Identification of Potential Inhibitors for the Treatment of Alkaptonuria Using an Integrated In Silico Computational Strategy
Abstract
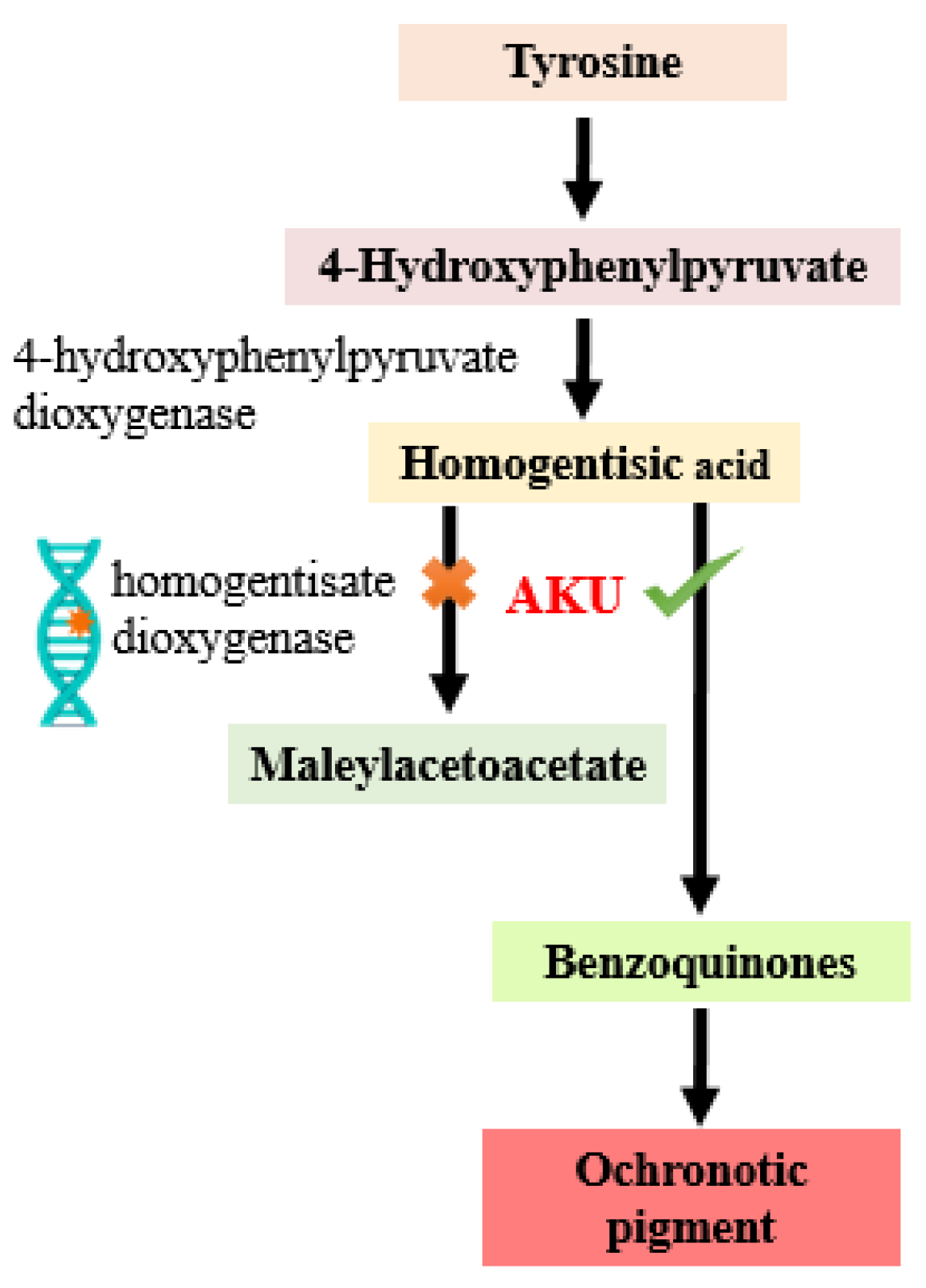
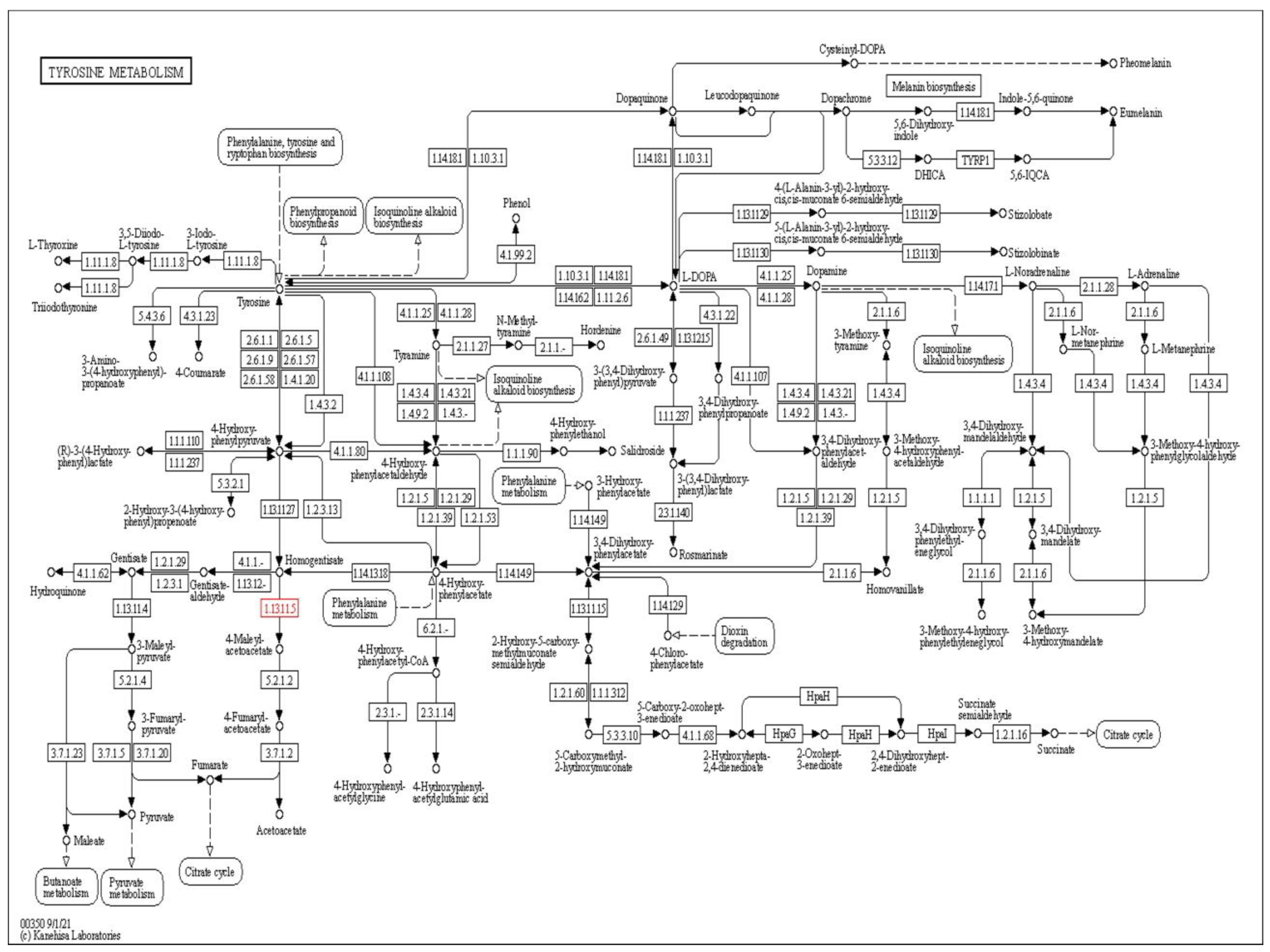
1. Introduction
2. Results and Discussion
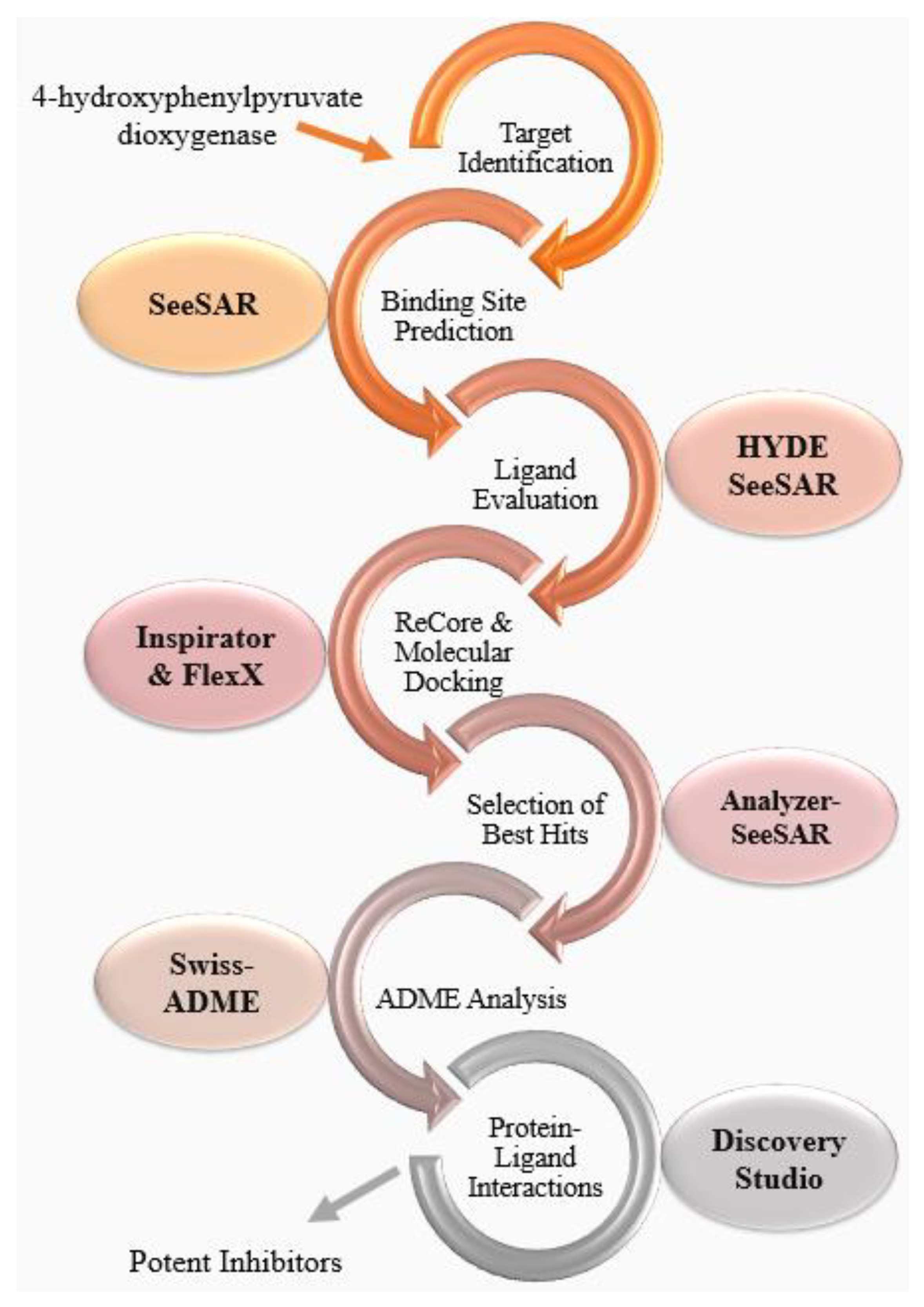
2.1. Target Identification
2.2. Binding Site Prediction
2.3. Ligand Evaluation
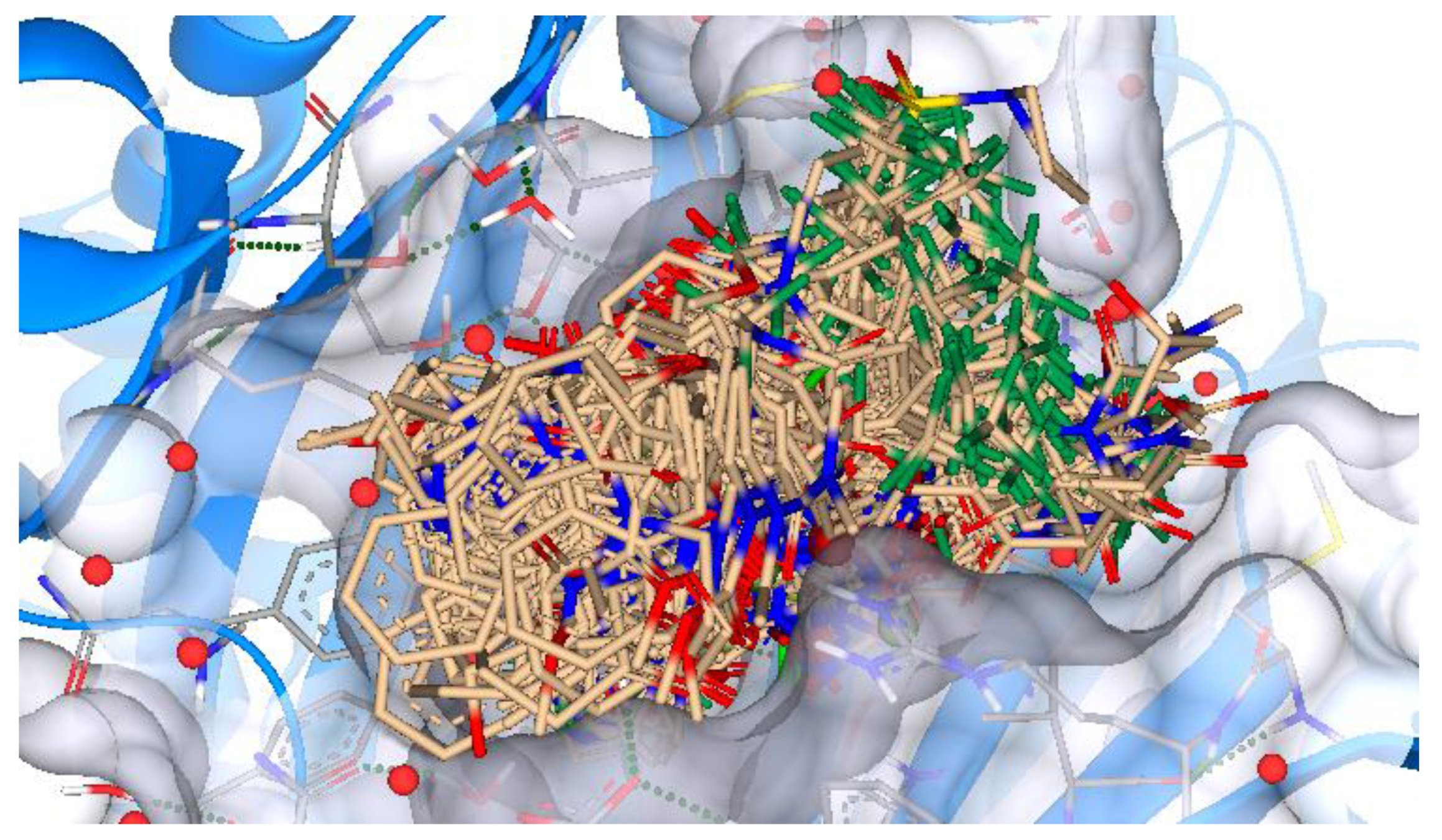
2.4. ReCore and Molecular Docking
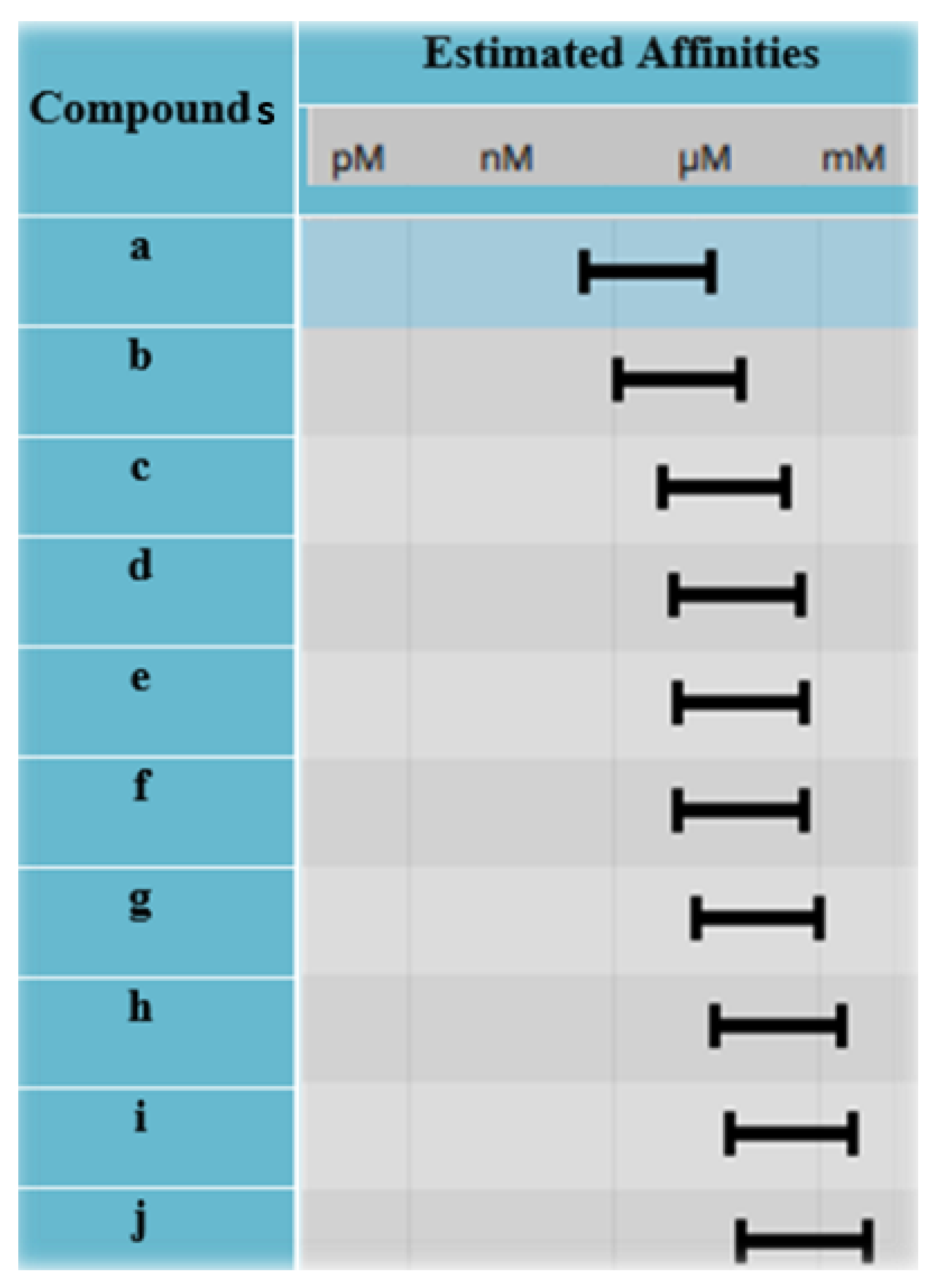
2.5. Selection of Best Hits
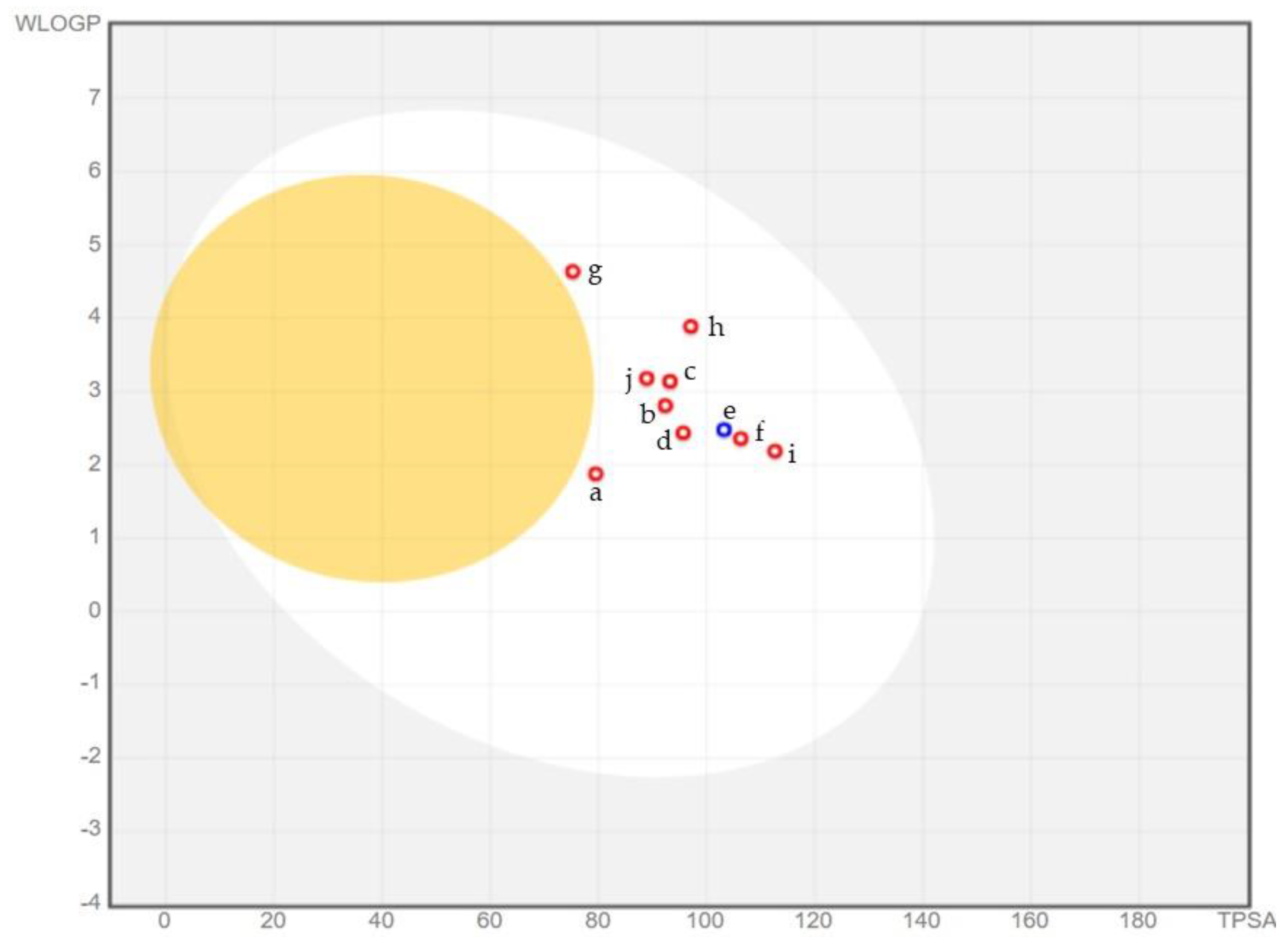
2.6. ADME Analysis
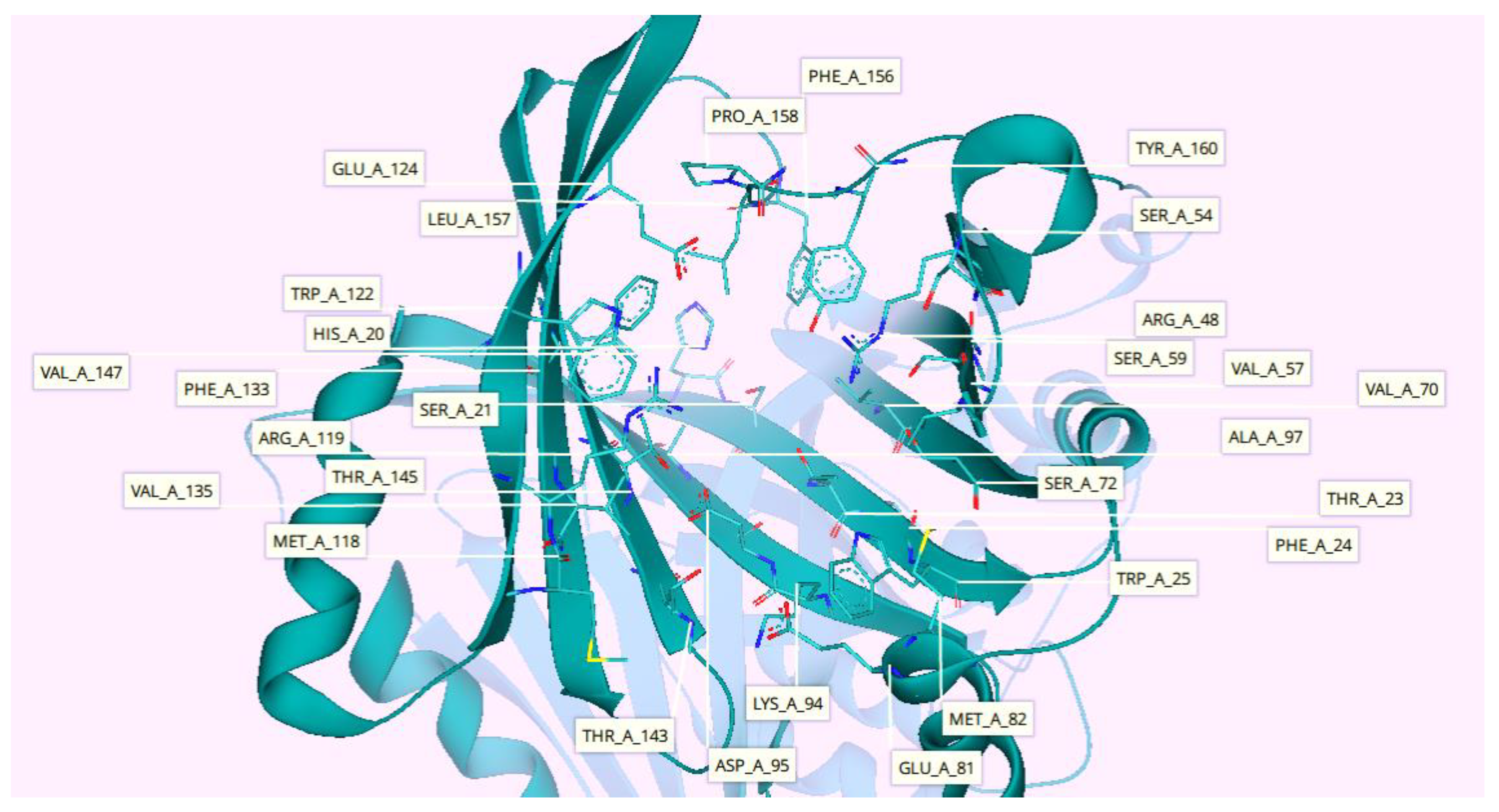
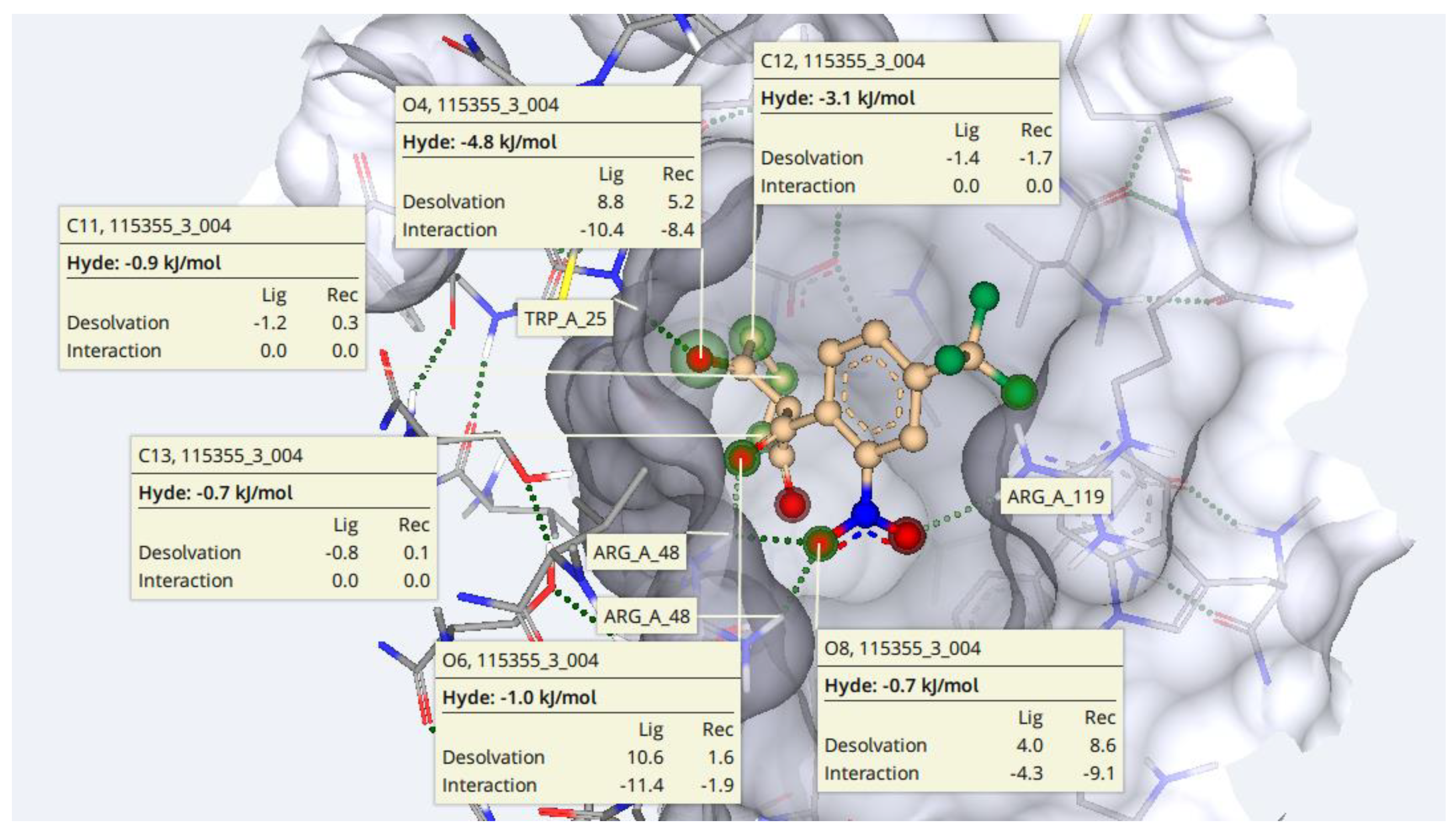
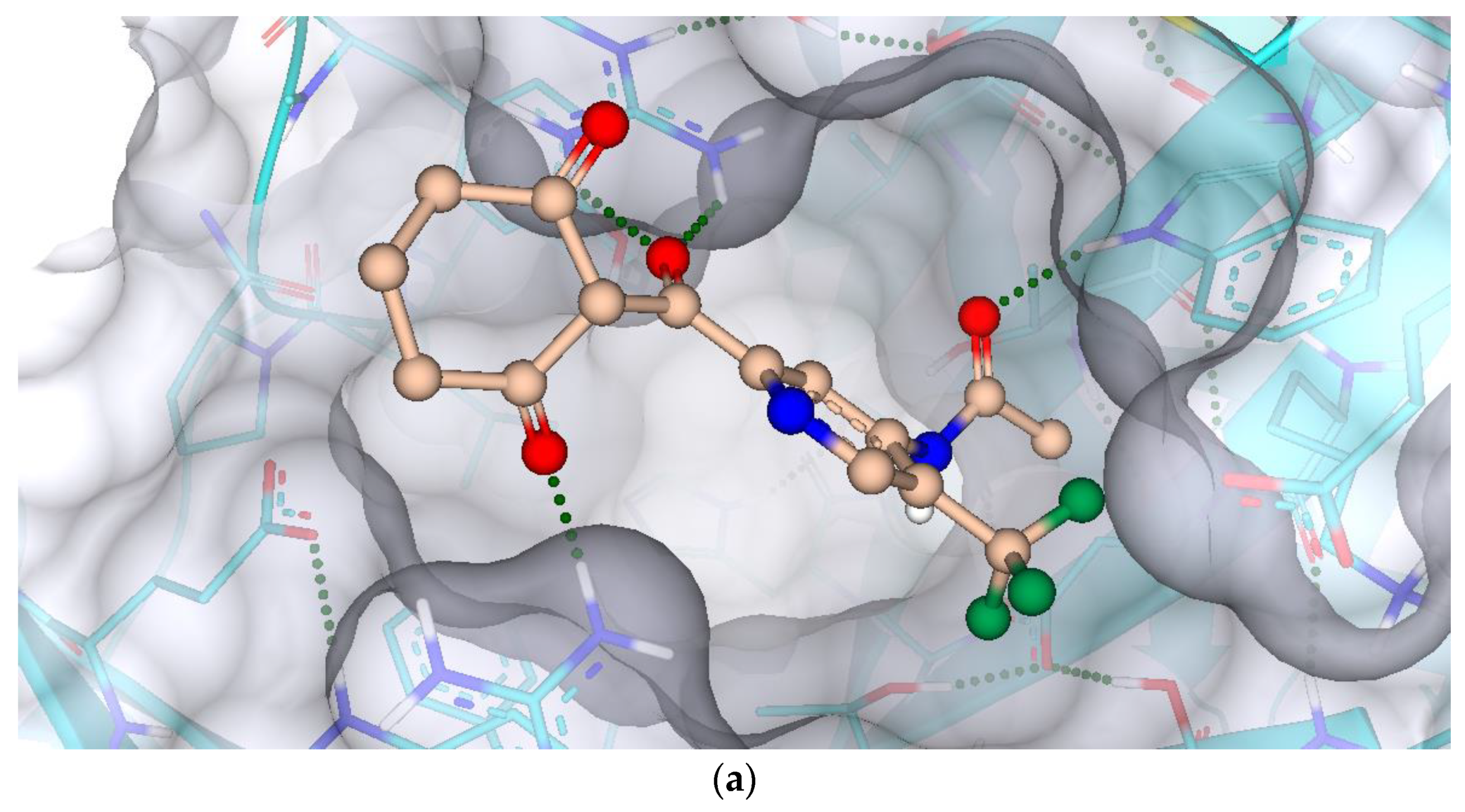
2.7. Protein-Ligand Interactions
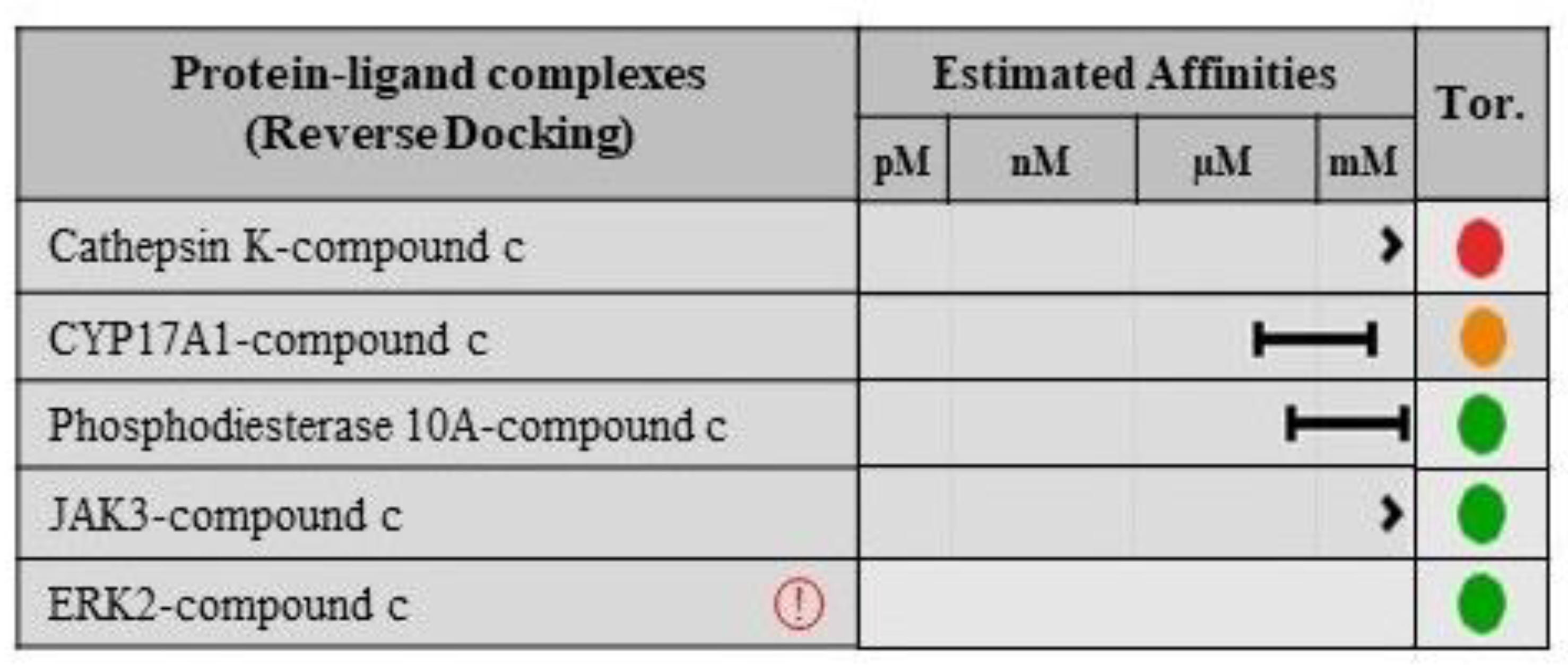
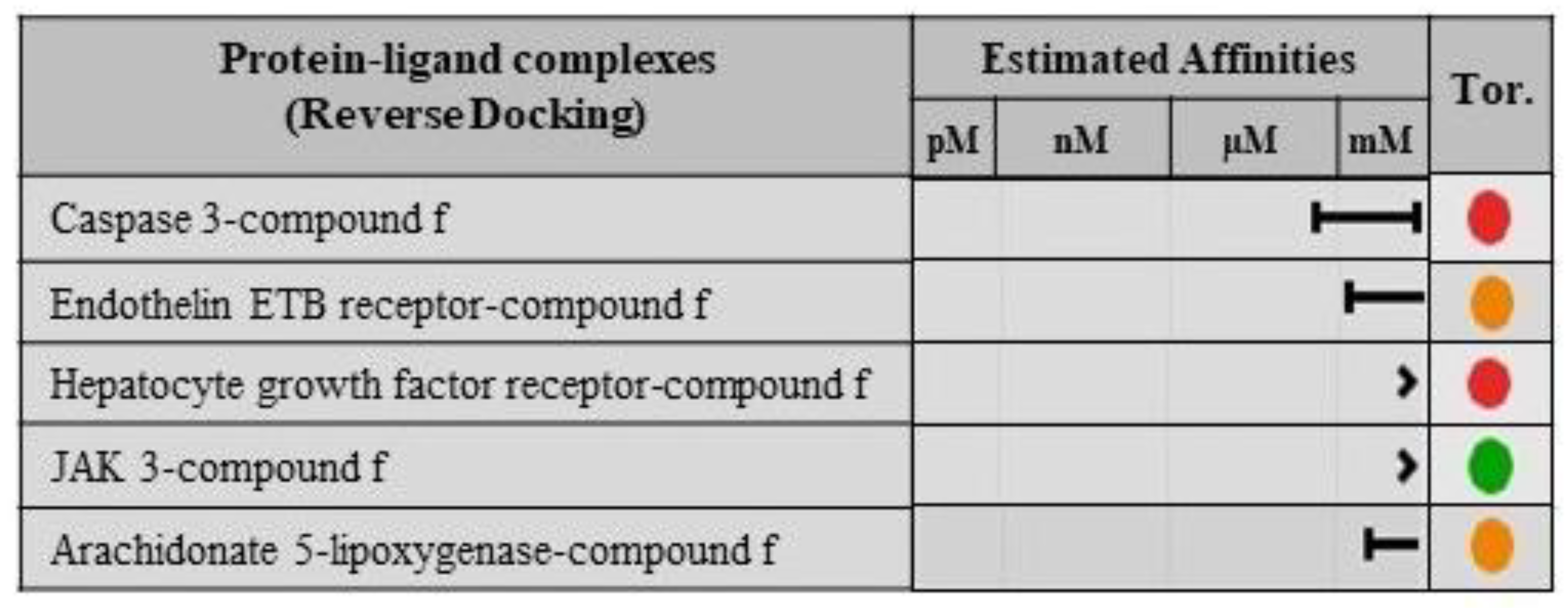
2.8. Validation of Ligand Specificity
3. Methodology
3.1. Target Identification
3.2. Binding Site Prediction
3.3. Ligand Evaluation
3.4. ReCore and Molecular Docking
3.5. Selection of Best Hits
3.6. ADME Analysis
3.7. Protein-Ligand Interactions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zatkova, A.; Ranganath, L.; Kadasi, L. Alkaptonuria: Current perspectives. Appl. Clin. Genet. 2020, 13, 37. [Google Scholar] [CrossRef]
- Davison, A.S.; Hughes, A.T.; Milan, A.M.; Sireau, N.; Gallagher, J.A.; Ranganath, L.R. Alkaptonuria–Many questions answered, further challenges beckon. Ann. Clin. Biochem. 2020, 57, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Rudebeck, M.; Scott, C.; Sireau, N.; Ranganath, L. A patient survey on the impact of alkaptonuria symptoms as perceived by the patients and their experiences of receiving diagnosis and care. JIMD Rep. 2020, 53, 71–79. [Google Scholar] [CrossRef]
- Arnold, G.L. Inborn errors of metabolism in the 21st century: Past to present. Ann. Transl. Med. 2018, 6, 467. [Google Scholar] [CrossRef] [PubMed]
- Grasso, D.; Geminiani, M.; Galderisi, S.; Iacomelli, G.; Peruzzi, L.; Marzocchi, B.; Santucci, A.; Bernini, A. Untargeted NMR Metabolomics Reveals Alternative Biomarkers and Pathways in Alkaptonuria. Int. J. Mol. Sci. 2022, 23, 15805. [Google Scholar] [CrossRef]
- Zatkova, A. An update on molecular genetics of alkaptonuria (AKU). J. Inherit. Metab. Dis. 2011, 34, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.S.; Milan, A.M.; Hughes, A.T.; Dutton, J.J.; Ranganath, L.R. Serum concentrations and urinary excretion of homogentisic acid and tyrosine in normal subjects. Clin. Chem. Lab. Med. 2015, 53, e81–e83. [Google Scholar] [CrossRef]
- Phornphutkul, C.; Introne, W.J.; Perry, M.B.; Bernardini, I.; Murphey, M.D.; Fitzpatrick, D.L.; Anderson, P.D.; Huizing, M.; Anikster, Y.; Gerber, L.H.; et al. Natural history of alkaptonuria. N. Engl. J. Med. 2002, 347, 2111–2121. [Google Scholar] [CrossRef]
- Donaldson, C.J.; Mitchell, S.L.; Riley III, L.H.; Kebaish, K.M. “As black as ink”: A case of alkaptonuria-associated myelopathy and a review of the literature. Spine 2019, 44, E53–E59. [Google Scholar] [CrossRef]
- Ranganath, L.R.; Norman, B.P.; Gallagher, J.A. Ochronotic pigmentation is caused by homogentisic acid and is the key event in alkaptonuria leading to the destructive consequences of the disease—A review. J. Inherit. Metab. Dis. 2019, 42, 776–792. [Google Scholar] [CrossRef]
- Arnoux, J.B.; Le Quan Sang, K.H.; Brassier, A.; Grisel, C.; Servais, A.; Wippf, J.; Dubois, S.; Sireau, N.; Job-Deslandre, C.; Ranganath, L.; et al. Old treatments for new insights and strategies: Proposed management in adults and children with alkaptonuria. J. Inherit. Metab. Dis. 2015, 38, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.F.; Ranganath, L. A quantitative assessment of alkaptonuria: Testing the reliability of two disease severity scoring systems. J. Inherit. Metab. Dis. 2011, 34, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Hannoush, H.; Introne, W.J.; Chen, M.Y.; Lee, S.J.; O’Brien, K.; Suwannarat, P.; Kayser, M.A.; Gahl, W.A.; Sachdev, V. Aortic stenosis and vascular calcifications in alkaptonuria. Mol. Genet. Metab. 2012, 105, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cao, L.; Fang, J.; Yu, X.; Dai, X.; Miao, X. Ochronotic arthritis and ochronotic Achilles tendon rupture in alkaptonuria: A 6 years follow-up case report in China. Medicine 2019, 98, e16837. [Google Scholar] [CrossRef] [PubMed]
- Tokuhara, Y.; Shukuya, K.; Tanaka, M.; Mouri, M.; Ohkawa, R.; Fujishiro, M.; Takahashi, T.; Okubo, S.; Yokota, H.; Kurano, M.; et al. Detection of novel visible-light region absorbance peaks in the urine after alkalization in patients with alkaptonuria. PLoS One 2014, 9, e86606. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.A.; Dillon, J.P.; Sireau, N.; Timmis, O.; Ranganath, L.R. Alkaptonuria: An example of a “fundamental disease”—A rare disease with important lessons for more common disorders. Semin. Cell. Dev. Biol. 2016, 52, 53–57. [Google Scholar] [CrossRef]
- Taylor, A.M.; Boyde, A.; Wilson, P.J.; Jarvis, J.C.; Davidson, J.S.; Hunt, J.A.; Ranganath, L.R.; Gallagher, J.A. The role of calcified cartilage and subchondral bone in the initiation and progression of ochronotic arthropathy in alkaptonuria. Arthritis Rheum. 2011, 63, 3887–3896. [Google Scholar] [CrossRef]
- Hughes, J.H.; Keenan, C.M.; Sutherland, H.; Edwards, H.R.; Wilson, P.J.; Ranganath, L.R.; Jarvis, J.C.; Bou-Gharios, G.; Gallagher, J.A. Anatomical Distribution of Ochronotic Pigment in Alkaptonuric Mice is Associated with Calcified Cartilage Chondrocytes at Osteochondral Interfaces. Calcif. Tissue Int. 2021, 108, 207–218. [Google Scholar] [CrossRef]
- Boyce, B.F.; Xing, L. The RANKL/RANK/OPG pathway. Curr. Osteoporos. Rep. 2007, 5, 98–104. [Google Scholar] [CrossRef]
- Schiavone, M.L.; Millucci, L.; Bernardini, G.; Giustarini, D.; Rossi, R.; Marzocchi, B.; Santucci, A. Homogentisic acid affects human osteoblastic functionality by oxidative stress and alteration of the Wnt/β-catenin signaling pathway. J. Cell. Physiol. 2020, 235, 6808–6816. [Google Scholar] [CrossRef]
- Vo, T.; Edwards, J.R. A black heart: Aortic valve ochronosis secondary to alkaptonuria causing aortic stenosis. J. Card. Surg. 2021, 36, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Kisa, P.T.; Gunduz, M.; Dorum, S.; Uzun, O.U.; Cakar, N.E.; Yildirim, G.K.; Erdol, S.; Hismi, B.O.; Tugsal, H.Y.; Ucar, U.; et al. Alkaptonuria in Turkey: Clinical and molecular characteristics of 66 patients. Eur. J. Med. Genet. 2021, 64, 104197. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Bauer, E.; Myung, G.; Fang, M.A. Musculoskeletal manifestations of alkaptonuria: A case report and literature review. Eur. J. Rheumatol. 2019, 6, 98. [Google Scholar] [CrossRef]
- Spiga, O.; Cicaloni, V.; Visibelli, A.; Davoli, A.; Paparo, M.A.; Orlandini, M.; Vecchi, B.; Santucci, A. Towards a precision medicine approach based on machine learning for tailoring medical treatment in alkaptonuria. Int. J. Mol. Sci. 2021, 22, 1187. [Google Scholar] [CrossRef] [PubMed]
- Braconi, D.; Millucci, L.; Spiga, O.; Santucci, A. Cell and tissue models of alkaptonuria. Drug Discov. Today Dis. Models 2020, 31, 3–10. [Google Scholar] [CrossRef]
- Gambassi, S.; Geminiani, M.; Thorpe, S.D.; Bernardini, G.; Millucci, L.; Braconi, D.; Orlandini, M.; Thompson, C.L.; Petricci, E.; Manetti, F.; et al. Smoothened-antagonists reverse homogentisic acid-induced alterations of Hedgehog signaling and primary cilium length in alkaptonuria. J. Cell. Physiol. 2017, 232, 3103–3111. [Google Scholar] [CrossRef]
- Introne, W.J.; Perry, M.; Chen, M. Alkaptonuria. 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1454/ (accessed on 15 January 2023).
- Ranganath, L.R.; Milan, A.M.; Hughes, A.T.; Khedr, M.; Davison, A.S.; Shweihdi, E.; Norman, B.P.; Hughes, J.H.; Bygott, H.; Luangrath, E.; et al. Homogentisic acid is not only eliminated by glomerular filtration and tubular secretion but also produced in the kidney in alkaptonuria. J. Inherit. Metab. Dis. 2020, 43, 737–747. [Google Scholar] [CrossRef]
- Al-Tarawneh, A.; Al-Limoun, M.; Khlaifat, A.M.; Tarawneh, I.; Mwafi, N.; Khleifat, K.; Alqaraleh, M.; Mizher, H. Bacterial quality of urinary tract in patients with alkaptonuria. Am. J. Med. Sci. 2023. [Google Scholar] [CrossRef]
- Chévez Barrios, P.; Font, R.L. Pigmented conjunctival lesions as initial manifestation of ochronosis. Arch. Ophthalmol. 2004, 122, 1060–1063. [Google Scholar] [CrossRef][Green Version]
- Kumar, A.; Karthikeyan, K.; Vyas, M.T. Osler’s sign revisited. Indian Dermatol. Online J. 2015, 6, 308–309. [Google Scholar] [CrossRef]
- Damarla, N.; Linga, P.; Goyal, M.; Tadisina, S.R.; Reddy, G.S.; Bommisetti, H. Alkaptonuria: A case report. Indian J. Ophthalmol. 2017, 65, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Skinsnes, O.K. Generalized ochronosis; report of an instance in which it was misdiagnosed as melanosarcoma, with resultant enucleation of an eye. Arch. Pathol. 1948, 45, 552–558. [Google Scholar]
- Ranganath, L.R.; Jarvis, J.C.; Gallagher, J.A. Recent advances in management of alkaptonuria (invited review; best practice article). J. Clin. Pathol. 2013, 66, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Ranganath, L.R.; Khedr, M.; Milan, A.M.; Davison, A.S.; Hughes, A.T.; Usher, J.L.; Taylor, S.; Loftus, N.; Daroszewska, A.; West, E.; et al. Nitisinone arrests ochronosis and decreases rate of progression of alkaptonuria: Evaluation of the effect of nitisinone in the United Kingdom National Alkaptonuria Centre. Mol. Genet. Metab. 2018, 125, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Khedr, M.; Cooper, M.S.; Hughes, A.T.; Milan, A.M.; Davison, A.S.; Norman, B.P.; Sutherland, H.; Jarvis, J.C.; Fitzgerald, R.; Markinson, L.; et al. Nitisinone causes acquired tyrosinosis in alkaptonuria. J. Inherit. Metab. Dis. 2020, 43, 1014–1023. [Google Scholar] [CrossRef]
- Moran, G.R. 4-Hydroxyphenylpyruvate dioxygenase. Arch. Biochem. Biophys. 2005, 433, 117–128. [Google Scholar] [CrossRef]
- Pilka, E.S.; Shafqat, N.; Cocking, R.; Bray, J.E.; Krojer, T.; Pike, A.C.W.; von Delft, F.; Yue, W.W.; Arrowsmith, C.H.; Weigelt, J.; et al. Crystal structure of human 4-hydroxyphenylpyruvate dioxygenase. RCSB Protein Data Bank (PDB ID: 3ISQ), 2009. [Google Scholar]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef]
- Schneider, N.; Lange, G.; Hindle, S.; Klein, R.; Rarey, M. A consistent description of HYdrogen bond and DEhydration energies in protein–ligand complexes: Methods behind the HYDE scoring function. J. Comput.-Aided Mol. Des. 2013, 27, 15–29. [Google Scholar] [CrossRef]
- Zaib, S.; Munir, R.; Younas, M.T.; Kausar, N.; Ibrar, A.; Aqsa, S.; Shahid, N.; Asif, T.T.; Alsaab, H.O.; Khan, I. Hybrid Quinoline-Thiosemicarbazone Therapeutics as a New Treatment Opportunity for Alzheimer’s Disease–Synthesis, In Vitro Cholinesterase Inhibitory Potential and Computational Modeling Analysis. Molecules 2021, 26, 6573. [Google Scholar] [CrossRef]
- Mishra, S.; Dahima, R. In vitro ADME studies of TUG-891, a GPR-120 inhibitor using SWISS ADME predictor. J. Drug Deliv. Ther. 2019, 9, 366–369. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Zaib, S.; Rana, N.; Hussain, N.; Alrbyawi, H.; Dera, A.A.; Khan, I.; Khalid, M.; Khan, A.; Al-Harrasi, A. Designing multi-epitope monkeypox virus-specific vaccine using immunoinformatics approach. J. Infect. Public Health. 2023, 16, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Altman, T.; Travers, M.; Kothari, A.; Caspi, R.; Karp, P.D. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinform. 2013, 14, 112. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The single global macromolecular structure archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, D.L.; Prokai, L.; Prokai-Tatrai, K. The Antagonist pGlu-βGlu-Pro-NH2 Binds to an Allosteric Site of the Thyrotropin-Releasing Hormone Receptor. Molecules 2021, 26, 5397. [Google Scholar] [CrossRef]
- Reulecke, I.; Lange, G.; Albrecht, J.; Klein, R.; Rarey, M. Towards an integrated description of hydrogen bonding and dehydration: Decreasing false positives in virtual screening with the HYDE scoring function. ChemMedChem 2008, 3, 885–897. [Google Scholar] [CrossRef]
- Maass, P.; Schulz-Gasch, T.; Stahl, M.; Rarey, M. Recore: A fast and versatile method for scaffold hopping based on small molecule crystal structure conformations. J. Chem. Inf. Model. 2007, 47, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Chouhan, U. Pharmacoinformatic Studies on 4-Thiazolyl-phenoxy tail containing indanyl acetic acid derivatives as PPAR-Pan agonists as potent anti-diabetic agent. Indian J. Pharm. Edu. Res. 2019, 53, s288–s298. [Google Scholar] [CrossRef]
- VanDrie, J.H. ReCore. J. Am. Chem. Soc. 2009, 131, 1617. [Google Scholar] [CrossRef]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A Fast Flexible Docking Method Using an Incremental Construction Algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.C.; Freitas, H.F.; Campos, J.M.; Kimani, N.M.; Silva, C.H.; Borges, R.S.; Pita, S.S.; Santos, C.B. Natural products-based drug design against SARS-CoV-2 Mpro 3CLpro. Int. J. Mol. Sci. 2021, 22, 11739. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Pantaleão, S.Q.; Fernandes, P.O.; Gonçalves, J.E.; Maltarollo, V.G.; Honorio, K.M. Recent advances in the prediction of pharmacokinetics properties in drug design studies: A review. ChemMedChem 2022, 17, e202100542. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, A.; Gupta, U. Molecular Docking studies on the Anti-fungal activity of Allium sativum (Garlic) against Mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1. 0.0. J. Antivir. Antiretrovir. 2021, 5, 028–032. [Google Scholar] [CrossRef]
- Baskaran, K.P.; Arumugam, A.; Kandasamy, R.; Alagarsamy, S. In silico method for prediction of maximum binding affinity and ligand-protein interaction studies on Alzheimer’s disease. Int. J. Res. Granthaalayah. 2020, 8, 362–370. [Google Scholar] [CrossRef]
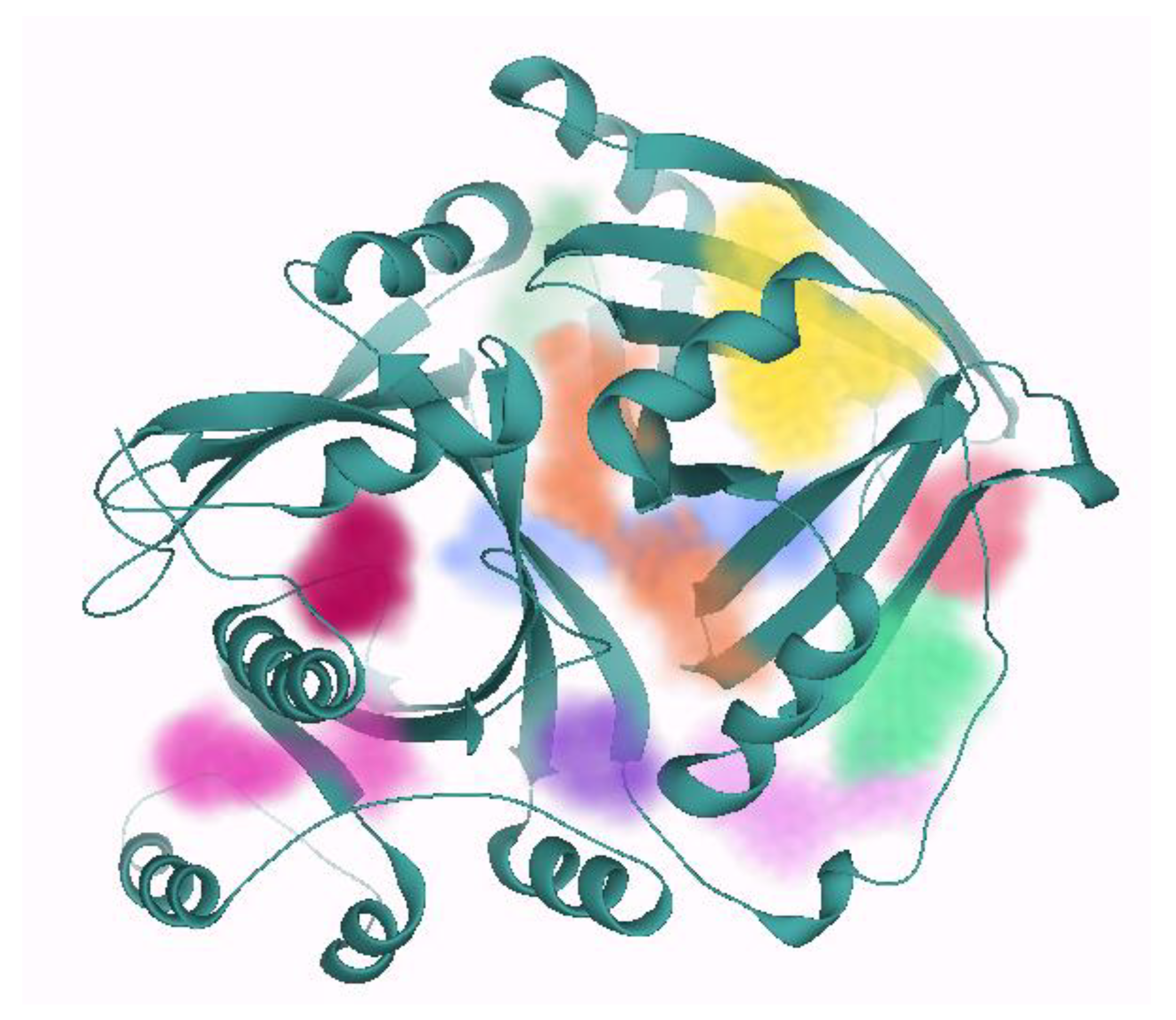


| Pocket ID | Number of Residues | DoGSiteScore | Number of Donors | Number of Acceptors | Hydrophobicity | Solvent Accessible Surface (Å2) | Total Volume of the Pocket (Å3) | |
|---|---|---|---|---|---|---|---|---|
| 1 | 23 | 0.42 | 11 | 14 | 0.71 | 283.32 | 538.27 | |
| 2 | 24 | 0.39 | 11 | 8 | 0.76 | 267.12 | 247.75 | |
| 3 | 17 | 0.34 | 4 | 12 | 0.77 | 194.40 | 164.16 | |
| 4 | 25 | 0.31 | 18 | 14 | 0.67 | 392.04 | 477.36 | |
| 5 | 16 | 0.31 | 5 | 9 | 0.76 | 158.76 | 170.86 | |
| 6 | 18 | 0.28 | 11 | 9 | 0.73 | 200.16 | 237.60 | |
| 7 | 9 | 0.16 | 3 | 6 | 0.74 | 86.04 | 112.10 | |
| 8 | 27 | 0.16 | 17 | 19 | 0.67 | 293.76 | 266.33 | |
| 9 | 12 | 0.12 | 6 | 8 | 0.69 | 129.60 | 192.89 | |
| 10 | 17 | 0.11 | 6 | 9 | 0.67 | 171.36 | 239.11 | |
| Compounds | Structures | Compounds | Structures |
|---|---|---|---|
| a | 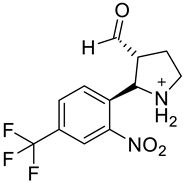 | b | 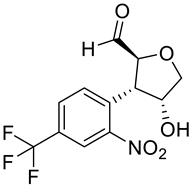 |
| c |  | d | 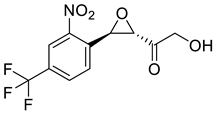 |
| e |  | f | 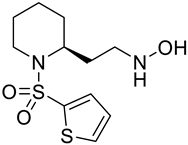 |
| g | 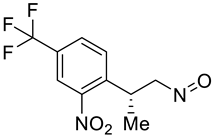 | h |  |
| i |  | j | 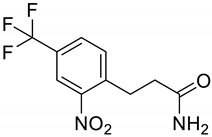 |
| Compounds | a | b | c | d | e | f | g | h | i | j |
|---|---|---|---|---|---|---|---|---|---|---|
| Formula | C12H12F3N2O3+ | C12H10F3NO5 | C15H13F3N2O4 | C11H8F3NO5 | C12H10F3N2O5+ | C11H18N2O3S2 | C10H9F3N2O3 | C14H10F3NO5 | C13H12F3NO6 | C10H9F3N2O3 |
| Molecular weight | 289.23 g/mol | 305.21 g/mol | 342.27 g/mol | 291.18 g/mol | 319.21 g/mol | 290.40 g/mol | 262.19 g/mol | 329.23 g/mol | 335.23 g/mol | 262.19 g/mol |
| Heavy atoms | 20 | 21 | 24 | 20 | 22 | 18 | 18 | 23 | 23 | 18 |
| Aromatic heavy atoms | 6 | 6 | 6 | 6 | 6 | 5 | 6 | 6 | 6 | 6 |
| Fraction Csp3 | 0.42 | 0.42 | 0.40 | 0.36 | 0.42 | 0.64 | 0.40 | 0.36 | 0.46 | 0.30 |
| Rotatable bonds | 4 | 4 | 5 | 5 | 4 | 5 | 5 | 4 | 4 | 5 |
| H-bond acceptors | 6 | 8 | 8 | 8 | 8 | 5 | 7 | 8 | 9 | 6 |
| H-bond donors | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 2 | 1 |
| Molar refractivity | 70.23 | 64.79 | 76.07 | 59.99 | 73.05 | 74.48 | 59.58 | 72.78 | 70.76 | 57.75 |
| TPSA (Å2) | 79.50 | 92.35 | 93.20 | 95.65 | 103.20 | 106.26 | 75.25 | 97.03 | 112.58 | 88.91 |
| Consensus Log Po/w | 1.13 | 1.20 | 1.76 | 1.14 | −0.17 | 1.54 | 2.56 | 1.99 | 0.99 | 1.62 |
| Class | Soluble | Soluble | Soluble | Soluble | Soluble | Soluble | Soluble | Soluble | Soluble | Soluble |
| GI absorption | High | High | High | High | High | High | High | High | High | High |
| BBB permeant | No | No | No | No | No | No | No | No | No | No |
| P-gp substrate | No | No | No | No | Yes | No | No | No | No | No |
| CYP1A2 inhibitor | No | No | No | No | No | No | Yes | No | No | Yes |
| CYP2C19 inhibitor | No | No | No | No | No | No | Yes | Yes | No | No |
| CYP2C9 inhibitor | No | No | No | No | No | No | No | No | No | No |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | No | No | No | No | No | No |
| Log Kp (skin permetion) | −6.90 cm/s | −7.27 cm/s | −7.73 cm/s | −7.20 cm/s | −7.35 cm/s | −7.06 cm/s | −6.07 cm/s | −6.67 cm/s | −7.22 cm/s | −6.66 cm/s |
| Lipinski | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation |
| Ghose | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Veber | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Egan | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Muegge | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| PAINS | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert |
| Lead-likeness | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Synthetic accessibility | 3.07 | 3.47 | 2.63 | 3.27 | 3.23 | 3.52 | 2.76 | 2.46 | 3.76 | 2.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaib, S.; Rana, N.; Hussain, N.; Ogaly, H.A.; Dera, A.A.; Khan, I. Identification of Potential Inhibitors for the Treatment of Alkaptonuria Using an Integrated In Silico Computational Strategy. Molecules 2023, 28, 2623. https://doi.org/10.3390/molecules28062623
Zaib S, Rana N, Hussain N, Ogaly HA, Dera AA, Khan I. Identification of Potential Inhibitors for the Treatment of Alkaptonuria Using an Integrated In Silico Computational Strategy. Molecules. 2023; 28(6):2623. https://doi.org/10.3390/molecules28062623
Chicago/Turabian StyleZaib, Sumera, Nehal Rana, Nadia Hussain, Hanan A. Ogaly, Ayed A. Dera, and Imtiaz Khan. 2023. "Identification of Potential Inhibitors for the Treatment of Alkaptonuria Using an Integrated In Silico Computational Strategy" Molecules 28, no. 6: 2623. https://doi.org/10.3390/molecules28062623
APA StyleZaib, S., Rana, N., Hussain, N., Ogaly, H. A., Dera, A. A., & Khan, I. (2023). Identification of Potential Inhibitors for the Treatment of Alkaptonuria Using an Integrated In Silico Computational Strategy. Molecules, 28(6), 2623. https://doi.org/10.3390/molecules28062623







