide A, a novel fungitoxic compound from Pseudomonas cepacia D-202. Tetrahedron 1996, 37, 1039–1042. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]
- Swain, D.M.; Yadav, S.K.; Tyagi, I.; Kumar, R.; Kumar, R.; Ghosh, S.; Das, J.; Jha, G. A prophage tail-like protein is deployed by Burkholderia bacteria to feed on fungi. Nat. Commun. 2017, 8, 404. [Google Scholar] [CrossRef]
- Kang, J.G.; Shin, S.Y.; Kim, M.J.; Bajpai, V.; Maheshwari, D.K.; Kang, S.C. Isolation and anti-fungal activities of 2-hydroxymethyl-chroman-4-one produced by Burkholderia sp. MSSP. J. Antibiot. 2004, 57, 726–731. [Google Scholar] [CrossRef]
- Kirinuki, T.; Ichiba, T.; Katamaya, K. General survey of action site of altericidins on metabolism of Alternaria kikuchiana and Ustilago maydis. J. Pestic. Sci. 1984, 9, 601–610. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Mawgoud, A.M.; Lepine, F.; Deziel, E. Rhamnolipids: Diversity of structures, microbial origins, and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef]
- Elshikh, M.; Funston, S.; Chebbi, A.; Ahmed, S.; Marchant, R.; Banat, I.M. Rhamnolipids from non-pathogenic Burkholderia thailandensis E264: Physicochemical characterization, antimicrobial and antibiofilm efficacy against oral hygiene related pathogens. N. Biotechnol. 2017, 36, 26–36. [Google Scholar] [CrossRef]
- Hormann, B.; Muller, M.M.; Syldatk, C.; Hausmann, R. Rhamnolipid production by Burkholderia plantarii DSM 9509T. Eur. J. Lipid Sci. Technol. 2010, 112, 674–680. [Google Scholar] [CrossRef]
- Costa, S.; Deziel, E.; Lepine, F. Characterization of rhamnolipid production by Burkholderia glumae. Lett. Appl. Microbiol. 2011, 53, 620–627. [Google Scholar] [CrossRef]
- Funston, S.J.; Tsaousi, K.; Rudden, M.; Smyth, T.J.; Stevenson, P.S.; Marchant, R.; Banat, I.M. Characterizing rhamnolipid production in Burkholderia thailandensis E264, a non-pathogenic producer. Appl. Microbiol. Biotechnol. 2016, 100, 7945–7956. [Google Scholar] [CrossRef]
- Haubler, S.; Nimtz, M.; Domke, T.; Wray, V.; Steinmetz, I. Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect. Immun. 1998, 66, 1588–1593. [Google Scholar]
- Tawfik, K.A.; Jeffs, P.; Bray, B.; Dubay, G.; Falkinham, J.O.; Mesbah, M.; Youssef, D.; Khalifa, S.; Schmidt, E.W. Burkholdines 1097 and 1229, potent antifungal peptides from Burkholderia ambifaria 2.2N. Org. Lett. 2010, 12, 664–666. [Google Scholar] [CrossRef]
- Lin, Z.; Falkinham, J.O.; Tawfik, K.A.; Jeffs, P.; Bray, B.; Dubay, G.; Cox, J.E.; Schmidt, E.W. Burkholdines from Burkholderia ambifaria: Antifungal agents and possible virulence factors. J. Nat. Prod. 2012, 75, 1518–1523. [Google Scholar] [CrossRef]
- Wang, M.; Tachibana, S.; Murai, Y.; Li, L.; Lau, S.Y.L.; Cao, M.; Zhu, G.; Hashimoto, M.; Hashidoko, Y. Indole-3-acetic acid produced by Burkholderia heleia acts as a phenylacetic acid antagonist to disrupt tropolone biosynthesis in Burkholderia plantarii. Sci. Rep. 2016, 6, 22596. [Google Scholar] [CrossRef] [PubMed]
- Azegami, K.; Nishiyama, K.; Watanabe, Y.; Suzuki, T.; Yoshida, M.; Nose, K.; Toda, S. Tropolone as a root growth-inhibitor produced by a plant pathogenic Pseudomonas sp. causing seedling blight of rice. Jpn. J. Phytopathol. 1985, 51, 315–317. [Google Scholar] [CrossRef]
- Wakimoto, S.; Hirayae, K.; Tsuchiya, K.; Kushima, Y.; Furuya, N.; Matsuyama, N. Production of antibiotics by plant pathogenic pseudomonads. Ann. Phytopathol. Soc. Jpn. 1986, 52, 835–842. [Google Scholar] [CrossRef]
- Abe, M.; Nakazawa, T. Characterization of hemolytic and antifungal substance, cepalycin, from Pseudomonas cepacia. Microbiol. Immunol. 1994, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, E.; Roberts, I.N.; Montecchia, M.S.; Gutierrez-Boem, F.H.; Gomez, F.M.; Ruiz, J.A. A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth. Microbiol. Res. 2018, 206, 50–59. [Google Scholar] [CrossRef]
- Guttenberger, N.; Blankenfeldt, W.; Breinbauer, R. Recent developments in the isolation, biological function, biosynthesis, and synthesis of phenazine natural products. Bioorg. Med. Chem. 2017, 25, 6149–6166. [Google Scholar] [CrossRef]
- Mullins, A.J.; Mahenthiralingam, E. The hidden genomic diversity, specialized metabolic capacity, and revised taxonomy of Burkholderia sensu lato. Front. Microbiol. 2021, 12, 726847. [Google Scholar] [CrossRef]
- Coulon, P.M.L.; Groleau, M.C.; Deziel, E. Potential of the Burkholderia cepacia complex to produce 4-hydroxy-3-methyl-2-alkyquinolines. Front. Cell. Infect. Microbiol. 2019, 9, 33. [Google Scholar] [CrossRef]
- Matthew, A.; Jenul, C.; Carlier, A.L.; Eberl, L. The role of siderophores in metal homeostasis of member of the genus Burkholderia. Environ. Microbiol. Rep. 2016, 8, 103–109. [Google Scholar] [CrossRef]
- Li, X.; Quan, C.S.; Fan, S.D. Antifungal activity of a novel compound from Burkholderia cepacia against plant pathogenic fungi. Lett. Appl. Microbiol. 2007, 45, 508–514. [Google Scholar] [CrossRef]



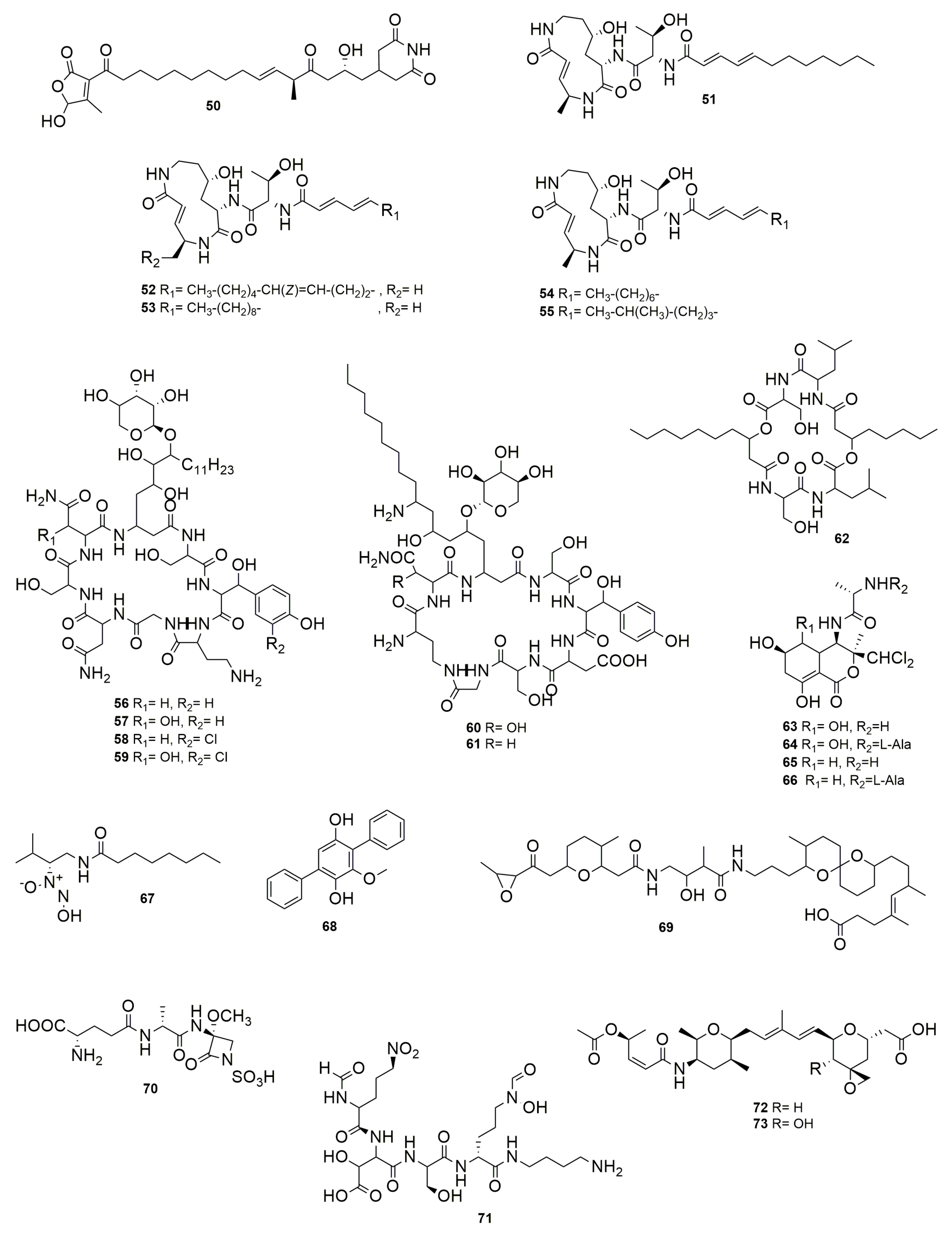

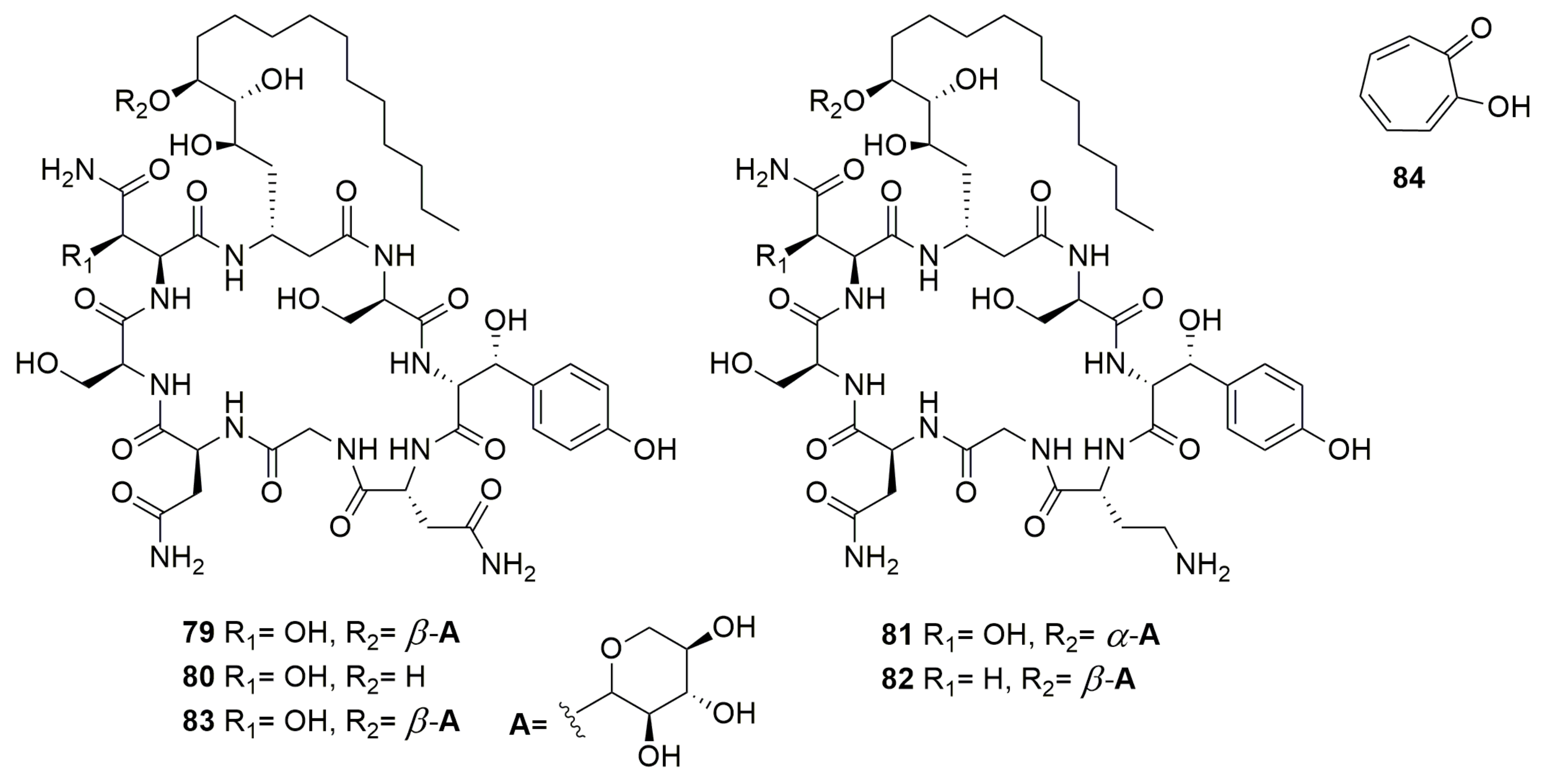
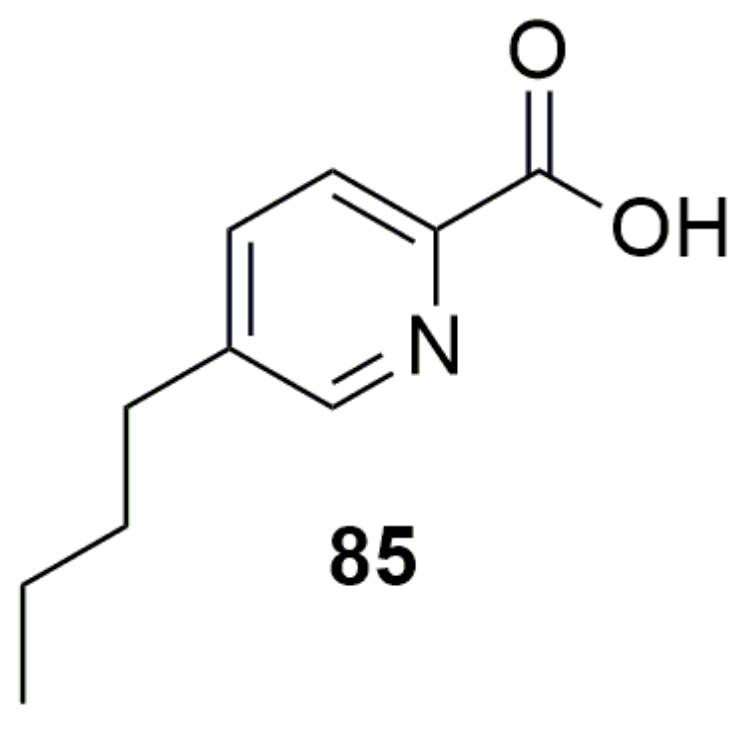
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Cisneros, M.; Morales-Ruíz, L.M.; Salazar-Gómez, A.; Rojas-Rojas, F.U.; Estrada-de los Santos, P. Compilation of the Antimicrobial Compounds Produced by Burkholderia Sensu Stricto. Molecules 2023, 28, 1646. https://doi.org/10.3390/molecules28041646
Rodríguez-Cisneros M, Morales-Ruíz LM, Salazar-Gómez A, Rojas-Rojas FU, Estrada-de los Santos P. Compilation of the Antimicrobial Compounds Produced by Burkholderia Sensu Stricto. Molecules. 2023; 28(4):1646. https://doi.org/10.3390/molecules28041646
Chicago/Turabian StyleRodríguez-Cisneros, Mariana, Leslie Mariana Morales-Ruíz, Anuar Salazar-Gómez, Fernando Uriel Rojas-Rojas, and Paulina Estrada-de los Santos. 2023. "Compilation of the Antimicrobial Compounds Produced by Burkholderia Sensu Stricto" Molecules 28, no. 4: 1646. https://doi.org/10.3390/molecules28041646




