Colorimetric Assaying of Exosomal Metabolic Biomarkers
Abstract
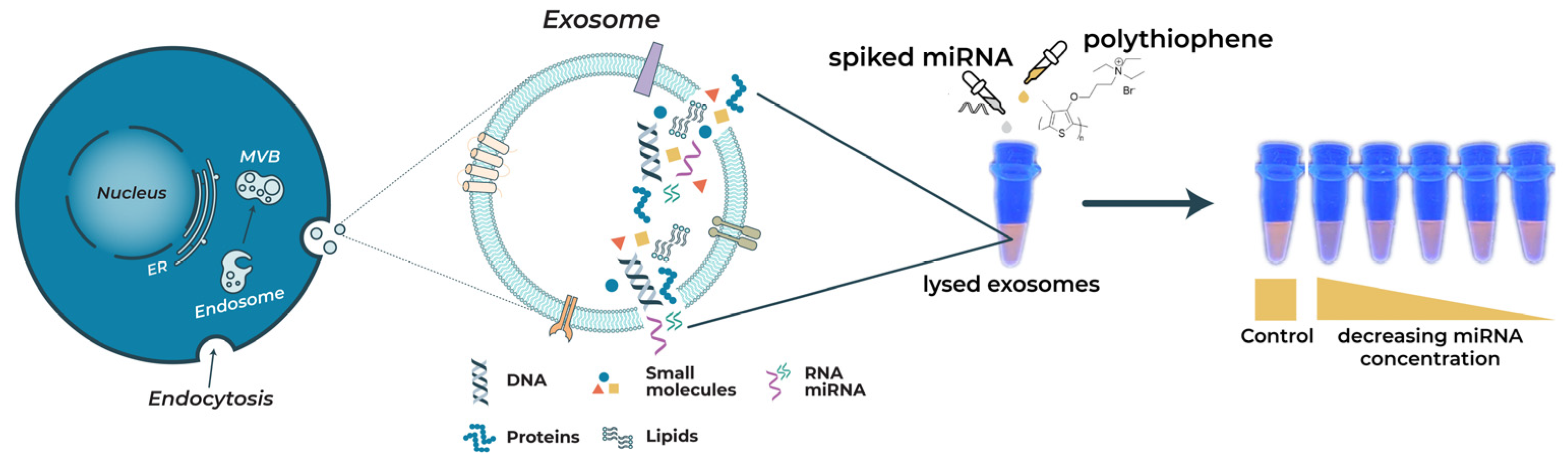
1. Introduction
2. Results and Discussion
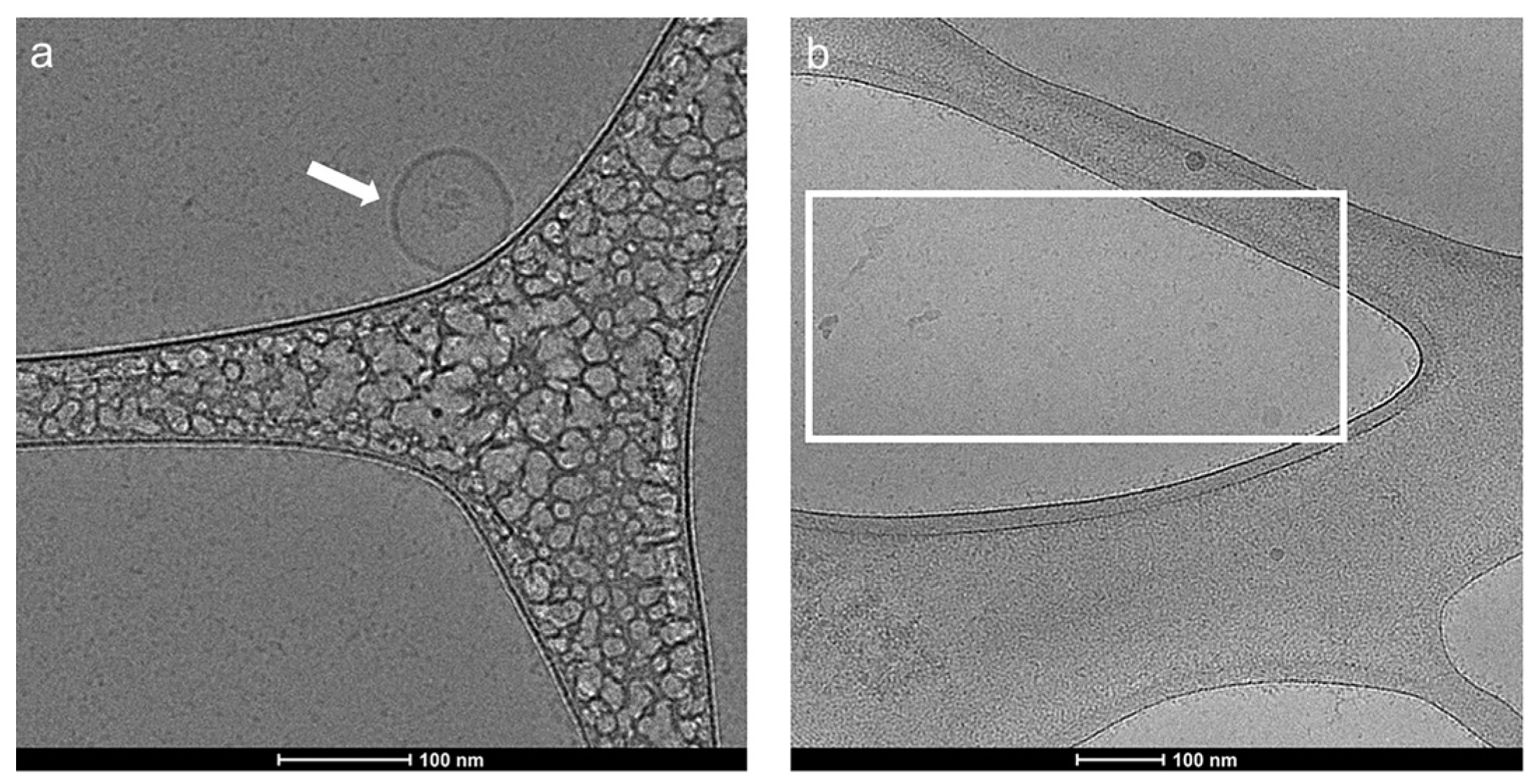
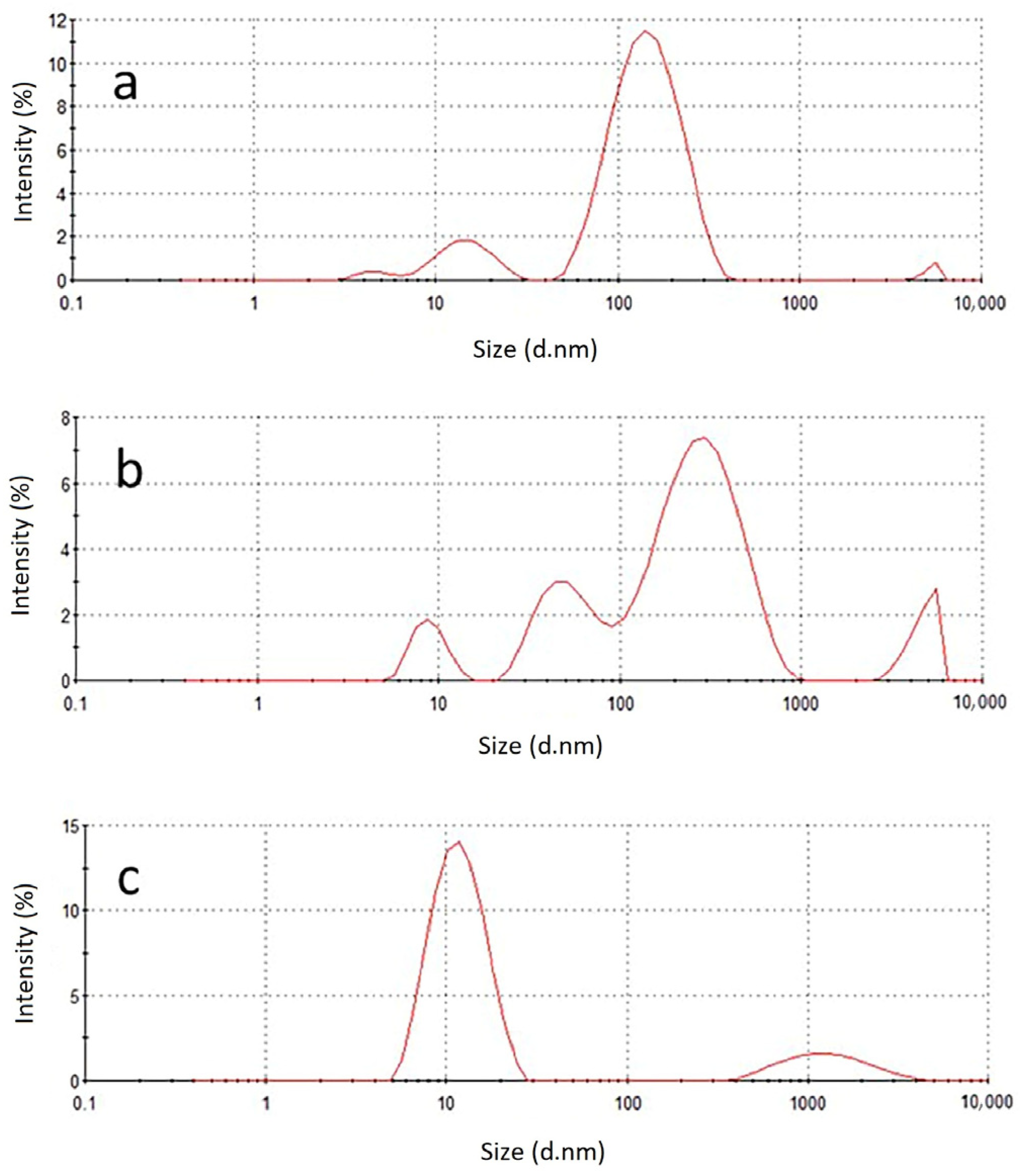
2.1. Characterization of Exosomes before and after Lysis with 0.1% Triton X-100
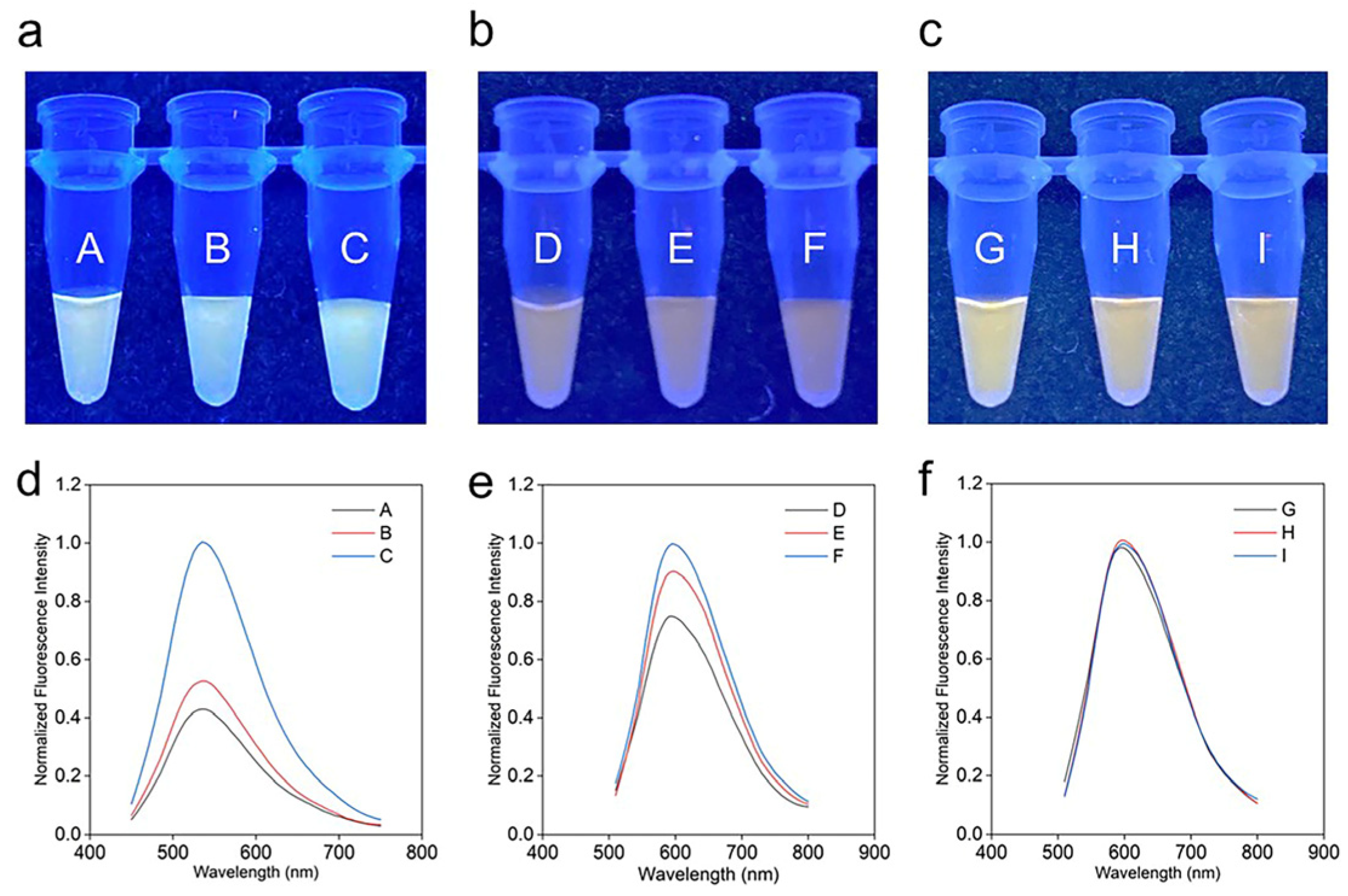
2.2. The Influence of Tween 20 (T20)
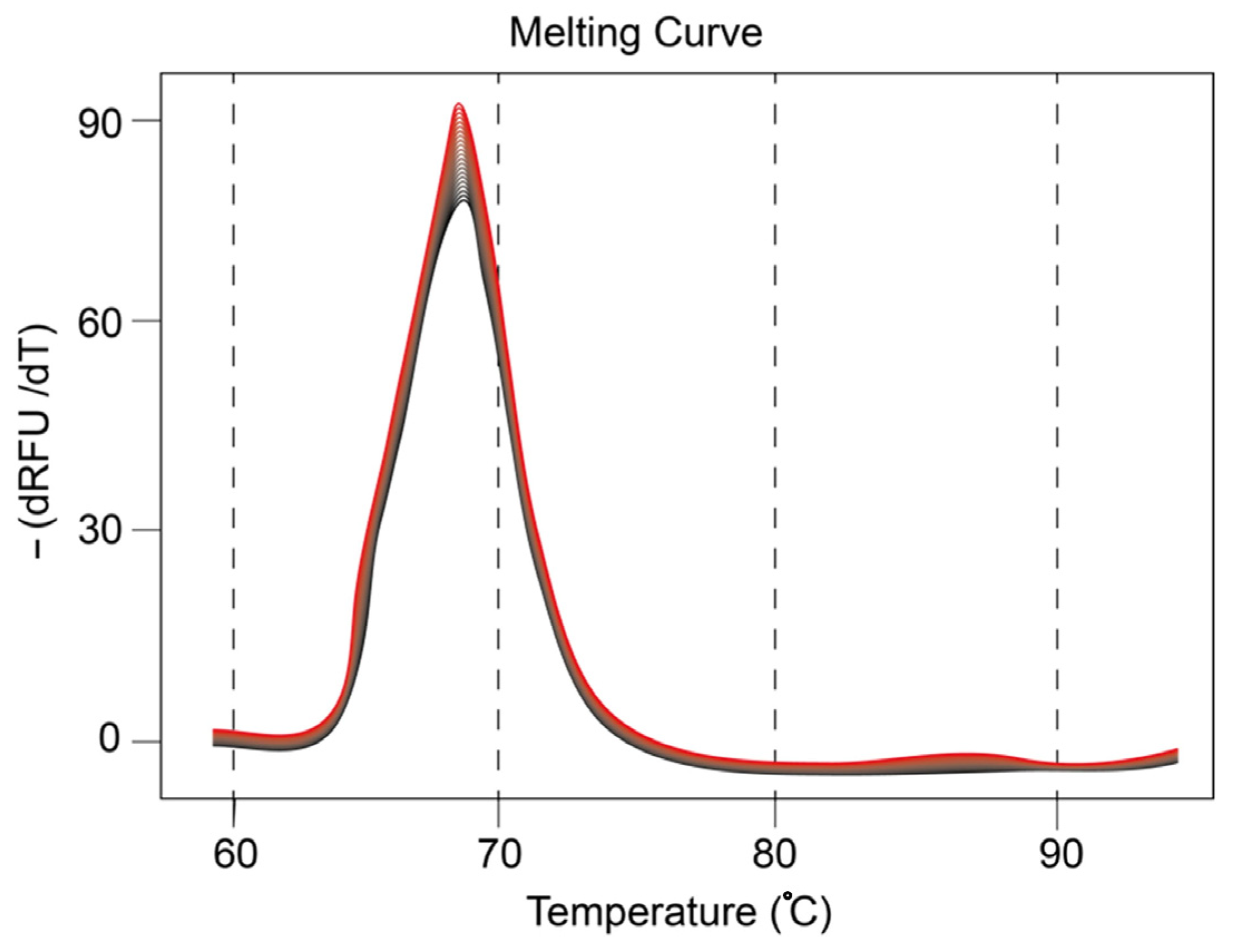
2.3. Presence of mir21 Exosomes Extracted from Cancerous Cells
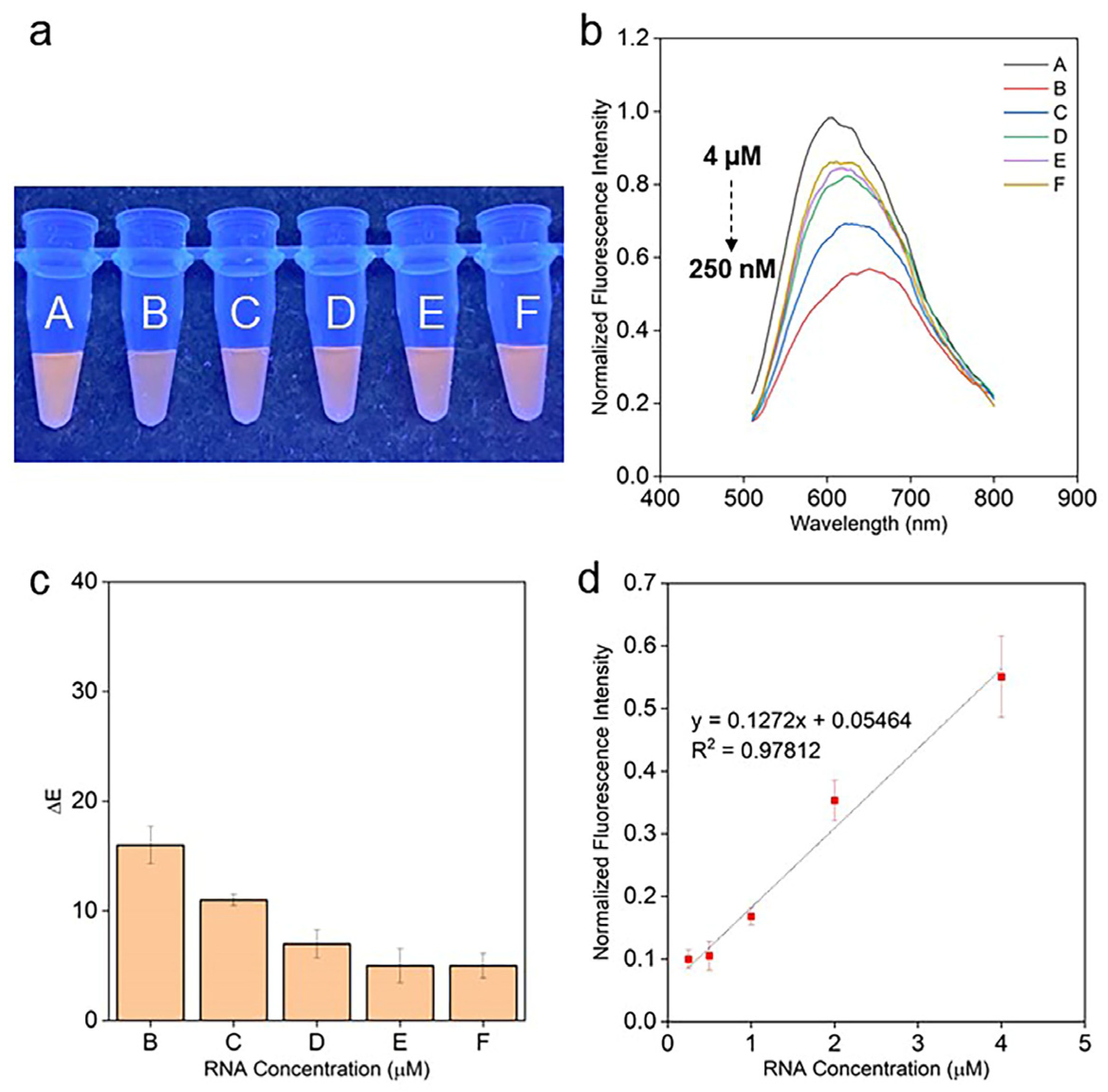
2.4. mir21 Assay
2.5. Comparative Analysis of LODs and Scope for Improvement
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Extraction and Purification of Exosomes from Human Plasma and Lysis of the Exosomes
3.3. Presence of mir21 Exosomes Extracted from Cancerous Cells
3.4. Synthesis of PT
3.5. Assay Preparation
3.6. RGB Analysis of the Colorimetric Assays
3.7. Fluorescence Spectra Analysis of the Assays
3.8. Cryogenic Electron Microscopy
3.9. Dynamic Light Scattering
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteomics 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Vidal, M.; Sainte-Marie, J.; Philippot, J.R.; Bienvenue, A. Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: Evidence precluding a role for “aminophospholipid translocase”. J. Cell. Physiol. 1989, 140, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Waldenström, A.; Gennebäck, N.; Hellman, U.; Ronquist, G. Cardiomyocyte Microvesicles Contain DNA/RNA and Convey Biological Messages to Target Cells. PLoS ONE 2012, 7, e34653. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Gopal, S.K.; Xu, R.; Simpson, R.J.; Chen, W. Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 2015, 40, 72–81. [Google Scholar] [CrossRef]
- Gangoda, L.; Boukouris, S.; Liem, M.; Kalra, H.; Mathivanan, S. Extracellular vesicles including exosomes are mediators of signal transduction: Are they protective or pathogenic? Proteomics 2015, 15, 260–271. [Google Scholar] [CrossRef]
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-Like Vesicles with Dipeptidyl Peptidase IV in Human Saliva. Biol. Pharm. Bull. 2008, 31, 1059–1062. [Google Scholar] [CrossRef]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.D.; Abdel-Bakky, M.S.; Gutwein, P.; Altevogt, P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007, 72, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef]
- Belting, M.; Christianson, H.C. Role of exosomes and microvesicles in hypoxia-associated tumour development and cardiovascular disease. J. Intern. Med. 2015, 278, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Bai, X.; Zhang, A.; Huang, J.; Xu, S.; Zhang, J. Role of Exosomes in Central Nervous System Diseases. Front. Mol. Neurosci. 2019, 12, 240. [Google Scholar] [CrossRef]
- Rastogi, S.; Sharma, V.; Bharti, P.S.; Rani, K.; Modi, G.P.; Nikolajeff, F.; Kumar, S. The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis. Int. J. Mol. Sci. 2021, 22, 440. [Google Scholar] [CrossRef]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar]
- Shao, B.; Xiao, Z. Recent achievements in exosomal biomarkers detection by nanomaterials-based optical biosensors—A review. Anal. Chim. Acta 2020, 1114, 74–84. [Google Scholar] [CrossRef]
- Alzhrani, G.N.; Alanazi, S.T.; Alsharif, S.Y.; Albalawi, A.M.; Alsharif, A.A.; Abdel-Maksoud, M.S.; Elsherbiny, N. Exosomes: Isolation, characterization, and biomedical applications. Cell Biol. Int. 2021, 45, 1807–1831. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Araujo-Filho, I.; Piffoux, M.; Nicolas-Boluda, A.; Grangier, A.; Boucenna, I.; Real, C.C.; Marques, F.L.N.; de Paula Faria, D.; do Rego, A.C.M.; et al. Local administration of stem cell-derived extracellular vesicles in a thermoresponsive hydrogel promotes a pro-healing effect in a rat model of colo-cutaneous post-surgical fistula. Nanoscale 2021, 13, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Dinh, P.C.; Paudel, D.; Brochu, H.; Popowski, K.D.; Gracieux, M.C.; Cores, J.; Huang, K.; Hensley, M.T.; Harrell, E.; Vandergriff, A.C.; et al. Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis. Nat. Commun. 2020, 11, 1064. [Google Scholar] [CrossRef]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; de Vries, M.; van Lent, P.L.; Koenders, M.I.; van den Berg, W.B.; van der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef]
- Parisse, P.; Rago, I.; Ulloa Severino, L.; Perissinotto, F.; Ambrosetti, E.; Paoletti, P.; Ricci, M.; Beltrami, A.P.; Cesselli, D.; Casalis, L. Atomic force microscopy analysis of extracellular vesicles. Eur. Biophys. J. 2017, 46, 813–820. [Google Scholar] [CrossRef]
- Qu, M.; Lin, Q.; Huang, L.; Fu, Y.; Wang, L.; He, S.; Fu, Y.; Yang, S.; Zhang, Z.; Zhang, L.; et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J. Control. Release 2018, 287, 156–166. [Google Scholar] [CrossRef]
- Zadka, L.; Buzalewicz, I.; Ulatowska-Jarza, A.; Rusak, A.; Kochel, M.; Ceremuga, I.; Dziegiel, P. Label-Free Quantitative Phase Imaging Reveals Spatial Heterogeneity of Extracellular Vesicles in Select Colon Disorders. Am. J. Pathol. 2021, 191, 2147–2171. [Google Scholar] [CrossRef]
- Wang, J.; Kao, Y.C.; Zhou, Q.; Wuethrich, A.; Stark, M.S.; Schaider, H.; Soyer, H.P.; Lin, L.L.; Trau, M. An Integrated Microfluidic-SERS Platform Enables Sensitive Phenotyping of Serum Extracellular Vesicles in Early Stage Melanomas. Adv. Funct. Mater. 2021, 32, 2010296. [Google Scholar] [CrossRef]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, 6232. [Google Scholar] [CrossRef]
- Roberg-Larsen, H.; Lund, K.; Seterdal, K.E.; Solheim, S.; Vehus, T.; Solberg, N.; Krauss, S.; Lundanes, E.; Wilson, S.R. Mass spectrometric detection of 27-hydroxycholesterol in breast cancer exosomes. J. Steroid Biochem. Mol. Biol. 2017, 169, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lin, Y.; Xiong, X.; Li, K.; Yao, Z.; Dong, H.; Jiang, Z.; Yu, D.; Yeung, S.J.; Zhang, H. Detection of Exosomal PD-L1 RNA in Saliva of Patients With Periodontitis. Front. Genet. 2019, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Fu, K.; Sun, H.; Rong, D.; Wang, H.; Cao, H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer 2018, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, M.; Ferreira Carlos, F.; Pedrosa, P.; Lopez, A.; Baptista, P.V. Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics 2016, 6, 43. [Google Scholar] [CrossRef]
- Denmark, D.J.; Bustos-Perez, X.; Swain, A.; Phan, M.-H.; Mohapatra, S.; Mohapatra, S.S. Readiness of Magnetic Nanobiosensors for Point-of-Care Commercialization. J. Electron. Mater. 2019, 48, 4749–4761. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Rocha-Santos, T.A.P.; Duarte, A.C.; Rocha-Santos, T.A.P. Advances in point-of-care technologies with biosensors based on carbon nanotubes. TrAC Trends Anal. Chem. 2013, 45, 24–36. [Google Scholar] [CrossRef]
- Dhinakaran, V.; Vigneswari, K.; Lavanya, M.; Varsha Shree, M. Chapter Ten-Point-of-care applications with graphene in human life. In Comprehensive Analytical Chemistry; Hussain, C.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 91, pp. 235–262. [Google Scholar]
- Abelha, T.F.; Dreiss, C.A.; Green, M.A.; Dailey, L.A. Conjugated polymers as nanoparticle probes for fluorescence and photoacoustic imaging. J. Mater. Chem. B 2020, 8, 592–606. [Google Scholar] [CrossRef]
- Yeasmin, S.; Ammanath, G.; Ali, Y.; Boehm, B.O.; Yildiz, U.H.; Palaniappan, A.; Liedberg, B. Colorimetric Urinalysis for On-Site Detection of Metabolic Biomarkers. ACS Appl. Mater. Interfaces 2020, 12, 31270–31281. [Google Scholar] [CrossRef]
- Ammanath, G.; Yeasmin, S.; Srinivasulu, Y.; Vats, M.; Cheema, J.A.; Nabilah, F.; Srivastava, R.; Yildiz, U.H.; Alagappan, P.; Liedberg, B. Flow-through colorimetric assay for detection of nucleic acids in plasma. Anal. Chim. Acta 2019, 1066, 102–111. [Google Scholar] [CrossRef]
- Yuana, Y.; Koning, R.I.; Kuil, M.E.; Rensen, P.C.; Koster, A.J.; Bertina, R.M.; Osanto, S. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J Extracell Vesicles 2013, 2, 21494. [Google Scholar] [CrossRef] [PubMed]
- Emelyanov, A.; Shtam, T.; Kamyshinsky, R.; Garaeva, L.; Verlov, N.; Miliukhina, I.; Kudrevatykh, A.; Gavrilov, G.; Zabrodskaya, Y.; Pchelina, S.; et al. Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid. PLoS ONE 2020, 15, e0227949. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.M.; Roberts, L.M.; Vidavsky, N.; Chiou, A.E.; Fischbach, C.; Paszek, M.J.; Estroff, L.A.; Kourkoutis, L.F. Direct comparison of optical and electron microscopy methods for structural characterization of extracellular vesicles. J. Struct. Biol. 2020, 210, 107474. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J. Cancer Res. Clin. Oncol. 2012, 138, 1659–1666. [Google Scholar] [CrossRef]
- Hill, B.R.T.; Vorhagen, F.W. Comparative Analysis of the Quantization of Color Spaces on the Basis of the CIELAB Color-Difference Formula. ACM Trans. Graph. 1997, 16, 109–154. [Google Scholar] [CrossRef]
- Baek, R.; Sondergaard, E.K.; Varming, K.; Jorgensen, M.M. The impact of various preanalytical treatments on the phenotype of small extracellular vesicles in blood analyzed by protein microarray. J. Immunol. Methods 2016, 438, 11–20. [Google Scholar] [CrossRef]
- Jayachandran, M.; Miller, V.M.; Heit, J.A.; Owen, W.G. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. J. Immunol. Methods 2012, 375, 207–214. [Google Scholar] [CrossRef]
- Onodi, Z.; Pelyhe, C.; Terezia Nagy, C.; Brenner, G.B.; Almasi, L.; Kittel, A.; Mancek-Keber, M.; Ferdinandy, P.; Buzas, E.I.; Giricz, Z. Isolation of High-Purity Extracellular Vesicles by the Combination of Iodixanol Density Gradient Ultracentrifugation and Bind-Elute Chromatography From Blood Plasma. Front. Physiol. 2018, 9, 1479. [Google Scholar] [CrossRef]
- Goyal, G.; Ammanath, G.; Palaniappan, A.; Liedberg, B. Stoichiometric Tuning of PNA Probes to Au(0.8)Ag(0.2) Alloy Nanoparticles for Visual Detection of Nucleic Acids in Plasma. ACS Sens. 2020, 5, 2476–2485. [Google Scholar] [CrossRef]
- Gómez-Polo, C.; Muñoz, M.P.; Lorenzo Luengo, M.C.; Vicente, P.; Galindo, P.; Martín Casado, A.M. Comparison of the CIELab and CIEDE2000 color difference formulas. J. Prosthet. Dent. 2016, 115, 65–70. [Google Scholar] [CrossRef]
- ColorMine.org. Available online: http://colormine.org/delta-e-calculator/cie2000 (accessed on 9 February 2023).
- Ho, J.C.S.; Steininger, C.; Hiew, S.H.; Kim, M.C.; Reimhult, E.; Miserez, A.; Cho, N.; Parikh, A.N.; Liedberg, B. Minimal Reconstitution of Membranous Web Induced by a Vesicle-Peptide Sol-Gel Transition. Biomacromolecules 2019, 20, 1709–1718. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, E.; Goyal, G.; Yildiz, U.H.; Boehm, B.O.; Palaniappan, A. Colorimetric Assaying of Exosomal Metabolic Biomarkers. Molecules 2023, 28, 1909. https://doi.org/10.3390/molecules28041909
Yan E, Goyal G, Yildiz UH, Boehm BO, Palaniappan A. Colorimetric Assaying of Exosomal Metabolic Biomarkers. Molecules. 2023; 28(4):1909. https://doi.org/10.3390/molecules28041909
Chicago/Turabian StyleYan, Evelias, Garima Goyal, Umit Hakan Yildiz, Bernhard O. Boehm, and Alagappan Palaniappan. 2023. "Colorimetric Assaying of Exosomal Metabolic Biomarkers" Molecules 28, no. 4: 1909. https://doi.org/10.3390/molecules28041909
APA StyleYan, E., Goyal, G., Yildiz, U. H., Boehm, B. O., & Palaniappan, A. (2023). Colorimetric Assaying of Exosomal Metabolic Biomarkers. Molecules, 28(4), 1909. https://doi.org/10.3390/molecules28041909





