Insights into the Formation of Intermolecular Complexes of Fluorescent Probe 10-N-Nonyl Acridine Orange with Cardiolipin and Phosphatidylglycerol in Bacterial Plasma Membrane by Molecular Modeling
Abstract
:1. Introduction
2. Results
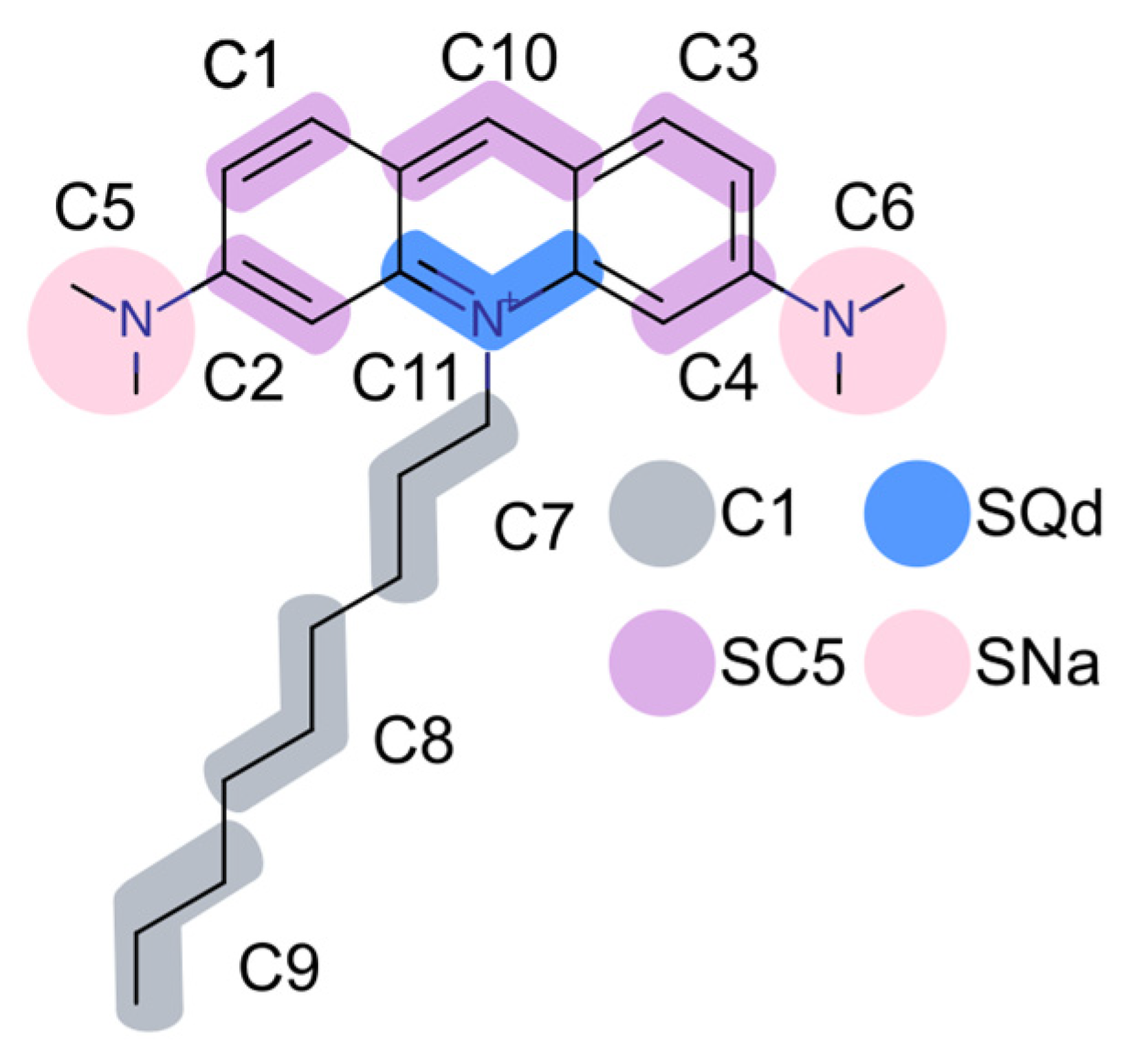
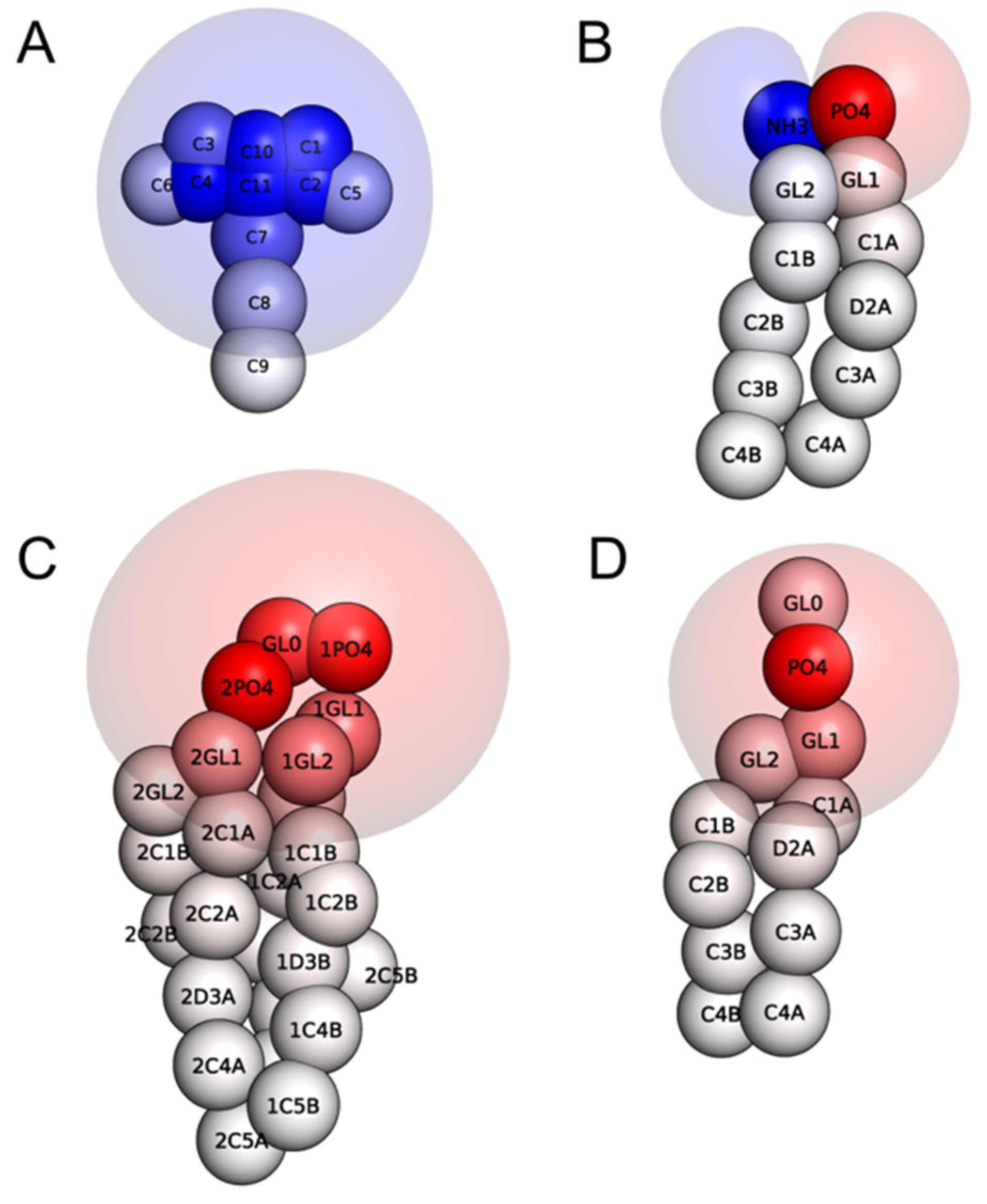
2.1. Electrostatic Charactersitcs of Coarse-Grained Model Components
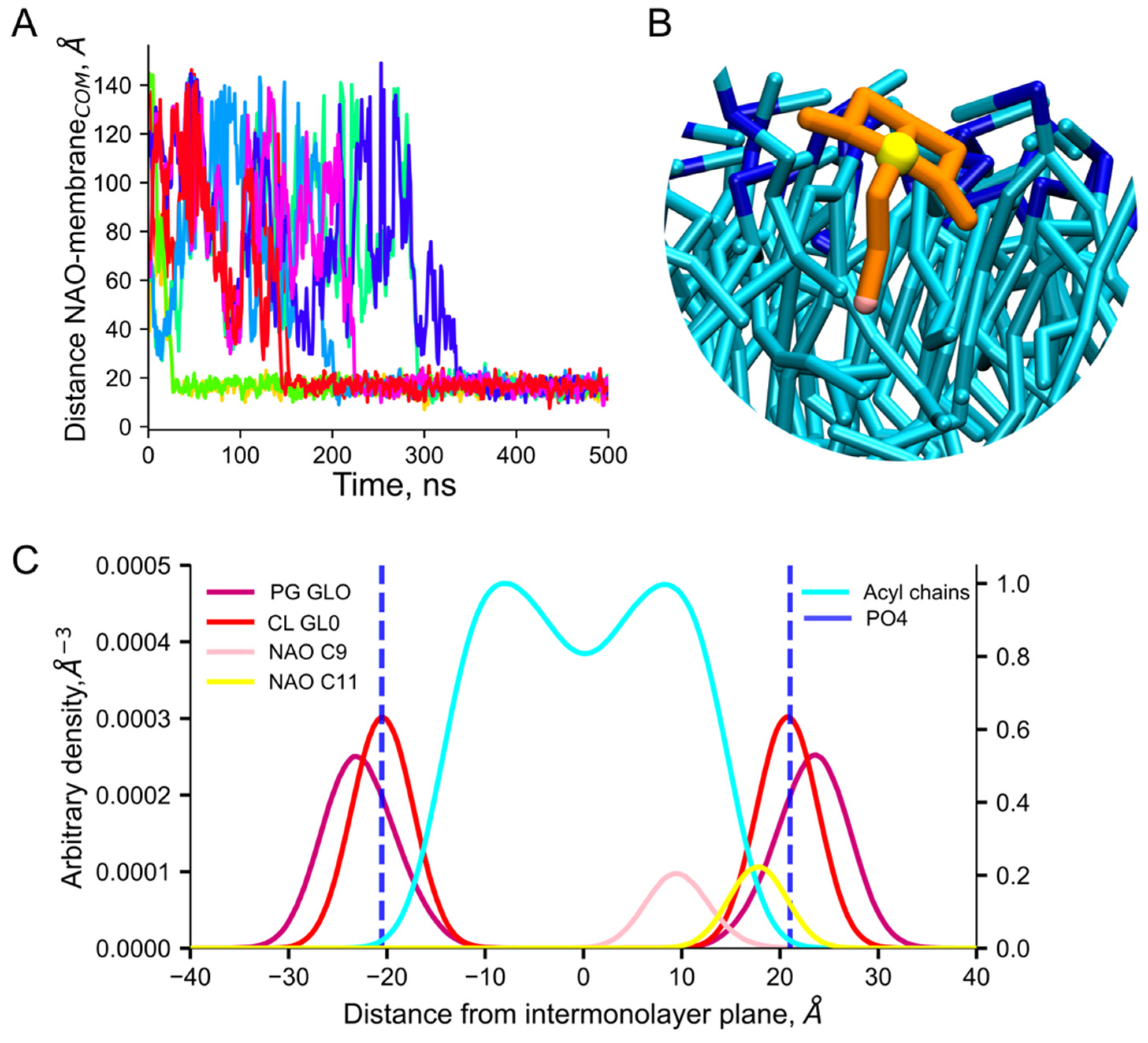
2.2. Insertion of NAO Molecules into the Model Bilayer
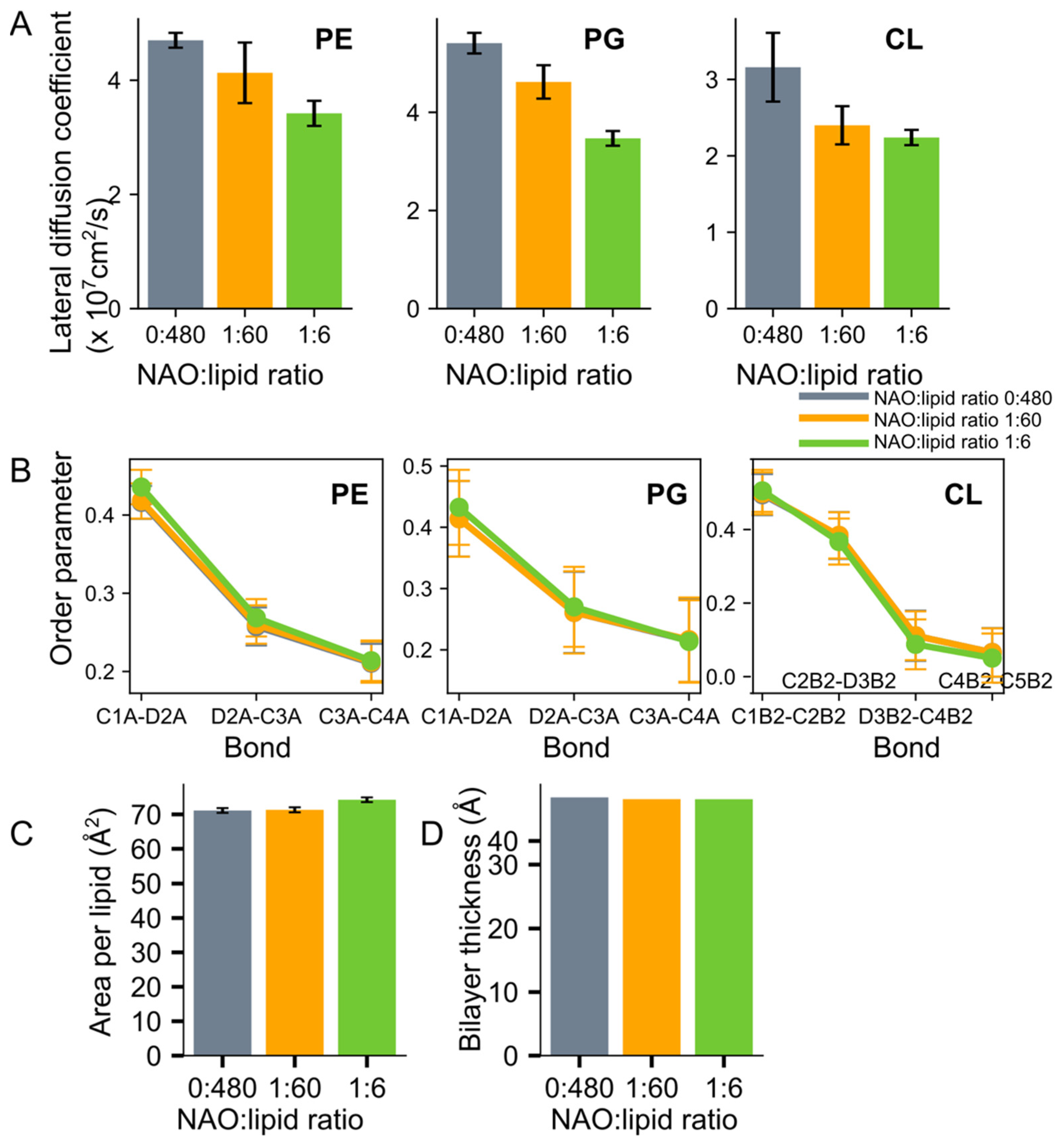
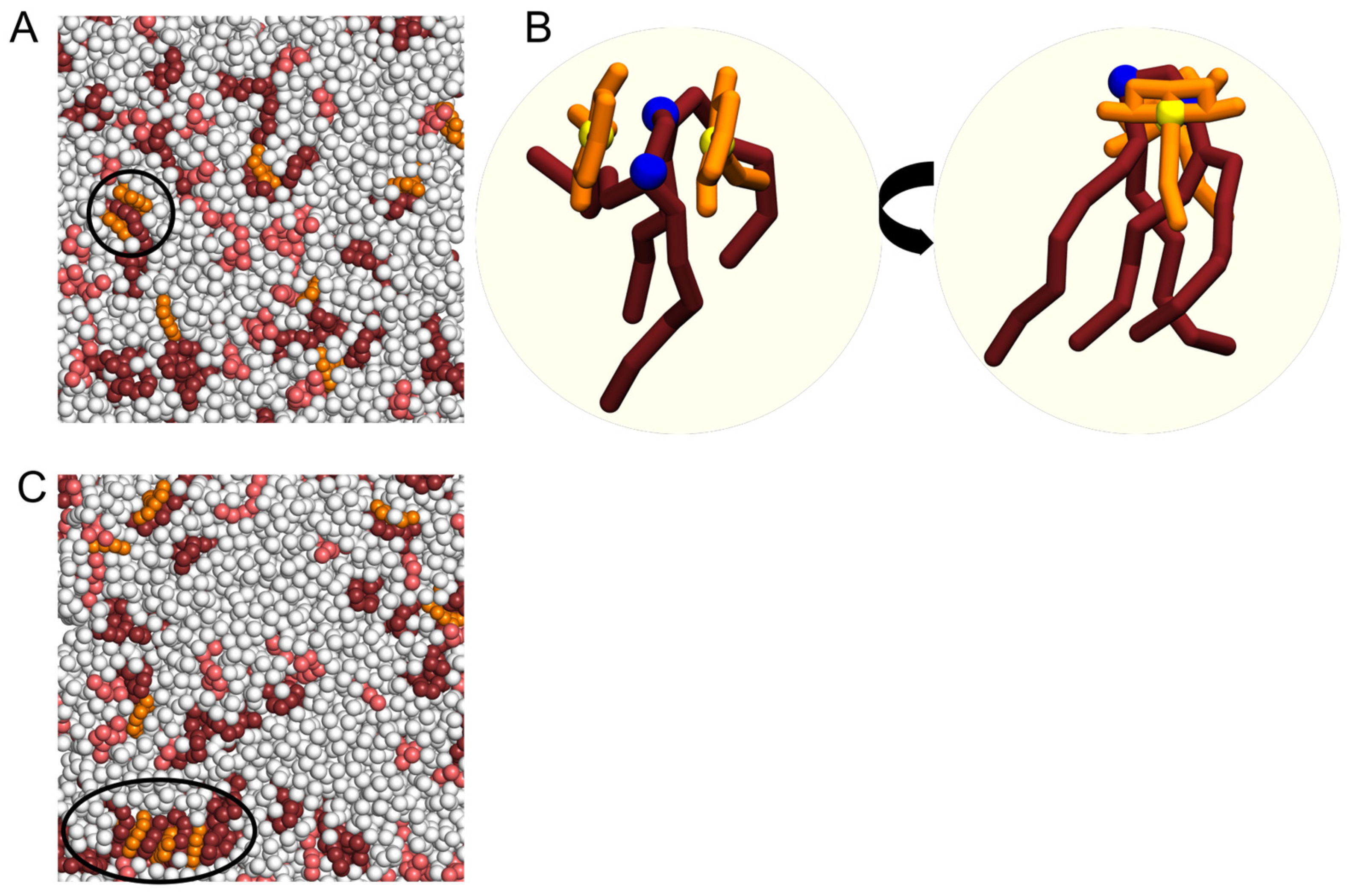
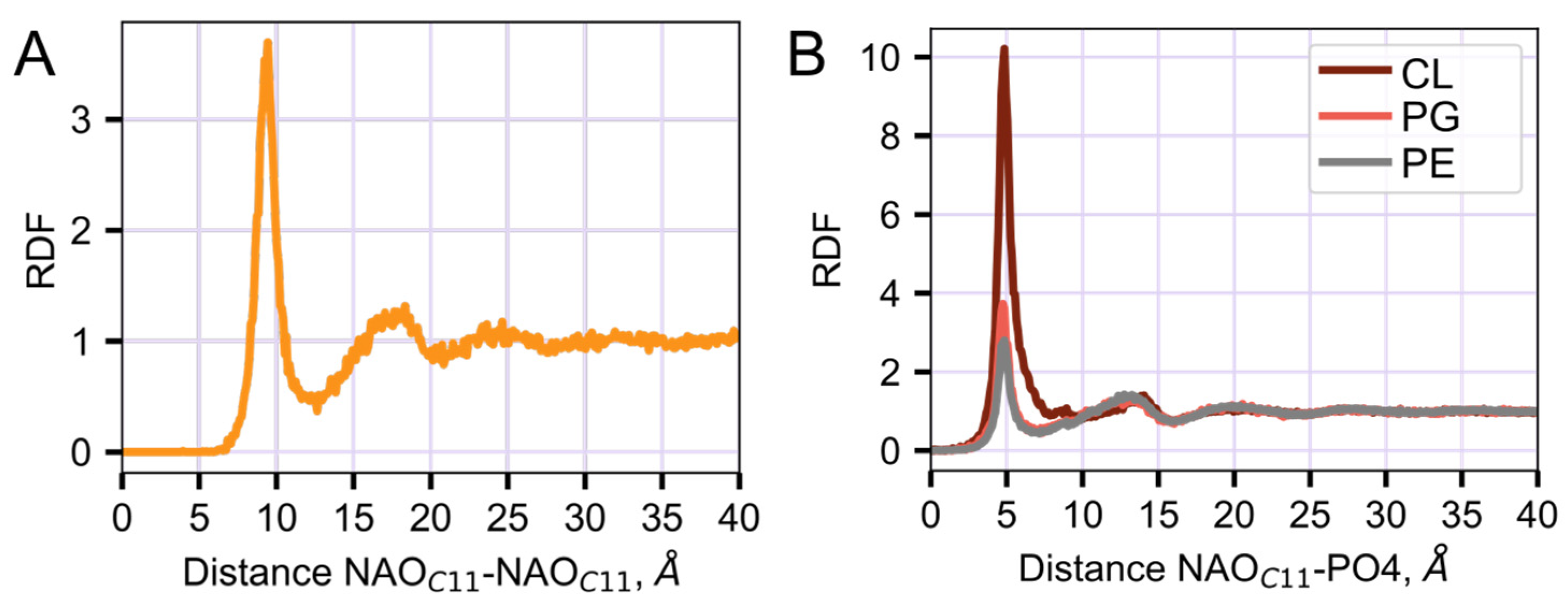
2.3. Interaction of NAO with Lipids Lead to the Formation of Complexes with Different Composition
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structuRes. and pathways. FEMS MicroBiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef]
- Strahl, H.; Errington, J. Bacterial Membranes: Structure, Domains, and Function. Annu. Rev. Microbiol. 2017, 71, 519–538. [Google Scholar] [CrossRef] [PubMed]
- López-Lara, I.M.; Geiger, O. Bacterial lipid diversity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017, 1862, 1287–1299. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Weibel, D.B. Organization and function of anionic phospholipids in bacteria. Appl. MicroBiol. Biotechnol. 2016, 100, 4255–4267. [Google Scholar] [CrossRef] [PubMed]
- Cronan, J.E. Bacterial membrane lipids: Where do we stand? Annu. Rev. Microbiol. 2003, 57, 203–224. [Google Scholar] [CrossRef]
- Dezanet, C.; Kempf, J.; Mingeot-Leclercq, M.-P.; Décout, J.-L. Amphiphilic Aminoglycosides as Medicinal Agents. Int. J. Mol. Sci. 2020, 21 Pt 19, 7411. [Google Scholar] [CrossRef]
- Tan, B.K.; Bogdanov, M.; Zhao, J.; Dowhan, W.; Raetz, C.R.H.; Guan, Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Nat. Acad. Sci. USA 2012, 109, 16504–16509. [Google Scholar] [CrossRef] [Green Version]
- Romantsov, T.; Gonzalez, K.; Sahtout, N.; Culham, D.E.; Coumoundouros, C.; Garner, J.; Kerr, C.H.; Chang, L.; Turner, R.J.; Wood, J.M. Cardiolipin synthase A colocalizes with cardiolipin and osmosensing transporter ProP at the poles of Escherichia coli cells. Mol. Microbiol. 2018, 107, 623–638. [Google Scholar] [CrossRef] [Green Version]
- Romantsov, T.; Stalker, L.; Culham, D.E.; Wood, J.M. Cardiolipin controls the osmotic stress response and the subcellular location of transporter ProP in Escherichia coli. J. Biol. Chem. 2008, 283, 12314–12323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romantsov, T.; Battle, A.R.; Hendel, J.L.; Martinac, B.; Wood, J.M. Protein localization in Escherichia coli cells: Comparison of the cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J. Bacteriol. 2010, 192, 912–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Muraih, J.K.; Tishbi, N.; Herskowitz, J.; Victor, R.L.; Silverman, J.; Uwumarenogie, S.; Taylor, S.D.; Palmer, M.; Mintzer, E. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J. Biol. Chem. 2014, 289, 11584–11591. [Google Scholar] [CrossRef] [Green Version]
- Verhaegh, R.; Becker, K.A.; Edwards, M.J.; Gulbins, E. Sphingosine kills bacteria by binding to cardiolipin. J. Biol. Chem. 2020, 295, 7686–7696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, K.; Kusaka, J.; Nishibori, A.; Hara, H. Lipid domains in bacterial membranes. Mol. Microbiol. 2006, 61, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Vanounou, S.; Parola, A.H.; Fishov, I. Phosphatidylethanolamine and phosphatidylglycerol are segregated into different domains in bacterial membrane. A study with pyrene-labelled phospholipids. Mol. Microbiol. 2003, 49, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Mileykovskaya, E.; Dowhan, W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000, 182, 1172–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mileykovskaya, E.; Dowhan, W. Role of membrane lipids in bacterial division-site selection. Curr. Opin. Microbiol. 2005, 8, 135–142. [Google Scholar] [CrossRef]
- Renner, L.D.; Weibel, D.B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Nat. Acad. Sci. USA 2011, 108, 6264–6269. [Google Scholar] [CrossRef] [Green Version]
- Elías-Wolff, F.; Lindén, M.; Lyubartsev, A.P.; Brandt, E.G. Curvature sensing by cardiolipin in simulated buckled membranes. Soft Matter. 2019, 15, 792–802. [Google Scholar] [CrossRef] [Green Version]
- Norris, V.; Woldringh, C.; Mileykovskaya, E. A hypothesis to explain division site selection in Escherichia coli by combining nucleoid occlusion and Min. FEBS Lett. 2004, 561, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakihara, T.; Takiguchi, N.; Uzawa, H.; Serizawa, R.; Kobayashi, T. Erylysin A inhibits cytokinesis in Escherichia coli by binding with cardiolipin. J. Biochem. 2021, 170, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhang, Z.; Tang, X.; Huang, S.; Li, H.; Peng, B.; Dong, C. Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci. Rep. 2016, 6, 30815. [Google Scholar] [CrossRef] [Green Version]
- Kaewsuya, P.; Danielson, N.D.; Ekhterae, D. Fluorescent determination of cardiolipin using 10-N-nonyl acridine orange. Anal. BioAnal. Chem. 2007, 387, 2775–2782. [Google Scholar] [CrossRef] [Green Version]
- Oliver, P.M.; Crooks, J.A.; Leidl, M.; Yoon, E.J.; Saghatelian, A.; Weibel, D.B. Localization of anionic phospholipids in Escherichia coli cells. J. Bacteriol. 2014, 196, 3386–3398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mileykovskaya, E.; Dowhan, W.; Birke, R.L.; Zheng, D.; Lutterodt, L.; Haines, T.H. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 2001, 507, 187–190. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.M. Perspective: Challenges and opportunities for the study of cardiolipin, a key player in bacterial cell structure and function. Curr. Genet. 2018, 64, 795–798. [Google Scholar] [CrossRef]
- Leung, C.W.T.; Hong, Y.; Hanske, J.; Zhao, E.; Chen, S.; Pletneva, E.V.; Tang, B.Z. Superior fluorescent probe for detection of cardiolipin. Anal. Chem. 2014, 86, 1263–1268. [Google Scholar] [CrossRef] [Green Version]
- Pyrshev, K.; Yesylevskyy, S.; Bogdanov, M. TTAPE-Me dye is not selective to cardiolipin and binds to common anionic phospholipids nonspecifically. Biophys. J. 2021, 120, 3776–3786. [Google Scholar] [CrossRef]
- Rigler, R. Fluorescence and single molecule analysis in cell biology. Biochem. Biophys. Res. Commun. 2010, 396, 170–175. [Google Scholar] [CrossRef]
- Oliveira, E.; Bértolo, E.; Núñez, C.; Pilla, V.; Santos, H.M.; Fernández-Lodeiro, J.; Fernández-Lodeiro, A.; Djafari, J.; Capelo, J.L.; Lodeiro, C. Green and Red Fluorescent Dyes for Translational Applications in Imaging and Sensing Analytes: A Dual-Color Flag. ChemistryOpen 2018, 7, 9–52. [Google Scholar] [CrossRef]
- Amado, A.M.; Ramos, A.P.; Silva, E.R.; Borissevitch, I.E. Quenching of acridine orange fluorescence by salts in aqueous solutions: Effects of aggregation and charge transfer. J. Lumin. 2016, 178, 288–294. [Google Scholar] [CrossRef]
- Shenderovich, I.G. The Partner Does Matter: The Structure of Heteroaggregates of Acridine Orange in Water. Molecules 2019, 24 Pt 15, 2816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaewsuya, P.; Miller, J.D.; Danielson, N.D.; Sanjeevi, J.; James, P.F. Comparison of N-alkyl acridine orange dyes as fluorescence probes for the determination of cardiolipin. Anal. Chim. Acta. 2008, 626, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.E.; Azizuddin, K.; Zhang, P.; Chiu, S.-M.; Lam, M.; Kenney, M.E.; Burda, C.; Oleinick, N.L. Targeting of mitochondria by 10-N-alkyl acridine orange analogues: Role of alkyl chain length in determining cellular uptake and localization. Mitochondrion 2008, 8, 237–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petit, J.M.; Maftah, A.; Ratinaud, M.H.; Julien, R. 10-N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur. J. Biochem. 1992, 209, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Lobasso, S.; Saponetti, M.S.; Polidoro, F.; Lopalco, P.; Urbanija, J.; Kralj-Iglic, V.; Corcelli, A. Archaebacterial lipid membranes as models to study the interaction of 10-N-nonyl acridine orange with phospholipids. Chem. Phys. Lipids. 2009, 157, 12–20. [Google Scholar] [CrossRef]
- Epand, R.M.; Epand, R.F. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept Sci. 2011, 17, 298–305. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [Green Version]
- Ileri Ercan, N.; Stroeve, P.; Tringe, J.W.; Faller, R. Molecular Dynamics Modeling of Methylene Blue-DOPC Lipid Bilayer Interactions. Langmuir 2018, 34, 4314–4323. [Google Scholar] [CrossRef] [Green Version]
- Orekhov, P.S.; Kholina, E.G.; Bozdaganyan, M.E.; Nesterenko, A.M.; Kovalenko, I.B.; Strakhovskaya, M.G. Molecular Mechanism of Uptake of Cationic Photoantimicrobial Phthalocyanine across Bacterial Membranes Revealed by Molecular Dynamics Simulations. J. Phys. Chem. B. 2018, 122, 3711–3722. [Google Scholar] [CrossRef]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An automated force field topology builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 2011, 7 Pt 12, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and Testing of the GROMOS Force-Field Versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ingólfsson, H.I.; Cheng, X.; Lee, J.; Marrink, S.J.; Im, W. CHARMM-GUI Martini Maker for Coarse-Grained Simulations with the Martini Force Field. J. Chem. Theory Comput. 2015, 11, 4486–4494. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, I.B.; Khrushchev, S.S.; Fedorov, V.A.; Riznichenko, G.Y.; Rubin, A.B. The role of electrostatic interactions in the process of diffusional encounter and docking of electron transport proteins. Dokl. Biochem. Biophys. 2016, 468, 183–186. [Google Scholar] [CrossRef] [PubMed]
- de Jong, D.H.; Baoukina, S.; Ingólfsson, H.I.; Marrink, S.J. Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 2016, 199, 1–7. [Google Scholar] [CrossRef]
- Yesylevskyy, S.O.; Schäfer, L.V.; Sengupta, D.; Marrink, S.J. Polarizable water model for the coarse-grained MARTINI force field. PLoS Comput. Biol. 2010, 6, e1000810. [Google Scholar] [CrossRef] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1 Pt 2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Kholina, E.G.; Kovalenko, I.B.; Bozdaganyan, M.E.; Strakhovskaya, M.G.; Orekhov, P.S. Cationic Antiseptics Facilitate Pore Formation in Model Bacterial Membranes. J. Phys. Chem. B 2020, 124, 8593–8600. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kholina, E.; Kovalenko, I.; Rubin, A.; Strakhovskaya, M. Insights into the Formation of Intermolecular Complexes of Fluorescent Probe 10-N-Nonyl Acridine Orange with Cardiolipin and Phosphatidylglycerol in Bacterial Plasma Membrane by Molecular Modeling. Molecules 2023, 28, 1929. https://doi.org/10.3390/molecules28041929
Kholina E, Kovalenko I, Rubin A, Strakhovskaya M. Insights into the Formation of Intermolecular Complexes of Fluorescent Probe 10-N-Nonyl Acridine Orange with Cardiolipin and Phosphatidylglycerol in Bacterial Plasma Membrane by Molecular Modeling. Molecules. 2023; 28(4):1929. https://doi.org/10.3390/molecules28041929
Chicago/Turabian StyleKholina, Ekaterina, Ilya Kovalenko, Andrew Rubin, and Marina Strakhovskaya. 2023. "Insights into the Formation of Intermolecular Complexes of Fluorescent Probe 10-N-Nonyl Acridine Orange with Cardiolipin and Phosphatidylglycerol in Bacterial Plasma Membrane by Molecular Modeling" Molecules 28, no. 4: 1929. https://doi.org/10.3390/molecules28041929
APA StyleKholina, E., Kovalenko, I., Rubin, A., & Strakhovskaya, M. (2023). Insights into the Formation of Intermolecular Complexes of Fluorescent Probe 10-N-Nonyl Acridine Orange with Cardiolipin and Phosphatidylglycerol in Bacterial Plasma Membrane by Molecular Modeling. Molecules, 28(4), 1929. https://doi.org/10.3390/molecules28041929






