Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins
Abstract
:1. Introduction
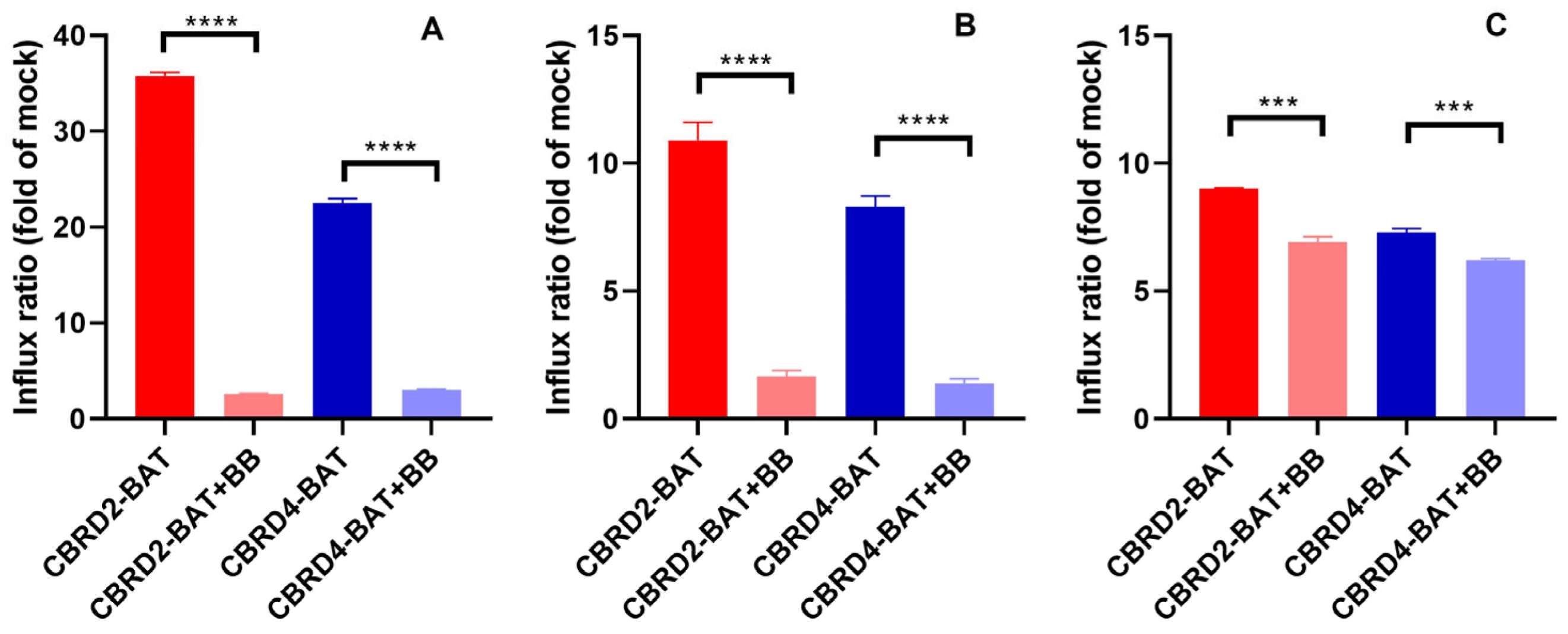
2. Results and Discussion
3. Materials and Methods
3.1. General
3.2. Cell Cultures
3.3. Western Blot Analysis
3.4. Transport Activity Measurements by Flow Cytometry
3.5. Effect of Dyes on Cell Viability
3.6. Live-Cell Labeling
3.7. Confocal and STED Imaging and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Liu, J.; Cui, Z. Fluorescent Labeling of Proteins of Interest in Live Cells: Beyond Fluorescent Proteins. Bioconj. Chem. 2020, 31, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Specht, E.A.; Braselmann, E.; Palmer, A.E. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol. 2017, 79, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Bayguinov, P.O.; Oakley, D.M.; Shih, C.C.; Geanon, D.J.; Joens, M.S.; Fitzpatrick, J.A.J. Modern Laser Scanning Confocal Microscopy. Curr. Protoc. Cytom. 2018, 85, e39. [Google Scholar] [CrossRef]
- Li, C.; Tebo, A.G.; Gautier, A. Fluorogenic Labeling Strategies for Biological Imaging. Int. J. Mol. Sci. 2017, 18, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Zhen, S.; Liu, H.; Chen, P.R. Illuminating biological processes through site-specific protein labeling. Chem. Soc. Rev. 2015, 44, 3405–3417. [Google Scholar] [CrossRef]
- Lang, K.; Chin, J.W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 2014, 114, 4764–4806. [Google Scholar] [CrossRef] [PubMed]
- Nikić, I.; Plass, T.; Schraidt, O.; Szymański, J.; Briggs, J.A.; Schultz, C.; Lemke, E.A. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed. 2014, 53, 2245–2249. [Google Scholar] [CrossRef]
- Chin, J.W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014, 83, 379–408. [Google Scholar] [CrossRef]
- Arsić, A.; Hagemann, C.; Stajković, N.; Schubert, T.; Nikić-Spiegel, I. Minimal genetically encoded tags for fluorescent protein labeling in living neurons. Nat. Commun. 2022, 13, 314. [Google Scholar] [CrossRef]
- Bessa-Neto, D.; Kuhlemann, A.; Beliu, G.; Pecoraro, V.; Doose, S.; Retailleau, N.; Chevrier, N.; Perrais, D.; Sauer, M.; Choquet, D. Bioorthogonal labeling of transmembrane proteins with non-canonical amino acids unveils masked epitopes in live neurons. Nat. Commun. 2021, 12, 6715. [Google Scholar] [CrossRef]
- Grimm, J.; English, B.; Chen, J.; Chen, J.; Slaughter, J.P.; Zhang, Z.; Revyakin, A.; Patel, R.; Macklin, J.J.; Normanno, D.; et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 2015, 12, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Wombacher, R.; Heidbreder, M.; van de Linde, S.; Sheetz, M.P.; Heilemann, M.; Cornish, V.W.; Sauer, M. Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods 2010, 7, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, M.; Zhang, W.; Han, Y.; Zhang, Y.-H. Simple and efficient delivery of cell-impermeable organic fluorescent probes into live cells for live-cell superresolution imaging. Light Sci. Appl. 2019, 8, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakos, É.; Tusnády, G.E.; Német, O.; Patik, I.; Magyar, C.; Németh, K.; Kele, P.; Özvegy-Laczka, C. Synergistic transport of a fluorescent coumarin probe marks coumarins as pharmacological modulators of Organic anion-transporting polypeptide, OATP3A1. Biochem. Pharmacol. 2020, 182, 114250. [Google Scholar] [CrossRef] [PubMed]
- Bakos, É.; Német, O.; Kucsma, N.; Tőkési, N.; Stieger, B.; Rushing, E.; Tőkés, A.-M.; Kele, P.; Tusnády, G.E.; Özvegy-Laczka, C. Cloning and characterization of a novel functional organic anion transporting polypeptide 3A1 isoform highly expressed in the human brain and testis. Front. Pharmacol. 2022, 13, 958023. [Google Scholar] [CrossRef]
- Patik, I.; Székely, V.; Német, O.; Szepesi, Á.; Kucsma, N.; Várady, G.; Szakács, G.; Bakos, É.; Özvegy-Laczka, C. Identification of novel cell-impermeant fluorescent substrates for testing the function and drug interaction of Organic Anion-Transporting Polypeptides, OATP1B1/1B3 and 2B1. Sci. Rep. 2018, 8, 2630. [Google Scholar] [CrossRef] [Green Version]
- Stieger, B.; Hagenbuch, B. Organic Anion Transporting Polypeptides. Curr. Top. Membr. 2014, 73, 205–232. [Google Scholar] [CrossRef] [Green Version]
- Kovacsics, D.; Patik, I.; Özvegy-Laczka, C. The role of organic anion transporting polypeptides in drug absorption, distribution, excretion and drug-drug interactions. Expert Opin. Drug Metab. Toxicol. 2017, 13, 409–424. [Google Scholar] [CrossRef] [Green Version]
- Shitara, Y.; Maeda, K.; Ikejiri, K.; Yoshida, K.; Horie, T.; Sugiyama, Y. Clinical significance of organic anion transporting polypeptides (OATPs) in drug disposition: Their roles in hepatic clearance and intestinal absorption. Biopharm. Drug Dispos. 2013, 34, 45–78. [Google Scholar] [CrossRef]
- McFeely, S.J.; Wu, L.; Ritchie, T.K.; Unadkat, J. Organic anion transporting polypeptide 2B1—More than a glass-full of drug interactions. Pharmacol. Ther. 2019, 196, 204–215. [Google Scholar] [CrossRef]
- Urquhart, B.L.; Kim, R.B. Blood-brain barrier transporters and response to CNS-active drugs. Eur. J. Clin. Pharmacol. 2009, 65, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhou, Z.; Tay-Sontheimer, J.; Levy, R.H.; Ragueneau-Majlessi, I. Intestinal Drug Interactions Mediated by OATPs: A Systematic Review of Preclinical and Clinical Findings. J. Pharm. Sci. 2017, 106, 2312–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamai, I.; Nezu, J.; Uchin, H.; Sai, Y.; Oku, A.; Shimane, M.; Tsuji, A. Molecular identification and characterization of novel members of the human organic anion transporter (OATP) family. Biochem. Biophys. Res. Commun. 2000, 273, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Suzuki, T.; Abe, M.; Asano, N.; Mizutamari, H.; Tanemoto, M.; Nishio, T.; Onogawa, T.; Toyohara, T.; Kasai, S.; et al. Molecular characterization of human and rat organic anion transporter OATP-D. Am. J. Physiol. Renal. Physiol. 2003, 285, F1188–F1197. [Google Scholar] [CrossRef] [Green Version]
- Wlcek, K.; Svoboda, M.; Riha, J.; Zakaria, S.; Olszewski, U.; Dvorak, Z.; Sellner, F.; Ellinger, I.; Jager, W.; Thalhammer, T. The analysis of organic anion transporting polypeptide (OATP) mRNA and protein patterns in primary and metastatic liver cancer. Cancer Biol. Ther. 2011, 11, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, N.; Miller, N.; Allen, C.; Bendayan, R. Expression of membrane transporters and metabolic enzymes involved in estrone-3-sulphate disposition in human breast tumour tissues. Breast Cancer Res. Treat. 2014, 145, 647–661. [Google Scholar] [CrossRef]
- Wei, S.C.; Tan, Y.Y.; Weng, M.T.; Lai, L.C.; Hsiao, J.H.; Chuang, E.Y.; Shun, C.T.; Wu, D.C.; Kao, A.W.; Chuang, C.S.; et al. SLCO3A1, A novel crohn’s disease-associated gene, regulates nf-κB activity and associates with intestinal perforation. PLoS ONE 2014, 19, e100515. [Google Scholar] [CrossRef] [Green Version]
- Török, G.; Cserép, G.B.; Telek, A.; Arany, D.; Váradi, M.; Homolya, L.; Kellermayer, M.; Kele, P.; Németh, K. Large Stokes-shift bioorthogonal probes for STED, 2P-STED and multi-color STED nanoscopy. Methods Appl. Fluoresc. 2021, 9, 015006. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kawai, A.; Sakurama, K.; Udo, N.; Yoshino, Y.; Saito, Y.; Tsukigawa, K.; Nishi, K.; Otagiri, M. Interaction of Benzbromarone with Subdomains IIIA and IB/IIA on Human Serum Albumin as the Primary and Secondary Binding Regions. Mol. Pharm. 2021, 18, 1061–1070. [Google Scholar] [CrossRef]
- Kozma, E.; Girona, G.E.; Paci, G.; Lemke, E.A.; Kele, P. Bioorthogonal double-fluorogenic siliconrhodamine probes for intracellular super-resolution microscopy. Chem. Commun. 2017, 53, 6696–6699. [Google Scholar] [CrossRef] [Green Version]
- Szatmári, Á.; Cserép, G.B.; Molnár, T.Á.; Söveges, B.; Biró, A.; Várady, G.; Németh, K.; Kele, P. A Genetically Encoded Isonitrile Lysine for Orthogonal Bioorthogonal Labeling Schemes. Molecules 2021, 26, 4988. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Németh, K.; László, Z.; Biró, A.; Szatmári, Á.; Cserép, G.B.; Várady, G.; Bakos, É.; Özvegy-Laczka, C.; Kele, P. Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins. Molecules 2023, 28, 2521. https://doi.org/10.3390/molecules28062521
Németh K, László Z, Biró A, Szatmári Á, Cserép GB, Várady G, Bakos É, Özvegy-Laczka C, Kele P. Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins. Molecules. 2023; 28(6):2521. https://doi.org/10.3390/molecules28062521
Chicago/Turabian StyleNémeth, Krisztina, Zsófia László, Adrienn Biró, Ágnes Szatmári, Gergely B. Cserép, György Várady, Éva Bakos, Csilla Özvegy-Laczka, and Péter Kele. 2023. "Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins" Molecules 28, no. 6: 2521. https://doi.org/10.3390/molecules28062521
APA StyleNémeth, K., László, Z., Biró, A., Szatmári, Á., Cserép, G. B., Várady, G., Bakos, É., Özvegy-Laczka, C., & Kele, P. (2023). Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins. Molecules, 28(6), 2521. https://doi.org/10.3390/molecules28062521





