MSF Enhances Human Antimicrobial Peptide β-Defensin (HBD2 and HBD3) Expression and Attenuates Inflammation via the NF-κB and p38 Signaling Pathways
Abstract
:1. Introduction
2. Results
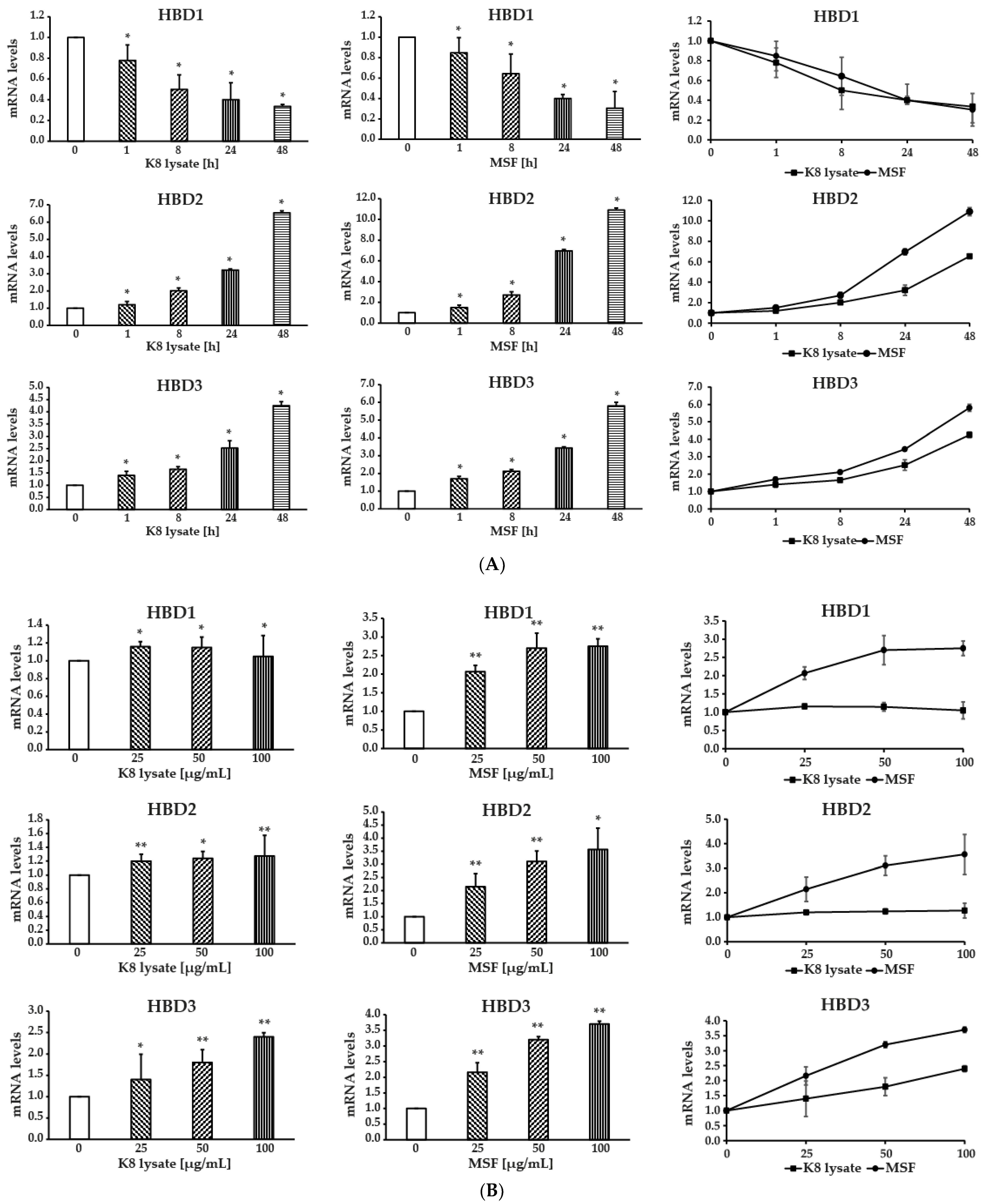
2.1. MSF Significantly Enhanced the Expression of Human β-Defensin-2 and β-Defensin-3
2.2. Effect of MSF on the PI3K, NF-κB, and MAPKs Signaling Pathways in HaCaT Cells
2.3. MSF Promoted the Expression of HBD2 and HBD3 through p38 Pathway
2.4. NF-κB Involved in the Induction of HBD3 and HBD2 Expression
2.5. MSF Inhibited S. aureus-Induced Inflammation in THP1 Cells
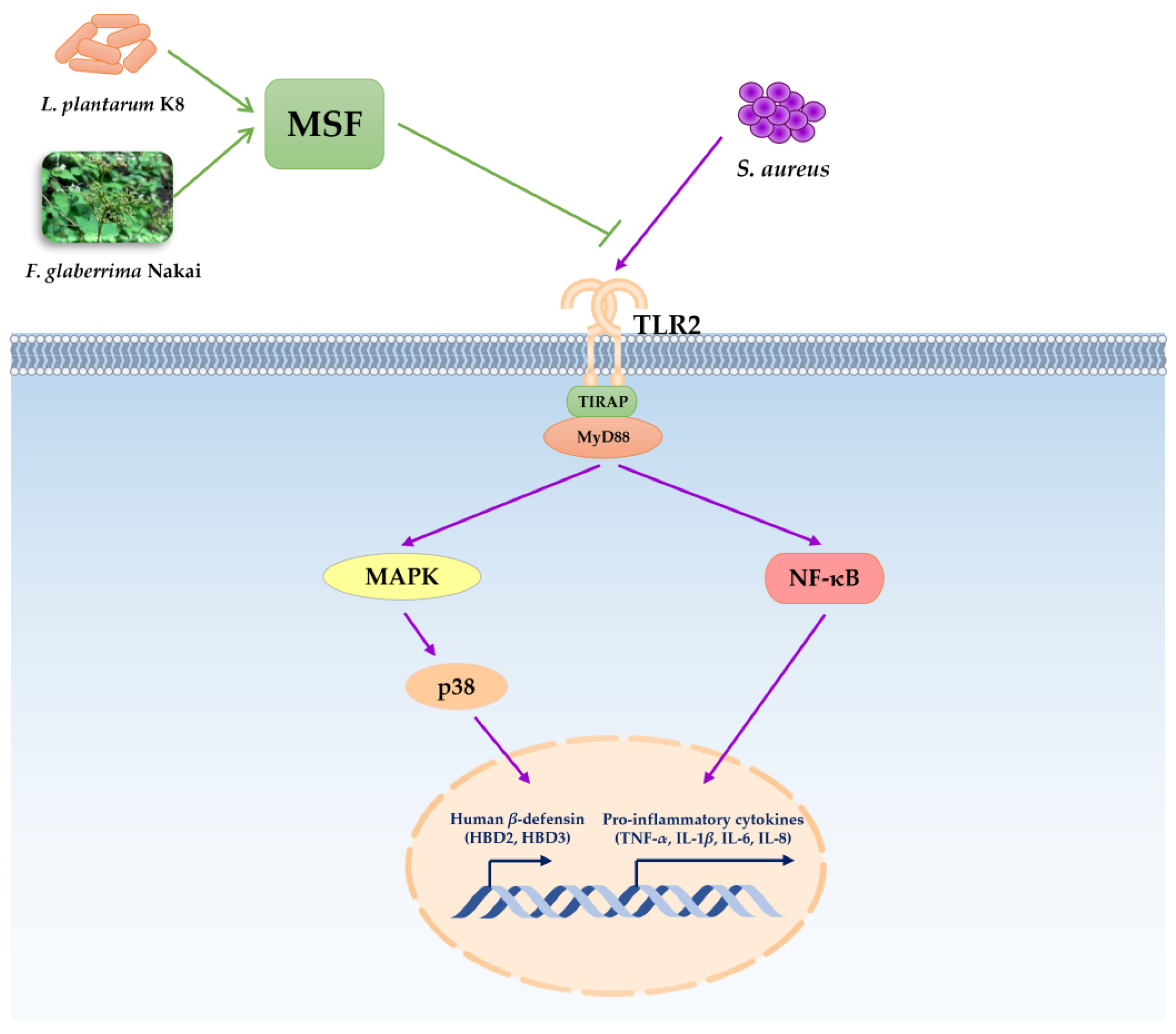
2.6. TLR2 Was Involved in Regulating Human β-Defensin and Inflammation of MSF
2.7. MSF Ameliorated S. Aureus-Induced Skin Inflammation in Mice
3. Discussion
4. Materials and Methods
4.1. Preparation of MSF
4.2. Cell Culture and Stimulation
4.3. S. aureus Preparation
4.4. Cell Viability
4.5. qRT-PCR Analysis
4.6. Western Blot Analysis
4.7. Animal Experiments
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| AKT | Protein kinase B |
| AMPs | Antimicrobial peptides |
| CCARM | Culture Collection of Antimicrobial Resistant Microbes |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DMSO | Dimethyl sulfoxide |
| ERK1/2 | Extracellular signal-regulated kinases 1 and 2 |
| FBS | Fetal bovine serum |
| FGE | F. glaberrima extracts |
| hBD1 | Human β-defensins 1 |
| hBD2 | Human β-defensins 2 |
| hBD3 | Human β-defensins 3 |
| HNP | Human neutrophil peptide |
| IL-1β | Interleukin 1 beta |
| IL-6 | Interleukin 6 |
| IL-8 | Interleukin 8 |
| IκB | Inhibitory kappa B alpha |
| JNK | c-Jun N-terminal kinase |
| MAPK | Mitogen-activated protein kinase |
| MSF | Miracle synergy material made using F. glaberrima |
| NF-κB | Nuclear Factor-kappa B |
| PBS | Phosphate-buffered saline |
| PI3K | Phosphoinositide 3-kinase |
| TLR2 | Toll-like receptor 2 |
| TNF-α | Tumor necrosis factor alpha |
| TSB | Tryptic soy broth |
References
- Clausen, M.-L.; Agner, T. Antimicrobial Peptides, Infections and the Skin Barrier. Curr. Probl. Dermatol 2016, 49, 38–46. [Google Scholar] [CrossRef]
- Jenei, A.; Kalló, G.; Dajnoki, Z.; Gáspár, K.; Szegedi, A.; Kapitány, A.; Csősz, É. Detection of Antimicrobial Peptides in Stratum Corneum by Mass Spectrometry. Int. J. Mol. Sci. 2021, 22, 4233. [Google Scholar] [CrossRef]
- Cui, J.; Chen, Y.; Wang, H.Y.; Wang, R.-F. Mechanisms and Pathways of Innate Immune Activation and Regulation in Health and Cancer. Hum. Vaccin Immunother. 2014, 10, 3270–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Lu, W. Defensins in Innate Immunity. Curr. Opin. Hematol. 2014, 21, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted Immune Functions of Human Defensins and Underlying Mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Sechet, E.; Telford, E.; Bonamy, C.; Sansonetti, P.J.; Sperandio, B. Natural Molecules Induce and Synergize to Boost Expression of the Human Antimicrobial Peptide β-Defensin-3. PNAS 2018, 115, E9869–E9878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koenderman, L.; Buurman, W.; Daha, M.R. The Innate Immune Response. Immunol. Lett. 2014, 162, 95–102. [Google Scholar] [CrossRef]
- Vane, J.R.; Botting, R.M. Anti-Inflammatory Drugs and Their Mechanism of Action. Inflamm. Res. 1998, 47, 78–87. [Google Scholar] [CrossRef]
- Choi, K.-H.; Hong, J.; Kim, K.-Y.; Kim, H.; Lee, S.; Lee, Y.; Chung, D.-K. Anti-Inflammatory Properties of MSF, a Lactiplantibacillus Plantarum K8 Lysate Fermented with Filipendula Glaberrima Extract. Appl. Sci. 2022, 12, 2602. [Google Scholar] [CrossRef]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus Plantarum–Nomad and Ideal Probiotic. Front. Microbiol. 2021, 12, 2911. [Google Scholar] [CrossRef]
- Jung, J.; Kim, H.; Lee, S.; Hong, M.; Hwang, D. Antioxidant and Anti-Inflammatory Activity of Filipendula Glaberrima Nakai Ethanolic Extract and Its Chemical Composition. Molecules 2022, 27, 4628. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [Green Version]
- Steubesand, N.; Kiehne, K.; Brunke, G.; Pahl, R.; Reiss, K.; Herzig, K.-H.; Schubert, S.; Schreiber, S.; Fölsch, U.R.; Rosenstiel, P.; et al. The Expression of the β-Defensins HBD-2 and HBD-3 Is Differentially Regulated by NF-κB and MAPK/AP-1 Pathways in an in Vitro Model of Candida Esophagitis. BMC Immunol. 2009, 10, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arthur, J.S.C.; Ley, S.C. Mitogen-Activated Protein Kinases in Innate Immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shepherd, E.G.; Nelin, L.D. MAPK Phosphatases—Regulating the Immune Response. Nat. Rev. Immunol. 2007, 7, 202–212. [Google Scholar] [CrossRef]
- Ghosh, S.; Hayden, M.S. New Regulators of NF-κB in Inflammation. Nat. Rev. Immunol. 2008, 8, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Zhang, J.; Zhang, S.; Cao, J.; Fu, Y.; Hu, X.; Zhao, J.; Gu, B.; Li, Q.; Zhang, K.; et al. TLR2, TLR4, and NLRP3 Mediated the Balance between Host Immune-Driven Resistance and Tolerance in Staphylococcus aureus-Infected Mice. Microb. Pathog. 2022, 169, 105671. [Google Scholar] [CrossRef]
- Kielian, T.; Esen, N.; Bearden, E.D. Toll-like Receptor 2 (TLR2) Is Pivotal for Recognition of S. aureus Peptidoglycan but Not Intact Bacteria by Microglia. Glia 2005, 49, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Kim, K. Inhibition of Proinflammatory Cytokines in Cutibacterium Acnes -Induced Inflammation in HaCaT Cells by Using Buddleja Davidii Aqueous Extract. Int. J. Inflam. 2020, 2020, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Guo, H.; Li, Y.; Meng, X.; Yan, L.; Zhang, D.; Wu, S.; Zhou, H.; Peng, L.; Xie, Q.; et al. Oleoylethanolamide Exerts Anti-Inflammatory Effects on LPS-Induced THP-1 Cells by Enhancing PPARα Signaling and Inhibiting the NF-κB and ERK1/2/AP-1/STAT3 Pathways. Sci. Rep. 2016, 6, 34611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.L.; Goh, C.C.; Ng, L.G. Imaging of Inflammatory Responses in the Mouse Ear Skin. Methods Mol. Biol. 2018, 1763, 87–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, J.-G.; Kim, K.-Y. Trichosanthes Kirilowii Extract Promotes Wound Healing through the Phosphorylation of ERK1/2 in Keratinocytes. Biomimetics 2022, 7, 154. [Google Scholar] [CrossRef] [PubMed]












| Primers | Forward | Reverse |
|---|---|---|
| hBD1 | GTCGCCATGAGAACTTCCTACC | CATTGCCCTCCACTGCTGAC |
| hBD2 | CCTGTTACCTGCCTTAAGAGTG | GAATCCGCATCAGCCACAG |
| hBD3 | CTTCTGTTTGCTTTGCTCTTCC | CACTTGCCGATCTGTTCCTC |
| hTNF-α | TGAGCACTGAAAGCATGATCC | ATCACTCCAAAGTGCAGCAG |
| hIL-1β | TCTTTGAAGAAGAGCCCGTCCTC | GGATCCACACTCTCCAGCTGCA |
| hIL-6 | CCTGAACCTTCCAAAGATGGC | CACCAGGCAAGTCTCCTCATT |
| hIL-8 | TCTGTGTGAAGGTGCAGTTTT | GGGGTGGAAAGGTTTGGAGTA |
| hTLR2 | TGTCTTGTGACCGCAATGGT | GTTGGACAGGTCAAGGCTTT |
| hTLR4 | CCCTGAGGCATTTAGGCAGCTA | AGGTAGAGAGGTGGCTTAGGCT |
| hGAPDH | GTGAAGGTCGGAGTCAACG | TGAGGTCAATGAAGGGGTC |
| Antibody | Source | Category No. |
|---|---|---|
| p-PI3K | Cell Signaling Technology | 4257S |
| PI3K | Cell Signaling Technology | 4228S |
| p-AKT | Cell Signaling Technology | 9271S |
| AKT | Cell Signaling Technology | 9272S |
| p-NF-κB | Cell Signaling Technology | 3033S |
| NF-κB | Santa Cruz Biotechnology | sc-8008 |
| p-p38 | Cell Signaling Technology | 9211S |
| p38 | Santa Cruz Biotechnology | sc-535 |
| p-ERK1/2 | Cell Signaling Technology | 9101S |
| ERK1/2 | Cell Signaling Technology | 9102S |
| p-JNK | Santa Cruz Biotechnology | sc-6254 |
| JNK | Santa Cruz Biotechnology | sc-7345 |
| HBD2 | Abcam | ab9871 |
| HBD3 | Abcam | ab172703 |
| GAPDH | Cell Signaling Technology | 14C10 |
| Left Ear | Right Ear | |||
|---|---|---|---|---|
| Compound | Pathogen | Compound | Pathogen | |
| Group 1 | None | None | PBS | None |
| Group 2 | None | None | None | S. aureus |
| Group 3 | K8 lysate | None | PBS | None |
| Group 4 | K8 lysate | S. aureus | PBS | S. aureus |
| Group 5 | MSF | None | PBS | None |
| Group 6 | MSF | S. aureus | PBS | S. aureus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, A.-T.; Kim, M.; Kim, Y.-E.; Kim, H.; Lee, S.; Lee, Y.; Kim, K.-Y. MSF Enhances Human Antimicrobial Peptide β-Defensin (HBD2 and HBD3) Expression and Attenuates Inflammation via the NF-κB and p38 Signaling Pathways. Molecules 2023, 28, 2744. https://doi.org/10.3390/molecules28062744
Nguyen A-T, Kim M, Kim Y-E, Kim H, Lee S, Lee Y, Kim K-Y. MSF Enhances Human Antimicrobial Peptide β-Defensin (HBD2 and HBD3) Expression and Attenuates Inflammation via the NF-κB and p38 Signaling Pathways. Molecules. 2023; 28(6):2744. https://doi.org/10.3390/molecules28062744
Chicago/Turabian StyleNguyen, Anh-Thu, Minho Kim, Ye-Eun Kim, Hangeun Kim, Sanghyun Lee, Yunji Lee, and Ki-Young Kim. 2023. "MSF Enhances Human Antimicrobial Peptide β-Defensin (HBD2 and HBD3) Expression and Attenuates Inflammation via the NF-κB and p38 Signaling Pathways" Molecules 28, no. 6: 2744. https://doi.org/10.3390/molecules28062744
APA StyleNguyen, A. -T., Kim, M., Kim, Y. -E., Kim, H., Lee, S., Lee, Y., & Kim, K. -Y. (2023). MSF Enhances Human Antimicrobial Peptide β-Defensin (HBD2 and HBD3) Expression and Attenuates Inflammation via the NF-κB and p38 Signaling Pathways. Molecules, 28(6), 2744. https://doi.org/10.3390/molecules28062744







