The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds-DNA: Theoretical Studies of Clustered DNA Damage
Abstract
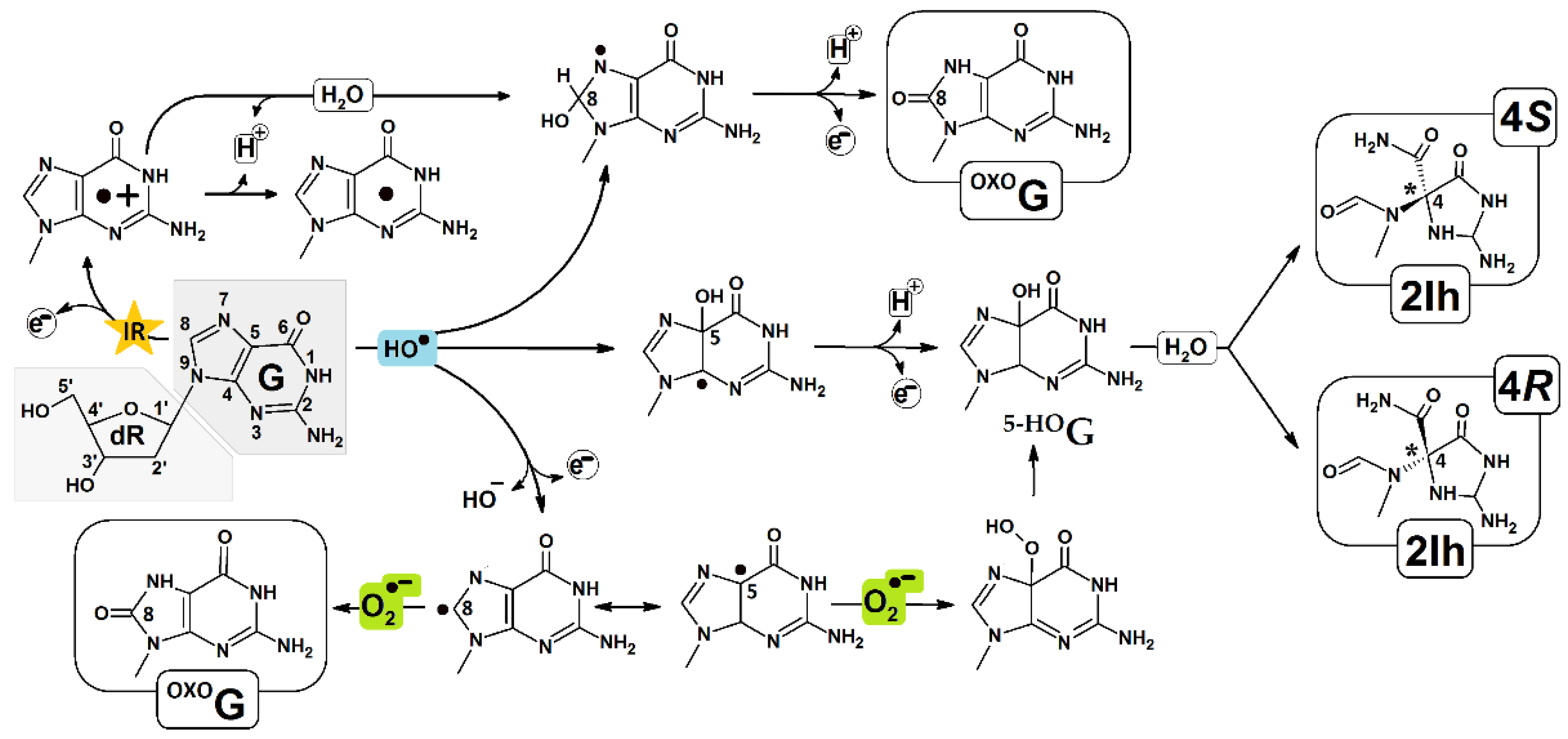
:1. Introduction
2. Results and Discussion
2.1. The Influence of Clustered DNA Damage Containing R/S-2Ih and OXOG on the Structure of Short Ds-oligos
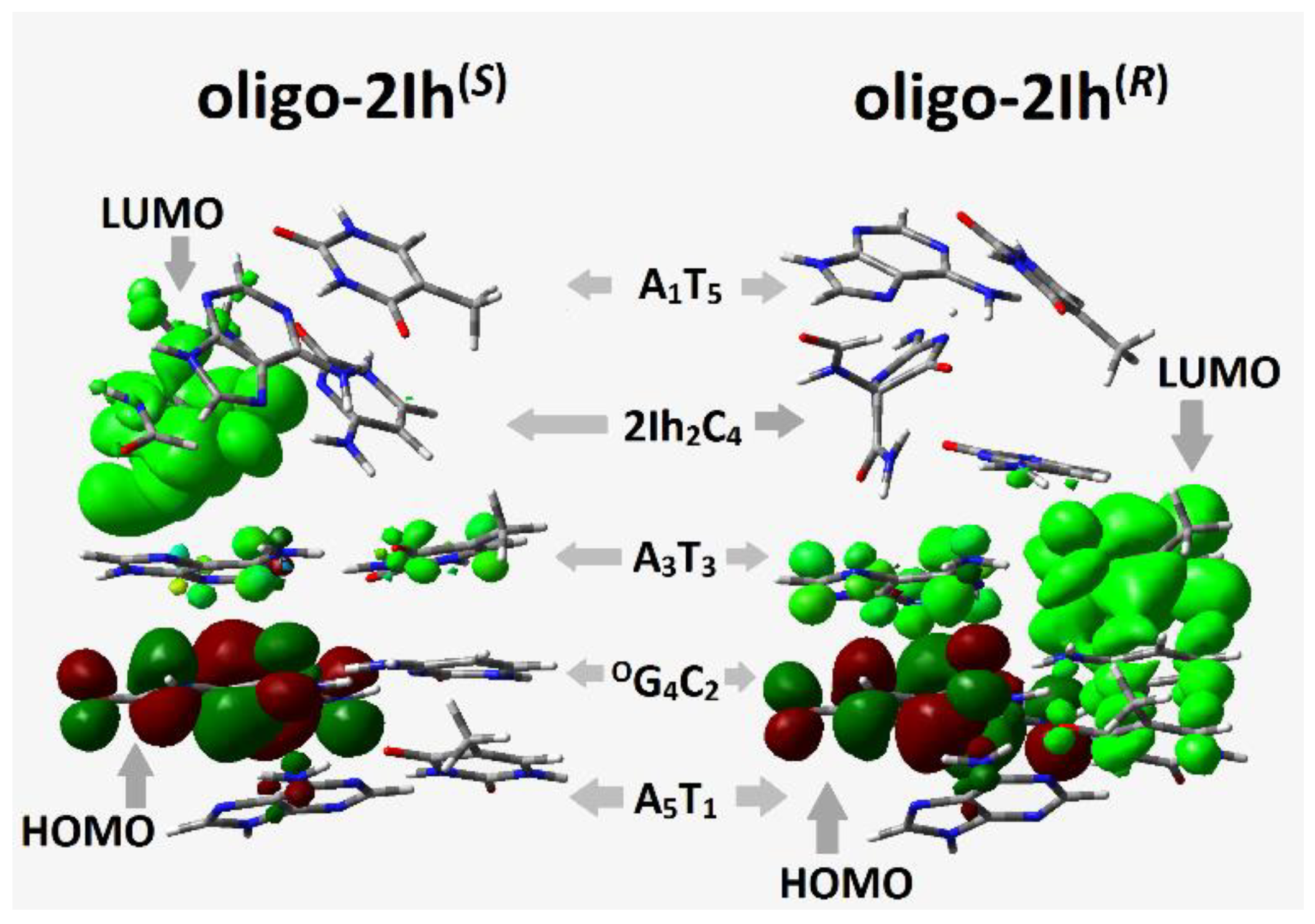
2.2. The Influence of (R/S)2Ih on the Electronic Properties of the Ds-oligonucleotides Containing Clustered DNA Damage
2.2.1. Charge and Spin Distribution
2.2.2. Electronic Properties of Oligo-2Ih(S) and Oligo-2Ih(R)
2.3. The Influence of (R)-2Ih and (S)-2Ih on Charge Migration through Ds-DNA Containing Clustered DNA Damage
3. Materials and Methods
4. Conclusions
- The ground-state molecule geometries were optimized using ONIOM methodology and the CPCM solvation model (M06-2x/D95**:M06-2x/sto-3G). All requested energies for theoretical studies were performed at the M06-2x/6-31++G** level of theory in the aqueous phase with equilibrated and non-equilibrated solvent-solute interactions.
- The spatial structure comparative analysis between cationic, anionic, and neutral forms showed that the presence of (R)-2Ih in the ds-oligo structure makes the double helix more sensitive to electron loss or adoption. Additionally (R)-2Ih::C forces significant changes in the geometry of the neighboring base pairs (5′-end site of 2Ih) with the standard DNA reference frame parameter. Buckle adopted a value at the level of −32° in comparison to −4° found in oligo-2Ih(S).
- Both diastereomers of 5-carboxamido-5-formamido-2-iminohydantoin exerted the same influence on the positive charge and spin distribution within the double helix containing the CDL. The radical cation in all the discussed cases settled exclusively on the OXOG:::C base pairs, as expected. A difference in the influence of (R) and (S)-2Ih on negative charge distribution was found when the non-equilibrated mode of the vertical anion was taken into consideration, i.e., in the oligo-2Ih(S), the negative charge and spin were mainly noted on the (S)-2Ih::C moiety while in the case of oligo-2Ih(R), no charge or spin were observed on the 2Ih::C base pair.
- A global electronic properties calculation revealed that oligo-2Ih(S) has a lower adiabatic ionization potential (5.53 eV) than oligo-2Ih(R) (5.67 eV) and a higher adiabatic electron affinity (2.09 eV) than oligo-2Ih(R) (2.0 eV). A careful analysis of the electronic properties of isolated base pairs gave these AIP values for 2Ih®::C (7.02 eV) and 2Ih(S)::C (6.94 eV) base pairs, while the lowest value was assigned for OXOG::C (6.62 eV) in both of the discussed ds-oligos. It should be pointed out that the AEA parameter adopted a higher absolute value in the case of 2Ih(R)::C (−1.4 eV) than in the case of 2Ih(S)::C (−1.30 eV).
- The comparative analysis of charge transfer rate, according to Marcus’s theory, elucidated that the presence of (S)-2Ih slows down the electron-hole and excess electron migration from the place of formation in comparison with (R)2Ih.
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fleming, A.M.; Burrows, C.J. Chemistry of ROS-mediated oxidation to the guanine base in DNA and its biological consequences. Int. J. Radiat. Biol. 2022, 98, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 2013, 54, 691–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angele, C.; Goodman, C.; Brumaghim, J.; Angele, C.; Goodman, C. Mechanisms, detection methods, and cellular consequences. Metallomics 2014, 6, 1358–1381. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Molecular radiobiology and the origins of the base excision repair pathway: An historical perspective. Int. J. Radiat. Biol. 2021, 1–26. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V.; Bietti, M.; Bernhard, K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2’deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J. Am. Chem. Soc. 2000, 122, 2373–2374. [Google Scholar] [CrossRef]
- Kino, K.; Kawada, T.; Hirao-Suzuki, M.; Morikawa, M.; Miyazawa, H. Products of oxidative guanine damage form base pairs with guanine. Int. J. Mol. Sci. 2020, 21, 7645. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Muller, J.G.; Dlouhy, A.C.; Burrows, C.J. Structural context effects in the oxidation of 8-Oxo-7,8-dihydro-2′-deoxyguanosine to hydantoin products: Electrostatics, base stacking, and base pairing. J. Am. Chem. Soc. 2012, 134, 15091–15102. [Google Scholar] [CrossRef] [Green Version]
- Van den Boogaard, W.M.C.; van den Heuvel-Eibrink, M.M.; Hoeijmakers, J.H.J.; Vermeij, W.P. Nutritional Preconditioning in Cancer Treatment in Relation to DNA Damage and Aging. Annu. Rev. Cancer Biol. 2020, 5, 161–179. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schär, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Fuss, J.O.; Tsai, C.L.; Ishida, J.P.; Tainer, J.A. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim. Biophys. Acta -Mol. Cell Res. 2015, 1853, 1253–1271. [Google Scholar] [CrossRef] [Green Version]
- Arnold, A.R.; Grodick, M.A.; Barton, J.K. DNA Charge Transport: From Chemical Principles to the Cell. Cell Chem. Biol. 2016, 23, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, B.M.; Bennett, P.V.; Sidorkina, O.; Laval, J. Clustered damages and total lesions induced in DNA by ionizing radiation: Oxidized bases and strand breaks. Biochemistry 2000, 39, 8026–8031. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2019. [Google Scholar]
- Alshykhly, O.R.; Fleming, A.M.; Burrows, C.J. 5-Carboxamido-5-formamido-2-iminohydantoin, in Addition to 8-oxo-7,8-Dihydroguanine, Is the Major Product of the Iron-Fenton or X-ray Radiation-Induced Oxidation of Guanine under Aerobic Reducing Conditions in Nucleoside and DNA Contexts. J. Org. Chem. 2015, 80, 6996–7007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hori, M.; Suzuki, T.; Minakawa, N.; Matsuda, A.; Harashima, H.; Kamiya, H. Mutagenicity of secondary oxidation products of 8-oxo-7,8-dihydro-2’-deoxyguanosine 5’-triphosphate (8-hydroxy-2’-deoxyguanosine 5’-triphosphate). Mutat. Res./Fundam. Mol. Mech. Mutagen. 2011, 714, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, G.P.; Besaratinia, A. Mutational spectra of human cancer. Hum. Genet. 2009, 125, 493–506. [Google Scholar] [CrossRef] [Green Version]
- Alshykhly, O.R.; Fleming, A.M.; Burrows, C.J. Guanine Oxidation Product 5-Carboxamido-5-formamido-2-iminohydantoin Induces Mutations When Bypassed by DNA Polymerases and Is a Substrate for Base Excision Repair. Chem. Res. Toxicol. 2015, 28, 1861–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, A.; Doublié, S.; Wallace, S.S. The Fpg/Nei family of DNA Glycosylases: Substrates, structures, and search for damage. Prog. Mol. Biol. Transl. Sci. 2012, 110, 71–91. [Google Scholar] [CrossRef] [Green Version]
- Genereux, J.C.; Barton, J.K. Mechanisms for DNA charge transport. Chem. Rev. 2010, 110, 1642–1662. [Google Scholar] [CrossRef] [Green Version]
- Olson, W.K.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Lu, X.J.; Neidle, S.; Shakked, Z.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Grodick, M.A.; Muren, N.B.; Barton, J.K. DNA Charge Transport within the Cell. Biochemistry 2015, 54, 962–973. [Google Scholar] [CrossRef] [Green Version]
- BIOVIA. Discovery Studio Visualizer; v16.1.0.15350; BIOVIA: San Diego, CA, USA, 2015. [Google Scholar]
- Arnittali, M.; Rissanou, A.N.; Harmandaris, V. Structure of Biomolecules Through Molecular Dynamics Simulations. Procedia Comput. Sci. 2019, 156, 69–78. [Google Scholar] [CrossRef]
- Tsuneda, T.; Song, J.W.; Suzuki, S.; Hirao, K. On Koopmans’ theorem in density functional theory. J. Chem. Phys. 2010, 133, 174101. [Google Scholar] [CrossRef]
- Close, D.M. Calculation of the Ionization Potentials of the DNA Bases in Aqueous Medium. J. Phys. Chem. A 2004, 108, 10376–10379. [Google Scholar] [CrossRef]
- Wagenknecht, H. Electron transfer processes in DNA: Mechanisms, biological relevance and applications in DNA analytics. Nat. Prod. Rep. 2006, 23, 973–1006. [Google Scholar] [CrossRef]
- Félix, M.; Voityuk, A.A. DFT performance for the hole transfer parameters in DNA p stacks. Int. J. Quantum Chem. 2011, 111, 191–201. [Google Scholar] [CrossRef]
- Oyler, N.A.; Adamowicz, L. Theoretical ab initio calculations of the electron affinity of thymine. Chem. Phys. Lett. 1994, 219, 223–227. [Google Scholar] [CrossRef]
- Oyler, N.A.; Adamowicz, L. Electron attachment to uracil. Theoretical ab initio study. J. Phys. Chem. 1993, 97, 11122–11123. [Google Scholar] [CrossRef]
- Shukla, L.I.; Adhikary, A.; Pazdro, R.; Becker, D.; Sevilla, M.D. Formation of 8-oxo-7,8-dihydroguanine-radicals in γ-irradiated DNA by multiple one-electron oxidations. Nucleic Acids Res. 2004, 32, 6565–6574. [Google Scholar] [CrossRef]
- Adhikary, A.; Khanduri, D.; Sevilla, M.D. Direct observation of the hole protonation state and hole localization site in DNA-oligomers. J. Am. Chem. Soc. 2009, 131, 8614–8619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhikary, A.; Kumar, A.; Munafo, S.A.; Khanduri, D.; Sevilla, M.D. Prototropic equilibria in DNA containing one-electron oxidized GC: Intra-duplex vs. duplex to solvent deprotonation. Phys. Chem. Chem. Phys. 2010, 12, 5353–5368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karwowski, B.T. Clustered DNA Damage: Electronic Properties and Their Influence on Charge Transfer. 7,8-Dihydro-8-Oxo-2′-Deoxyguaosine Versus 5′,8-Cyclo-2′-Deoxyadenosines: A Theoretical Approach. Cells 2020, 9, 424. [Google Scholar] [CrossRef] [Green Version]
- Karwowski, B.T. The Influence of Single, Tandem, and Clustered DNA Damage on the Electronic Properties of the Double Helix: A Theoretical Study. Molecules 2020, 25, 3126. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Adhikary, A.; Sevilla, M.D.; Close, D.M. One-electron oxidation of ds(5′-GGG-3′) and ds(5′-G(8OG)G-3′) and the nature of hole distribution: A density functional theory (DFT) study. Phys. Chem. Chem. Phys. 2020, 22, 5078–5089. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, X.B.; Vorpagel, E.R.; Wang, L.S. Direct experimental observation of the low ionization potentials of guanine in free oligonucleotides by using photoelectron spectroscopy. Proc. Natl. Acad. Sci. USA 2004, 101, 17588–17592. [Google Scholar] [CrossRef] [Green Version]
- Diamantis, P.; Tavernelli, I.; Rothlisberger, U. Redox Properties of Native and Damaged DNA from Mixed Quantum Mechanical/Molecular Mechanics Molecular Dynamics Simulations. J. Chem. Theory Comput. 2020, 16, 6690–6701. [Google Scholar] [CrossRef]
- Breslauer, K.J.; Frank, R.; Blocker, H.; Marky, L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 1986, 83, 3746–3750. [Google Scholar] [CrossRef] [Green Version]
- Zewail, A.H. Femtochemistry: Atomic-scale dynamics of the chemical bond. J. Phys. Chem. A 2000, 104, 5660–5694. [Google Scholar] [CrossRef]
- Lewis, F.D.; Liu, J.; Weigel, W.; Rettig, W.; Kurnikov, I.V.; Beratan, D.N. Donor-bridge-acceptor energetics determine the distance dependence of electron tunneling in DNA. Proc. Natl. Acad. Sci. USA 2002, 99, 12536–12541. [Google Scholar] [CrossRef] [Green Version]
- Siriwong, K.; Voityuk, A.A. Electron transfer in DNA. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 780–794. [Google Scholar] [CrossRef]
- Marcus, R.A. Electron Transfer Reactions in Chemistry: Theory and Experiment (Nobel Lecture). Angew. Chemie Int. Ed. Engl. 1993, 32, 1111–1121. [Google Scholar] [CrossRef]
- Marcus, R.A. Electron transfer reactions in chemistry. Theory and experiment. Rev. Mod. Phys. 1993, 65, 599–610. [Google Scholar] [CrossRef] [Green Version]
- Karwowski, B.T. The AT Interstrand Cross-Link: Structure, Electronic Properties, and Influence on Charge Transfer in dsDNA. Mol. Ther. -Nucleic Acids 2018, 13, 665–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voityuk, A.A. Estimates of electronic coupling for excess electron transfer in DNA. J. Chem. Phys. 2005, 123, 034903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rösch, N.; Voityuk, A.A. Quantum Chemical Calculation of Donor–Acceptor Coupling for Charge Transfer in DNA. Biosynthesis 2012, 237, 37–72. [Google Scholar] [CrossRef]
- Dreuw, A.; Head-Gordon, M. Single-reference ab initio methods for the calculation of excited states of large molecules. Chem. Rev. 2005, 105, 4009–4037. [Google Scholar] [CrossRef]
- Gu, J.; Xie, Y.; Schaefer, H.F. Electron attachment to nucleotides in aqueous solution. ChemPhysChem 2006, 7, 1885–1887. [Google Scholar] [CrossRef]
- Gu, J.; Xie, Y.; Schaefer, H.F. Electron attachment to DNA single strands: Gas phase and aqueous solution. Nucleic Acids Res. 2007, 35, 5165–5172. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Wang, J.; Leszczynski, J. Electron attachment-induced DNA single-strand breaks at the pyrimidine sites. Nucleic Acids Res. 2010, 38, 5280–5290. [Google Scholar] [CrossRef] [Green Version]
- Plumley, J.A.; Dannenberg, J.J. A comparison of the behavior of functional/basis set combinations for hydrogen-bonding in the water dimer with emphasis on basis set superposition error. J. Comput. Chem. 2011, 32, 1519–1527. [Google Scholar] [CrossRef]
- Zhao, Y.; Pu, J.; Lynch, B.J.; Truhlar, D.G. Tests of second-generation and third-generation density functionals for thermochemical kinetics. Phys. Chem. Chem. Phys. 2004, 6, 673. [Google Scholar] [CrossRef]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. Theochem 1999, 461–462, 1–21. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Journal, T.; Chemistry, P. 5’,8-cyclopurine-2’-deoxynucleosides: Molecular structure and charge distribution–DFT study in gaseous and aqueous phase. J. Mol. Struct. Theochem 2009, 915, 73–78. [Google Scholar]
- Si, D. Quantum Mechanical and Molecular Mechanical Study of Solvent Effects. Ph.D. Differtration, University of Nebraska, Lincoln, NE, USA, 2011. [Google Scholar]
- Cave, R.J.; Newton, M.D. Generalization of the Mulliken-Hush treatment for the calculation of electron transfer matrix elements. Chem. Phys. Lett. 1996, 249, 15–19. [Google Scholar] [CrossRef]
- Miertus, S.; Tomasi, J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 1982, 65, 239–245. [Google Scholar] [CrossRef]
- Mabesoone, M.F.J.; Palmans, A.R.A.; Meijer, E.W. Solute-Solvent Interactions in Modern Physical Organic Chemistry: Supramolecular Polymers as a Muse. J. Am. Chem. Soc. 2020, 142, 19781–19798. [Google Scholar] [CrossRef] [PubMed]
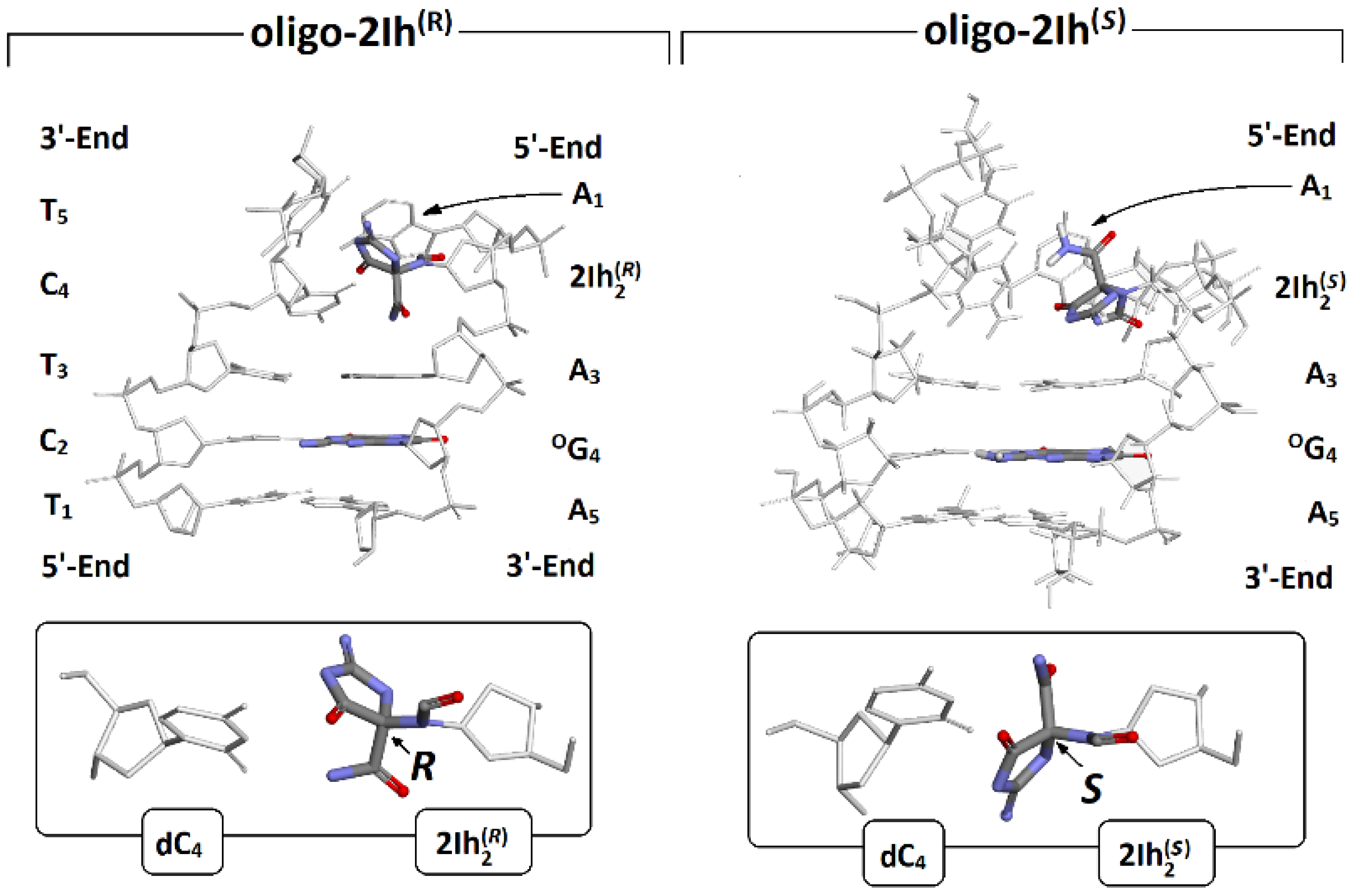


| ds-DNA | Base Pair | Structural Parameters | Spatial Geometry Comparison (RMSD Analysis) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buckle | Opening | HB-1 | HB-2 | HB-3 | ||||||
| oligo-2h(R) | A1::T5 | −32.84 | 5.61 | 3.06 | 2.80 | Form | Overlap | oligo-2Ih(S) | oligo-2Ih(R) | |
| A3::T3 | 11.05 | 7.32 | 3.10 | 2.77 | Anion versus Neutral | Total | 0.25 | 0.34 | ||
| °G4:::C2 | −0.89 | −1.92 | 2.83 | 2.91 | 2.89 | Base | 0.16 | 0.24 | ||
| oligo-2h(S) | A1::T5 | −4.22 | 0.52 | 2.95 | 2.84 | Frame | 0.31 | 0.41 | ||
| A3::T3 | 11.81 | 4.74 | 3.05 | 2.80 | Cation versus Neutral | Total | 0.36 | 0.40 | ||
| °G4:::C2 | −4.43 | −1.45 | 2.83 | 2.90 | 2.87 | Base | 0.25 | 0.31 | ||
| Canonical ds-oligo | A::T | 5.61 | −0.87 | 2.92 | 2.85 | Frame | 0.44 | 0.47 | ||
| G:::C | −0.51 | −1.50 | 2.88 | 2.88 | 2.86 | |||||
| Base Pair | Oligo-2Ih(S) X = (S)2Ih | Oligo-2Ih(R) X = (R)2Ih | ||||||
|---|---|---|---|---|---|---|---|---|
| VIP | AIP | VEA | AEA | VIP | AIP | VEA | AEA | |
| A1::T5 | 6.67 | 6.67 | −1.42 | −1.42 | 6.73 | 6.74 | −1.41 | −1.40 |
| X2::C4 | 6.93 | 6.94 | −1.30 | −1.30 | 7.03 | 7.02 | −1.37 | −1.40 |
| A3::T3 | 6.65 | 6.62 | −1.39 | −1.40 | 6.79 | 6.64 | −1.38 | −1.37 |
| OG4:::C2 | 5.93 | 5.53 | −1.49 | −1.93 | 5.94 | 5.54 | −1.48 | −1.93 |
| A5::T1 | 6.66 | 6.62 | −1.43 | −1.40 | 6.65 | 6.62 | −1.44 | −1.41 |
| ds-oligo | 6.65(NE), 5.99(EQ) | 5.51 | −1.07(NE), −1.40(EQ) | −2.09 | 6.70(NE), 6.04(EQ) | 5.67 | −1.00(NE), −1.68(EQ) | −2.00 |
| ds-BP | 6.53(NE), 5.90(EQ) | 5.50 | −0.65(NE), −1.37(EQ) | −2.09 | 6.56(NE), 5.94(EQ) | 5.57 | −0.78(NE), −1.31(EQ) | −1.84 |
| Excess Electron Transfer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oligo-2Ih(S) | Oligo-2Ih(R) | ||||||||||
| X = (S)2Ih | λ | G | Ea | V12 | kHT | X = (R)2Ih | λ | G | Ea | V12 | kHT |
| A1T5←X2C4 | −0.02 | −0.12 | −0.27 | 0.08 | --- | A1T5←X2C4 | 0.04 | −0.01 | 0.01 | 0.04 | 8.82 × 1013 |
| X2C4→A3T3 | 0.00 | −0.10 | 6.84 | 0.04 | 0.00 | X2C4→A3T3 | 0.04 | −0.03 | 0.00 | 2.26 | 3.97 × 1017 |
| A3T3→OG4C2 | 0.44 | −0.53 | 0.00 | 0.04 | 3.47 × 1013 | A3T3→OG4C2 | 0.46 | −0.55 | 0.01 | 0.05 | 4.87 × 1013 |
| OG4C2←A5T1 | 0.43 | −0.53 | 0.00 | 0.06 | 8.24 × 1013 | OG4C2←A5T1 | 0.43 | −0.52 | 0.00 | 0.06 | 7.27 × 1013 |
| A1T5←A3T3 | −0.01 | −0.02 | −0.02 | 0.09 | --- | A1T5←A3T3 | −0.02 | −0.02 | −0.02 | 0.03 | --- |
| X2C4→OG4C2 | 0.43 | −0.63 | 0.02 | 0.11 | 1.2 × 1014 | X2C4→OG4C2 | 0.43 | −0.53 | 0.01 | 0.003 | 1.9 × 1011 |
| A3T3→A5T1 | −0.03 | −0.002 | −0.01 | 0.07 | --- | A3T3→A5T1 | −0.02 | −0.04 | −0.04 | 0.07 | --- |
| Electron-Hole Transfer | |||||||||||
| A1T5←X2C4 | 0.00 | −0.28 | 33.56 | 0.19 | 0.00 | A1T5←X2C4 | 0.00 | −0.28 | −5.82 | 0.10 | --- |
| X2C4→ A3T3 | 0.03 | −0.32 | 0.81 | 0.23 | 1.06 × 102 | X2C4→A3T3 | 0.16 | −0.38 | 0.08 | 0.11 | 2.13 × 1013 |
| A3T3→OG4C2 | 0.38 | −1.09 | 0.32 | 0.35 | 1.13 × 1010 | A3T3→OG4C2 | 0.39 | −1.10 | 0.32 | 0.42 | 1.82 × 1010 |
| OG4C2← A5T1 | 0.37 | −1.09 | 0.35 | 0.37 | 4.72 × 109 | OG4C2←A5T1 | 0.36 | −1.08 | 0.35 | 0.35 | 4.21 × 109 |
| A1T5→ A3T3 | 0.03 | −0.05 | 0.002 | 0.19 | 3.1 × 1015 | A1T5→A3T3 | 0.16 | −0.10 | 0.01 | 0.03 | 3.5 × 1013 |
| X2C4→OG4C2 | 0.39 | −1.41 | 0.66 | 0.53 | 4.6 × 104 | X2C4→OG4C2 | 0.41 | −1.48 | 0.69 | 0.04 | 1.0 × 102 |
| A3T3← A5T1 | 0.03 | −0.003 | 0.01 | 0.01 | 2.9 × 1012 | A3T3→A5T1 | 0.02 | −0.02 | 0.00 | 0.07 | 5.8 × 1014 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwowski, B.T. The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds-DNA: Theoretical Studies of Clustered DNA Damage. Molecules 2023, 28, 2180. https://doi.org/10.3390/molecules28052180
Karwowski BT. The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds-DNA: Theoretical Studies of Clustered DNA Damage. Molecules. 2023; 28(5):2180. https://doi.org/10.3390/molecules28052180
Chicago/Turabian StyleKarwowski, Boleslaw T. 2023. "The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds-DNA: Theoretical Studies of Clustered DNA Damage" Molecules 28, no. 5: 2180. https://doi.org/10.3390/molecules28052180
APA StyleKarwowski, B. T. (2023). The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds-DNA: Theoretical Studies of Clustered DNA Damage. Molecules, 28(5), 2180. https://doi.org/10.3390/molecules28052180





