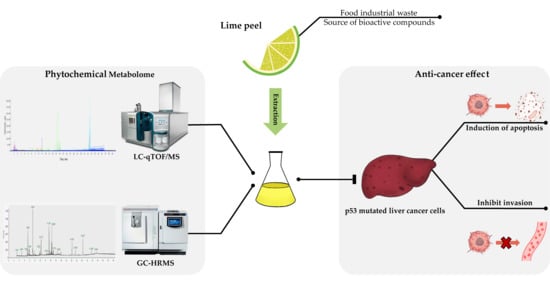
Metabolomic Analysis of Phytochemical Compounds from Ethanolic Extract of Lime (Citrus aurantifolia) Peel and Its Anti-Cancer Effects against Human Hepatocellular Carcinoma Cells
Abstract
:1. Introduction
2. Results
2.1. LC-qTOF/MS Screening and Identification
2.2. GC-HRMS Screening and Identification
2.3. The Concentration of Hesperidin and Limonin in the Extract
2.4. Cytotoxicity on PLC/PRF/5 Hepatoma Cells
2.5. Anti-Invasion Effect on PLC/PRF/5 Hepatoma Cells
2.6. Induction of Apoptosis in PLC/PRF/5 Hepatoma Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Preparation of Extract
4.3. Metabolomics Analysis
4.3.1. Using LC-MS/MS
4.3.2. Using GC-HRMS Analysis
4.4. Quantitative Measurement of Hesperidin and Limonin
4.5. Cell Culture
4.6. Cell Viability Assay
4.7. Cell Invasion Assay
4.8. Cell Apoptosis Assay
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- International Agency for Research on Cancer, World Health Organization. Globocans 2020: Liver. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/11-Liver-fact-sheet.pdf (accessed on 9 February 2023).
- World Cancer Research Fund. Worldwide Cancer Data by Cancer Type. Available online: https://www.wcrf.org/cancer-trends/worldwide-cancer-data/ (accessed on 9 February 2023).
- National Cancer Institute. Cancer Stat Facts: Liver and Intrahepatic Bile Duct Cancer. Available online: https://seer.cancer.gov/statfacts/html/livibd.html (accessed on 9 February 2023).
- Mittal, S.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma: Consider the Population. J. Clin. Gastroenterol. 2013, 47, S2–S6. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- Caron de Fromentel, C.; Levrero, M. p53 functional loss, stemness and hepatocellular carcinoma. Hepatoma Res. 2020, 6, 80. [Google Scholar] [CrossRef]
- Choudhari, A.S.; Mandave, P.C.; Deshpande, M.; Ranjekar, P.; Prakash, O. Phytochemicals in Cancer Treatment: From Preclinical Studies to Clinical Practice. Front. Pharmacol. 2020, 10, 1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahbandeh, M. Leading Fresh Lemon and Lime Producers Worldwide in 2020/2021. Available online: https://www.statista.com/statistics/1045016/world-lemons-and-limes-major-producers/ (accessed on 18 November 2021).
- Lau, K.Q.; Sabran, M.R.; Shafie, S.R. Utilization of Vegetable and Fruit By-products as Functional Ingredient and Food. Front Nutr. 2021, 8, 661693. [Google Scholar] [CrossRef]
- Mahato, N.; Sinha, M.; Sharma, K.; Koteswararao, R.; Cho, M.H. Modern Extraction and Purification Techniques for Obtaining High Purity Food-Grade Bioactive Compounds and Value-Added Co-Products from Citrus Wastes. Foods 2019, 8, 523. [Google Scholar] [CrossRef] [Green Version]
- Nogata, Y.; Sakamoto, K.; Shiratsuchi, H.; Ishii, T.; Yano, M.; Ohta, H. Flavonoid composition of fruit tissues of citrus species. Biosci. Biotechnol. Biochem. 2006, 70, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, V.; Tuli, H.S.; Thakral, F.; Singhal, P.; Aggarwal, D.; Srivastava, S.; Pandey, A.; Sak, K.; Varol, M.; Khan, M.A.; et al. Molecular mechanisms of action of hesperidin in cancer: Recent trends and advancements. Exp. Biol. Med. 2020, 245, 486–497. [Google Scholar] [CrossRef] [Green Version]
- Ghorbani, A.; Nazari, M.; Jeddi-Tehrani, M.; Zand, H. The citrus flavonoid hesperidin induces p53 and inhibits NF-κB activation in order to trigger apoptosis in NALM-6 cells: Involvement of PPARγ-dependent mechanism. Eur. J. Nutr. 2012, 51, 39–46. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, C.; Luo, T.; Wang, J.; Tang, Y.; Chen, Z.; Yu, L. Limonin: A Review of Its Pharmacology, Toxicity, and Pharmacokinetics. Molecules 2019, 24, 3679. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Liu, J.; Zhao, W. By blocking hexokinase-2 phosphorylation, limonin suppresses tumor glycolysis and induces cell apoptosis in hepatocellular carcinoma. Onco Targets 2018, 11, 3793–3803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langeswaran, K.; Gowthamkumar, S.; Vijayaprakash, S.; Revathy, R.; Balasubramanian, M.P. Influence of limonin on Wnt signalling molecule in HepG2 cell lines. J. Nat. Sci. Biol. Med. 2013, 4, 126–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banjerdpongchai, R.; Wudtiwai, B.; Khaw-On, P.; Rachakhom, W.; Duangnil, N.; Kongtawelert, P. Hesperidin from Citrus seed induces human hepatocellular carcinoma HepG2 cell apoptosis via both mitochondrial and death receptor pathways. Tumour Biol. 2016, 37, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Phucharoenrak, P.; Muangnoi, C.; Trachootham, D. A Green Extraction Method to Achieve the Highest Yield of Limonin and Hesperidin from Lime Peel Powder (Citrus aurantifolia). Molecules 2022, 27, 820. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, S.; Kaul, R.; Sofi, S.A.; Bashir, N.; Nazir, F.; Ahmad Nayik, G. Citrus peel as a source of functional ingredient: A review. J. Saudi Soc. Agric. Sci. 2018, 17, 351–358. [Google Scholar] [CrossRef] [Green Version]
- Arias, B.Á.; Ramón-Laca, L. Pharmacological properties of citrus and their ancient and medieval uses in the Mediterranean region. J. Ethnopharmacol. 2005, 97, 89–95. [Google Scholar] [CrossRef]
- Zhou, J.L.; Qi, L.W.; Li, P. Herbal medicine analysis by liquid chromatography/time-of-flight mass spectrometry. J. Chromatogr. A 2009, 1216, 7582–7594. [Google Scholar] [CrossRef] [PubMed]
- Gomathi, D.; Kalaiselvi, M.; Ravikumar, G.; Devaki, K.; Uma, C. GC-MS analysis of bioactive compounds from the whole plant ethanolic extract of Evolvulus alsinoides (L.) L. J. Food Sci. Technol. 2015, 52, 1212–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pallavi, M.; Ramesh, C.; Krishna, V.; Parveen, S.; Nanjunda Swamy, L. Quantitative phytochemical analysis and antioxidant activities of some Citrus fruits of South India. Asian J. Pharm. Clin. Res. 2017, 10, 198. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Yang, X.; Chen, B.; Chao, Y.X.; Hu, P.J.; Cai, Y.; Wu, B.; Wei, M. Identification and determination of chemical constituents of Citrus reticulata semen through ultrahigh-performance liquid chromatography combined with Q Exactive Orbitrap tandem mass spectrometry. J. Sep. Sci. 2019, 43, 438–451. [Google Scholar] [CrossRef]
- Yang, M.; Jiang, Z.; Wen, M.; Wu, Z.; Zha, M.; Xu, W.; Zhang, L. Chemical Variation of Chenpi (Citrus Peels) and Corresponding Correlated Bioactive Compounds by LC-MS Metabolomics and Multibioassay Analysis. Front. Nutr. 2022, 9, 825381. [Google Scholar] [CrossRef]
- Miao, W.; Liu, X.; Li, N.; Bian, X.; Zhao, Y.; He, J.; Zhou, T.; Wu, J.-L. Polarity-extended composition profiling via LC-MS-based metabolomics approaches: A key to functional investigation of Citrus aurantium L. Food Chem. 2023, 405, 134988. [Google Scholar] [CrossRef] [PubMed]
- Aripin, D.; Julaeha, E.; Dardjan, M.; Cahyanto, A. Chemical composition of Citrus spp. and oral antimicrobial effect of Citrus spp. peels essential oils against Streptococcus mutans. Padjadjaran J. Dent. 2015, 27, 1–11. [Google Scholar] [CrossRef]
- Kamal, G.M.; Anwar, F.; Hussain, A.; Sarri, N.; Ashraf, M. Yield and chemical composition of Citrus essential oils as affected by drying pretreatment of peels. Int. Food Res. J. 2011, 18, 1275–1282. [Google Scholar]
- Sutedja, A.M.; Yanase, E.; Batubara, I.; Fardiaz, D.; Lioe, H.N. Thermal Stability of Anisoyl Kaempferol Glycosides in Jack Bean (Canavalia ensiformis (L.) DC) and Their Effect on α-Glucosidase Inhibition. J. Agric. Food Chem. 2022, 70, 2695–2700. [Google Scholar] [CrossRef] [PubMed]
- Yusoff, Z.; Muhammad, Z.; Kasuan, N.; Fazalul Rahiman, M.H.; Taib, M.N. Effect of temperature on Kaffir lime oil by using hydro-diffusion steam distillation system. Malays. J. Anal. Sci. 2013, 17, 326–339. [Google Scholar]
- Berger, R.G. Flavours and Fragrances: Chemistry, Bioprocessing and Sustainability; Springer: Berlin, Germany; New York, NY, USA, 2007; p. 648. [Google Scholar]
- Pan, M.-H.; Ghai, G.; Ho, C.-T. Food bioactives, apoptosis, and cancer. Mol. Nutr. Food Res. 2008, 52, 43–52. [Google Scholar] [CrossRef]
- Shi, Y.-S.; Zhang, Y.; Li, H.-T.; Wu, C.-H.; El-Seedi, H.R.; Ye, W.-K.; Wang, Z.-W.; Li, C.-B.; Zhang, X.-F.; Kai, G.-Y. Limonoids from Citrus: Chemistry, anti-tumor potential, and other bioactivities. J. Funct. Foods 2020, 75, 104213. [Google Scholar] [CrossRef]
- Koolaji, N.; Shammugasamy, B.; Schindeler, A.; Dong, Q.; Dehghani, F.; Valtchev, P. Citrus Peel Flavonoids as Potential Cancer Prevention Agents. Curr. Dev. Nutr. 2020, 4, nzaa025. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.; Khan, H.; Aschner, M.; Mirzae, H.; Küpeli Akkol, E.; Capasso, R. Anticancer Potential of Furanocoumarins: Mechanistic and Therapeutic Aspects. Int. J. Mol. Sci. 2020, 21, 5622. [Google Scholar] [CrossRef]
- Daemer, R.J.; Feinstone, S.M.; Alexander, J.J.; Tully, J.G.; London, W.T.; Wong, D.C.; Purcell, R.H. PLC/PRF/5 (Alexander) hepatoma cell line: Further characterization and studies of infectivity. Infect. Immun. 1980, 30, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Garg, S.; Zaneveld, L.J.D.; Singla, A.K. Chemistry and pharmacology of the citrus bioflavonoid hesperidin. Phytother. Res. 2001, 15, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Zhang, Z.; Liu, C.; Li, D.; Huang, W.; Liu, C.; Jiang, N. Hesperetin and Hesperidin Improved β-Carotene Incorporation Efficiency, Intestinal Cell Uptake, and Retinoid Concentrations in Tissues. J. Agric. Food Chem. 2019, 67, 3363–3371. [Google Scholar] [CrossRef] [PubMed]
- Bulusu, K.C.; Guha, R.; Mason, D.J.; Lewis, R.P.; Muratov, E.; Kalantar Motamedi, Y.; Cokol, M.; Bender, A. Modelling of compound combination effects and applications to efficacy and toxicity: State-of-the-art, challenges and perspectives. Drug Discov. Today 2016, 21, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef] [Green Version]
- Liebl, M.C.; Hofmann, T.G. The Role of p53 Signaling in Colorectal Cancer. Cancers 2021, 13, 2125. [Google Scholar] [CrossRef]
- Bae, J.R.; Park, W.H.; Suh, D.H.; No, J.H.; Kim, Y.B.; Kim, K. Role of limonin in anticancer effects of Evodia rutaecarpa on ovarian cancer cells. BMC Complement. Med. 2020, 20, 94. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, Y.; Gao, J.; De, Y. Hesperidin inhibits ovarian cancer cell viability through endoplasmic reticulum stress signaling pathways. Oncol. Lett. 2017, 14, 5569–5574. [Google Scholar] [CrossRef] [Green Version]
- Aktary, Z.; Kulak, S.; Mackey, J.; Jahroudi, N.; Pasdar, M. Plakoglobin interacts with the transcription factor p53 and regulates the expression of 14-3-3σ. J. Cell Sci. 2013, 126 Pt 14, 3031–3042. [Google Scholar] [CrossRef] [Green Version]
- Alaee, M.; Padda, A.; Mehrabani, V.; Churchill, L.; Pasdar, M. The physical interaction of p53 and plakoglobin is necessary for their synergistic inhibition of migration and invasion. Oncotarget 2016, 7, 26898–26915. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Prives, C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 2007, 26, 2220–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozenberg, J.M.; Zvereva, S.; Dalina, A.; Blatov, I.; Zubarev, I.; Luppov, D.; Bessmertnyi, A.; Romanishin, A.; Alsoulaiman, L.; Kumeiko, V.; et al. The p53 family member p73 in the regulation of cell stress response. Biol. Direct. 2021, 16, 23. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). Monographs on the Evaluation of Carcinogenic Risks to Humans, Volume 63. Dry Cleaning, Some Chlorinated Solvents and Other Industrial Chemicals; IARC: Lyon, France, 1995; pp. 3194–3407.
- Carthew, P.; DiNovi, M.; Setzer, R.W. Application of the margin of exposure (MoE) approach to substances in food that are genotoxic and carcinogenic: Example: Furan (CAS No. 110-00-9). Food Chem. Toxicol. 2010, 48 (Suppl. 1), S69–S74. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Chao, Y.; Liu, M.; Yang, Y.; Zhang, D.; Wang, K.; Tao, Y.; Zhang, J.-Y.; Li, Y.; Wei, M. Evaluation of dynamic changes in the bioactive components in Citri Reticulatae Pericarpium (Citrus reticulata ‘Chachi’) under different harvesting and drying conditions. J. Sci. Food Agric. 2020, 101, 3280–3289. [Google Scholar] [CrossRef]
- Seok, Y.J.; Her, J.Y.; Kim, Y.G.; Kim, M.Y.; Jeong, S.Y.; Kim, M.K.; Lee, J.Y.; Kim, C.I.; Yoon, H.J.; Lee, K.G. Furan in Thermally Processed Foods—A Review. Toxicol. Res. 2015, 31, 241–253. [Google Scholar] [CrossRef]
- Gong, M.; Zhou, Z.; Liu, S.; Zhu, S.; Li, G.; Zhong, F.; Mao, J. Formation pathways and precursors of furfural during Zhenjiang aromatic vinegar production. Food Chem. 2021, 354, 129503. [Google Scholar] [CrossRef]
- Nateghpour, B.; Kavoosi, G.; Mirakhorli, N. Amino acid profile of the peel of three citrus species and its effect on the combination of amino acids and fatty acids Chlorella vulgaris. J. Food Compos. Anal. 2021, 98, 103808. [Google Scholar] [CrossRef]
- Kim, M.K.; Kim, M.Y.; Lee, K.-G. Determination of furan levels in commercial orange juice products and its correlation to the sensory and quality characteristics. Food Chem. 2016, 211, 654–660. [Google Scholar] [CrossRef]
- Randhawa, M.; Javed, M.S.; Ahmad, Z.; Amjad, A.; Khan, A.; Shah, F.u.H.; Filza, F. Amassing of Hydroxymethylfurfural, 2- Furfural and 5- Methyl furfural in orange (Citrus reticulata) juice during storage. Food Sci. Technol. 2019, 40, 382–386. [Google Scholar] [CrossRef] [Green Version]
- RaneZabAnishKumar, P.; Bhaskar, A. Determination of Bioactive components from the Ethanolic Peel extract of Citrus reticulata by Gas chromatography—MassSpectrometry. Int. J. Drug Dev. Res. 2012, 4, 166–174. [Google Scholar]
- Al-Nuri, M.; Abu-Reidah, I.M.; Alhajeh, A.A.; Omar, G.; Adwan, G.; Warad, I. GC-MS-Based Metabolites Profiling, In Vitro Antioxidant, Anticancer, and Antimicrobial Properties of Different Solvent Extracts from the Botanical Parts of Micromeria fruticosa (Lamiaceae). Processes 2022, 10, 1016. [Google Scholar] [CrossRef]
- Wang, X.; Xia, Y.; Liu, L.; Liu, M.; Gu, N.; Guang, H.; Zhang, F. Comparison of MTT assay, flow cytometry, and RT-PCR in the evaluation of cytotoxicity of five prosthodontic materials. J. Biomed. Mater. Res. Part B Appl. Biomater. 2010, 95B, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Justus, C.R.; Leffler, N.; Ruiz-Echevarria, M.; Yang, L.V. In vitro cell migration and invasion assays. J. Vis. Exp. 2014, 88, 51046. [Google Scholar] [CrossRef] [Green Version]
- Lam-Ubol, A.; Fitzgerald, A.L.; Ritdej, A.; Phonyiam, T.; Zhang, H.; Myers, J.N.; Huang, P.; Trachootham, D. Sensory acceptable equivalent doses of β-phenylethyl isothiocyanate (PEITC) induce cell cycle arrest and retard the growth of p53 mutated oral cancer in vitro and in vivo. Food Funct. 2018, 9, 3640–3656. [Google Scholar] [CrossRef]










| No. | RT (min) | Compounds | Mode | Molecular Mass | Precursor Mass | Molecular Formula | Library Score | Relative Area (%) |
|---|---|---|---|---|---|---|---|---|
| 1. | 0.93 | D-Aspartic acid | M+ | 133.0382 | 134.0449 | C4H7NO4 | 90.3 | 0.10 |
| 2. | 0.97 | N-Oxalylglycine | [M + CH3OH + H]+ | 115.0272 | 148.0602 | C4H5NO5 | 100.0 | 0.06 |
| 3. | 1.05 | Benzothiazole, 2-methylmercapto- | M+ | 181.0507 | 182.0575 | C8H7NS2 | 99.2 | 0.04 |
| 4. | 1.05 | Melibiose | [M + NH4]+ | 342.1164 | 360.1497 | C12H22O11 | 86.3 | 0.10 |
| 5. | 1.20 | 3-Pyridinecarboxaldehyde | M+ | 107.0378 | 108.0445 | C6H5NO | 85.7 | 0.10 |
| 6. | 1.28 | Synephrine | M+ | 167.0588 | 168.0655 | C9H13NO2 | 83.7 | 0.14 |
| 7. | 1.57 | 2,5-Dihydroxy benzaldehyde | M+ | 137.9961 | 139.0028 | C7H6O3 | 82.3 | 0.08 |
| 8. | 1.57 | trans-Aconitic acid | M+ | 174.0170 | 175.0237 | C6H6O6 | 72.1 | 0.85 |
| 9. | 1.57 | Citric acid | [M + NH4]+ | 192.0279 | 210.0611 | HOC (CH2CO2H)2 | 76.3 | 0.64 |
| 10. | 3.34 | Adenosine | M+ | 267.0976 | 268.1044 | C10H13N5O4 | 100.0 | 0.40 |
| 11. | 4.14 | 2,3-Desisopropylidenetopiramate | M+ | 321.0845 | 322.0913 | C9H17NO8S | 89.3 | 0.30 |
| 12. | 4.28 | Phenprobamate | M+ | 165.0794 | 166.0861 | C10H13NO2 | 99.7 | 0.22 |
| 13. | 5.30 | Poly-L-asparagine | M+ | 114.1026 | 115.0387 | C4H6 H2O2 | 98.9 | 0.03 |
| 14. | 6.49 | Stachydrine | M+ | 143.1732 | 144.0799 | C7H13NO2 | 83.6 | 0.02 |
| 15. | 7.32 | Isoorientin | M+ | 448.1002 | 449.1069 | C21H20O11 | 99.1 | 0.04 |
| 16. | 7.40 | Lactobionic acid | M+ | 380.1087 | 381.1155 | C12H22O12 | 97.9 | 0.07 |
| 17. | 7.83 | Engeletin | M+ | 435.1177 | 435.1244 | C21H22O10 | 80.0 | 0.06 |
| 18. | 7.91 | trans-3-Coumaric acid | [M + H]+ | 146.0374 | 147.0441 | C9H7O3 | 96.9 | 0.25 |
| 19. | 7.91 | cis-3-Coumaric acid | M+ | 164.0476 | 165.0543 | C9H7O3 | 80.1 | 0.05 |
| 20. | 7.91 | Quercetin | M+ | 302.0433 | 303.0501 | C15H10O7 | 97.9 | 0.03 |
| 21. | 7.99 | Isovitexin | [M + H]+ | 432.1077 | 433.1144 | C21H20O10 | 98.0 | 0.80 |
| 22. | 8.11 | Rutin | M+ | 610.4776 | 611.4843 | C27H30O16 | 99.6 | 0.02 |
| 23. | 8.54 | Quercetin 3′-methyl ether | M+ | 316.0587 | 317.0654 | C16H12O7 | 98.9 | 0.09 |
| 24. | 8.54 | Isorhoifolin | M+ | 578.1632 | 579.1699 | C27H30O14 | 100.0 | 0.19 |
| 25. | 8.54 | Narirutin | M+ | 580.1779 | 581.1846 | C27H32O14 | 92.0 | 0.09 |
| 26. | 8.54 | Narcissoside | M+ | 624.1702 | 625.1769 | C28H32O16 | 97.9 | 0.66 |
| 27. | 8.58 | Bisdemethoxy curcumin | M+ | 308.0900 | 309.0967 | C19H16O4 | 71.3 | 0.14 |
| 28. | 8.83 | Diosmin | M+ | 608.1755 | 609.1823 | C28H32O15 | 99.9 | 0.62 |
| 29. | 8.92 | Hesperetin | M+ | 302.0799 | 303.0867 | C16H14O6 | 99.5 | 0.69 |
| 30. | 8.92 | Hesperidin * | M+ | 610.1898 | 611.1966 | C28H34O15 | 88.8 | 4.98 |
| 31. | 8.92 | Neohesperidin | M+ | 610.2029 | 611.2097 | C28H34O15 | 82.4 | 4.29 |
| 32. | 8.92 | Hyperin | [M + CH3OH + H]+ | 432.1057 | 465.1387 | C21H19O12 | 91.7 | 0.30 |
| 33. | 9.32 | Isorhamnetin 3-O-glucoside | M+ | 478.1113 | 479.1181 | C22H22O12 | 98.9 | 0.11 |
| 34. | 9.32 | Syringetin 3-glucoside | M+ | 508.1228 | 509.1296 | C23H24O13 | 93.8 | 0.57 |
| 35. | 9.76 | Mono-2-ethylhexyl phthalate | M+ | 278.1169 | 279.1237 | C16H21O4 | 82.8 | 0.04 |
| 36. | 9.84 | Scoparone | M+ | 206.0261 | 207.0328 | C11H10O4 | 92.5 | 0.06 |
| 37. | 10.02 | Heraclenin | M+ | 286.0855 | 287.0922 | C16H14O5 | 95.0 | 0.68 |
| 38. | 10.48 | 2′,6′-Dihydroxy-4- methoxychalcone-4′- O-neohesperidoside | M+ | 594.1943 | 595.2010 | C28H34O14 | 79.3 | 0.04 |
| 39. | 10.60 | Oxypeucedanin hydrate | M+ | 304.1034 | 305.1102 | C16H16O6 | 98.1 | 4.24 |
| 40. | 10.83 | Byakangelicol | M+ | 316.0955 | 317.1022 | C17H16O6 | 93.0 | 0.45 |
| 41. | 11.32 | 7-Methoxycoumarin | M+ | 176.0489 | 177.0556 | C10H8O3 | 85.1 | 1.22 |
| 42. | 12.07 | Angelicin | M+ | 186.0316 | 187.0383 | C11H6O3 | 98.3 | 0.03 |
| 43. | 13.06 | 5,7-Dimethoxycoumarin | M+ | 206.0661 | 207.0729 | C11H10O4 | 79.2 | 14.62 |
| 44. | 13.38 | Isopimpinellin | [M + NH4]+ | 246.0529 | 264.0862 | C13H10O5 | 100.0 | 0.07 |
| 45. | 13.98 | Limonin * | M+ | 470.2057 | 471.2125 | C26H30O8 | 95.3 | 5.01 |
| 46. | 14.83 | Nomilin | [M + H]+ | 514.2206 | 515.2273 | C28H34O9 | 91.8 | 0.24 |
| 47. | 15.88 | Obacunone | M+ | 454.1990 | 455.2058 | C26H30O7 | 86.1 | 0.03 |
| 48. | 16.62 | Monolinolenin (9c,12c,15c) | M+ | 352.2618 | 353.2685 | C21H36O4 | 75.7 | 0.04 |
| 49. | 18.98 | 1-Palmitoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine | M+ | 453.2863 | 454.2931 | C21H44NO7P | 88.0 | 0.66 |
| 50. | 19.47 | 1-Palmitoyl-sn-glycero-3-phosphocholine | M+ | 495.3338 | 496.3405 | C26H52NO8P | 94.4 | 1.40 |
| 51. | 19.74 | Esculetin | M+ | 178.0269 | 179.0336 | C9H6O4 | 70.1 | 0.04 |
| 52. | 20.08 | 1-Oleoyl-sn-glycero-3-phosphocholine | M+ | 521.3492 | 522.3560 | C26H52NO7P | 97.2 | 1.20 |
| 53. | 20.63 | Cinnamic acid | M+ | 148.0167 | 149.0234 | C9H8O2 | 79.6 | 1.45 |
| 54. | 21.13 | 8-Allyloxypsoralen | M+ | 242.0583 | 243.0650 | C14H10O4 | 90.2 | 0.06 |
| 55. | 21.17 | 3-Propyl- 2-methylpyrazine | M+ | 136.1255 | 137.1322 | C8H12N2 | 81.3 | 0.49 |
| 56. | 21.17 | Xanthotoxol | M+ | 202.0275 | 203.0342 | C11H6O4 | 95.3 | 1.20 |
| 57. | 22.51 | Bergaptol | M+ | 202.0350 | 203.0417 | C11H6O4 | 85.8 | 14.30 |
| 58. | 22.55 | Bergamotin | M+ | 338.1583 | 339.1651 | C21H22O4 | 82.2 | 8.35 |
| 59. | 22.66 | 5-Geranoxy- 7-methoxy-coumarin | M+ | 328.1823 | 329.1890 | C20H24O4 | 91.7 | 24.36 |
| 60. | 23.61 | 8-O-Acetylharpagide | M+ | 406.0821 | 407.0888 | C17H26O11 | 95.0 | 0.17 |
| 61. | 23.75 | Oleamide | M+ | 281.2732 | 282.2799 | C18H35NO | 76.9 | 2.39 |
| 62. | 25.67 | Hexadecyltrimethylammonium cation | M+ | 284.2878 | 284.2945 | C16H33N(CH3)3 | 98.5 | 0.03 |
| No. | RT (min) | Compounds | Molecular Mass | Molecular Formula | SI | RSI | Relative Area (%) |
|---|---|---|---|---|---|---|---|
| 1. | 3.0932 | 1,1-Diethoxyethane | 118.1742 | C6H14O2 | 622 | 884 | 1.06 |
| 2. | 4.8921 | Furfural | 96.0841 | C5H4O2 | 752 | 888 | 1.36 |
| 3. | 7.1966 | α-pinene | 136.2340 | C10H16 | 776 | 866 | 1.43 |
| 4. | 7.2959 | Furan | 68.0740 | C4H4O | 849 | 911 | 9.35 |
| 5. | 7.7484 | 5-Methyl furfural | 110.1106 | C6H6O2 | 803 | 905 | 1.71 |
| 6. | 7.900 | 1-Methylcyclopentanol | 100.1589 | C6H12O2 | 689 | 792 | 2.04 |
| 7. | 9.1655 | D-Limonene | 136.2340 | C10H16 | 867 | 871 | 20.92 |
| 8. | 9.7108 | 3-Carene | 136.2340 | C10H16 | 792 | 842 | 1.67 |
| 9. | 11.8210 | Terpinen-4-ol | 154.2493 | C10H18O | 757 | 832 | 1.05 |
| 10. | 12.0373 | α-Terpineol | 154.2493 | C10H18O | 817 | 897 | 1.99 |
| 11. | 12.3772 | 5-Hydroxymethy furfural | 126.1100 | C6H6O3 | 736 | 758 | 4.59 |
| 12. | 15.3505 | Caryophyllene | 204.3511 | C15H24 | 888 | 896 | 7.60 |
| 13. | 15.4631 | Cis-α-Bergamotene | 204.3511 | C15H24 | 876 | 899 | 6.34 |
| 14. | 15.8405 | Aristolochene | 204.3511 | C15H24 | 762 | 835 | 1.71 |
| 15. | 16.3901 | α-Farnesene | 204.3511 | C15H24 | 733 | 747 | 6.31 |
| 16. | 16.4895 | Aromandendrene | 204.3511 | C15H24 | 867 | 881 | 8.61 |
| 17. | 17.0126 | 6,9-Guaiadene | 204.3511 | C15H24 | 815 | 857 | 1.47 |
| 18. | 17.0899 | Selina-3,7(11)-diene | 204.3511 | C15H24 | 823 | 879 | 1.38 |
| 19. | 24.1909 | Citraptene | 206.1947 | C11H10O4 | 878 | 896 | 7.11 |
| 20. | 25.5329 | Bergapten | 216.1895 | C12H8O4 | 840 | 937 | 3.12 |
| 21. | 28.3627 | Isopimpinellin | 246.2155 | C13H10O5 | 860 | 873 | 8.43 |
| 22. | 31.6207 | Palmitic acid β-monoglyceride | 330.5026 | C19H38O4 | 667 | 789 | 0.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phucharoenrak, P.; Muangnoi, C.; Trachootham, D. Metabolomic Analysis of Phytochemical Compounds from Ethanolic Extract of Lime (Citrus aurantifolia) Peel and Its Anti-Cancer Effects against Human Hepatocellular Carcinoma Cells. Molecules 2023, 28, 2965. https://doi.org/10.3390/molecules28072965
Phucharoenrak P, Muangnoi C, Trachootham D. Metabolomic Analysis of Phytochemical Compounds from Ethanolic Extract of Lime (Citrus aurantifolia) Peel and Its Anti-Cancer Effects against Human Hepatocellular Carcinoma Cells. Molecules. 2023; 28(7):2965. https://doi.org/10.3390/molecules28072965
Chicago/Turabian StylePhucharoenrak, Pakkapong, Chawanphat Muangnoi, and Dunyaporn Trachootham. 2023. "Metabolomic Analysis of Phytochemical Compounds from Ethanolic Extract of Lime (Citrus aurantifolia) Peel and Its Anti-Cancer Effects against Human Hepatocellular Carcinoma Cells" Molecules 28, no. 7: 2965. https://doi.org/10.3390/molecules28072965
APA StylePhucharoenrak, P., Muangnoi, C., & Trachootham, D. (2023). Metabolomic Analysis of Phytochemical Compounds from Ethanolic Extract of Lime (Citrus aurantifolia) Peel and Its Anti-Cancer Effects against Human Hepatocellular Carcinoma Cells. Molecules, 28(7), 2965. https://doi.org/10.3390/molecules28072965