Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors
Abstract
1. Introduction
2. Results
2.1. G3BP1 Is Specifically Targeted by PV 3Cpro for Cleavage
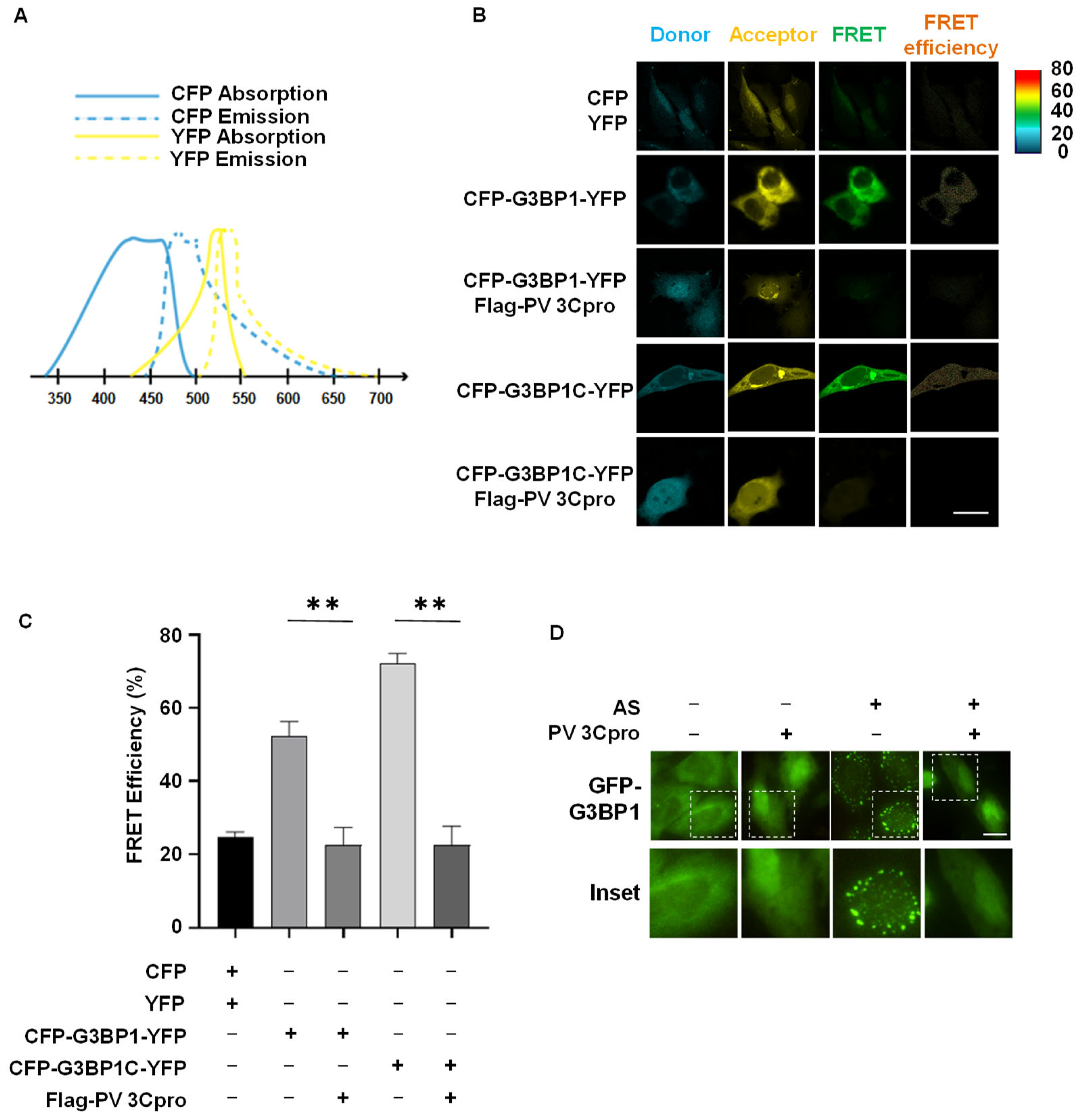
2.2. Establishment of FRET and SG Dual-Based System to Monitor PV 3Cpro Activity in Living Cells
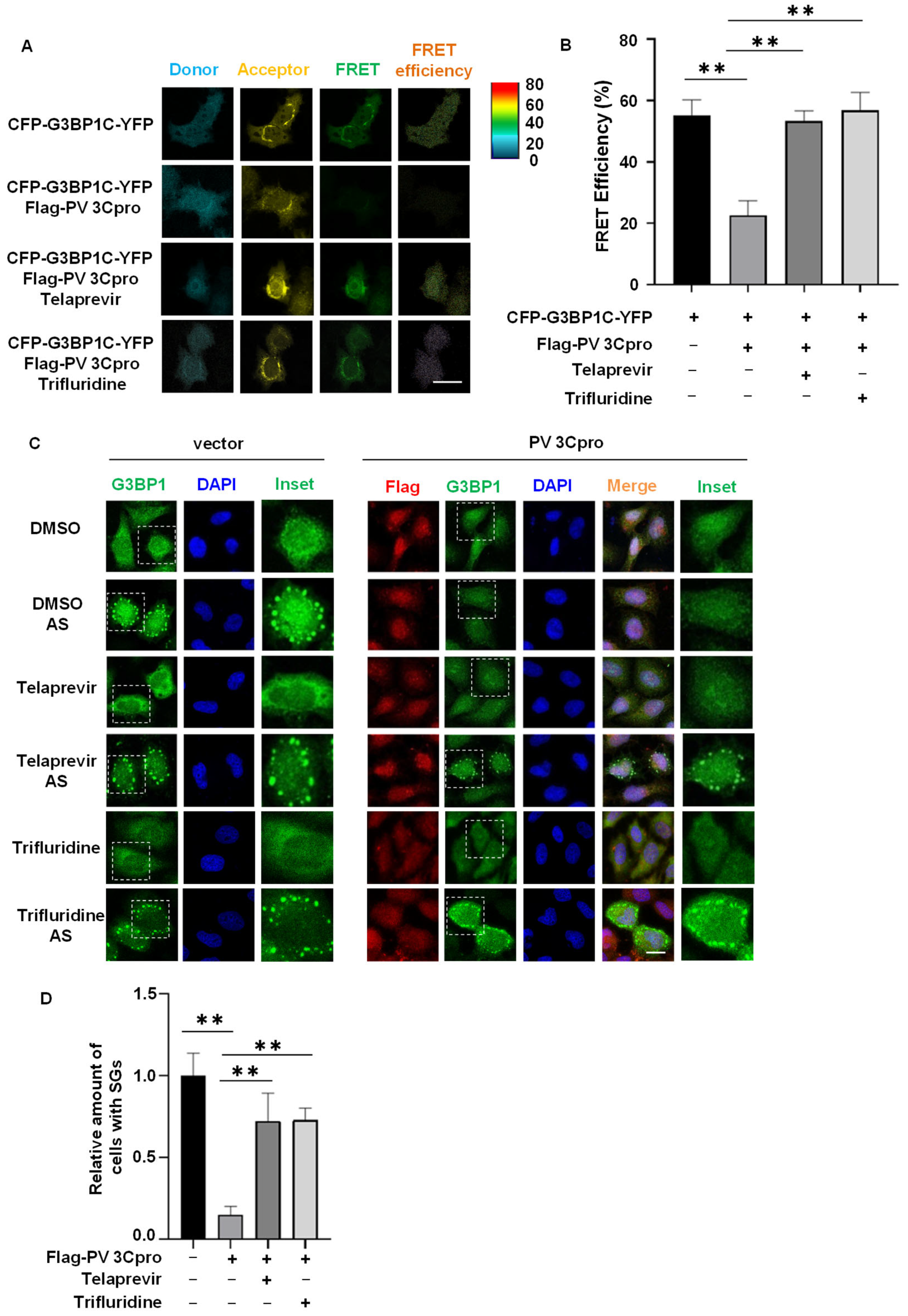
2.3. Drug Screen Identified Telaprevir and Trifluridine as PV 3Cpro Inhibitors
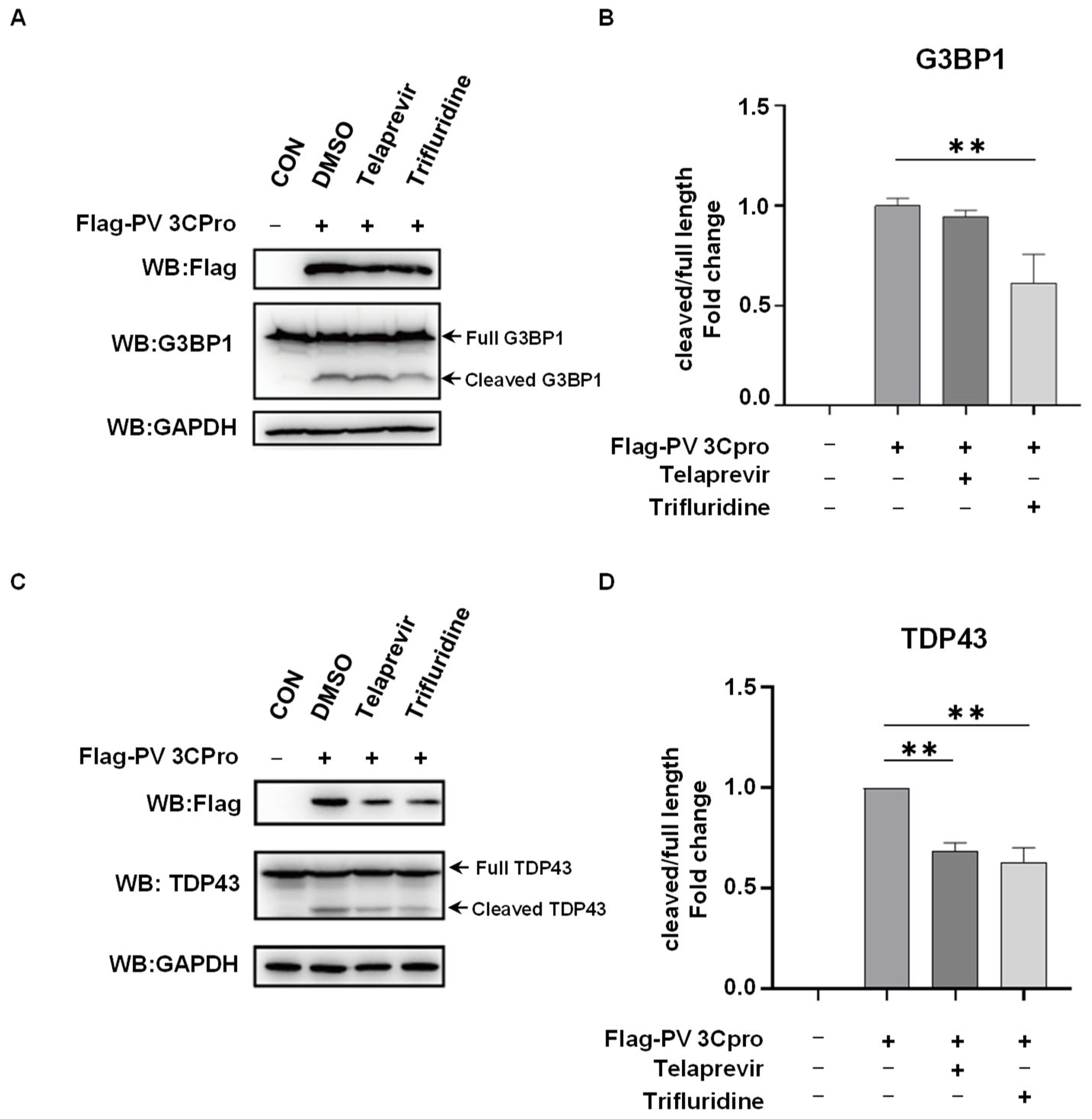
2.4. Telaprevir and Trifluridine Counteract PV 3Cpro-Mediated Physiological Events
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Plasmids and Reagents
4.3. SG Induction and Quantification
4.4. Immunofluorescence
4.5. Protein Extraction and Western Blotting
4.6. The Fluorescence Resonance Energy Transfer (FRET) Assay
4.7. RNA Extraction, Reverse Transcription, and Quantitative PCR
4.8. Viral Infection and Flow Cytometry Analysis
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Laitinen, O.H.; Svedin, E.; Kapell, S.; Nurminen, A.; Hytönen, V.P.; Flodström-Tullberg, M. Enteroviral proteases: Structure, host interactions and pathogenicity. Rev. Med. Virol. 2016, 26, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lovell, S.; Tiew, K.C.; Mandadapu, S.R.; Alliston, K.R.; Battaile, K.P.; Groutas, W.C.; Chang, K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012, 86, 11754–11762. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Chen, S.; Cheng, A.; Wang, M. Roles of the Picornaviral 3C Proteinase in the Viral Life Cycle and Host Cells. Viruses 2016, 8, 82. [Google Scholar] [CrossRef]
- Yi, J.; Peng, J.; Yang, W.; Zhu, G.; Ren, J.; Li, D.; Zheng, H. Picornavirus 3C—A protease ensuring virus replication and subverting host responses. J. Cell Sci. 2021, 134, jcs253237. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.S.; Stobart, C.C.; Luo, H. Innate immune evasion mediated by picornaviral 3C protease: Possible lessons for coronaviral 3C-like protease? Rev. Med. Virol. 2021, 31, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Tsu, B.V.; Fay, E.J.; Nguyen, K.T.; Corley, M.R.; Hosuru, B.; Dominguez, V.A.; Daugherty, M.D. Running with Scissors: Evolutionary Conflicts Between Viral Proteases and the Host Immune System. Front. Immunol. 2021, 12, 769543. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.; Fielding, B.C.; Gamieldien, J. Potential Broad Spectrum Inhibitors of the Coronavirus 3CLpro: A Virtual Screening and Structure-Based Drug Design Study. Viruses 2015, 7, 6642–6660. [Google Scholar] [CrossRef]
- Jagdeo, J.M.; Dufour, A.; Klein, T.; Solis, N.; Kleifeld, O.; Kizhakkedathu, J.; Luo, H.; Overall, C.M.; Jan, E. N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection. J. Virol. 2018, 92, e02211-17. [Google Scholar] [CrossRef]
- Rhoden, E.; Liu, H.M.; Wang-Chern, S.W.; Oberste, M.S. Anti-poliovirus activity of protease inhibitor AG-7404, and assessment of in vitro activity in combination with antiviral capsid inhibitor compounds. Antivir. Res. 2013, 98, 186–191. [Google Scholar] [CrossRef]
- Yang, H.; Xie, W.; Xue, X.; Yang, K.; Ma, J.; Liang, W.; Zhao, Q.; Zhou, Z.; Pei, D.; Ziebuhr, J.; et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005, 3, e324. [Google Scholar]
- De Palma, A.M.; Purstinger, G.; Wimmer, E.; Patick, A.K.; Andries, K.; Rombaut, B.; De Clercq, E.; Neyts, J. Potential use of antiviral agents in polio eradication. Emerg. Infect. Dis. 2008, 14, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.T.; Cheng, Y.H.; Liu, Y.N.; Liao, N.C.; Lu, W.W.; Kung, S.H. Real-time monitoring of human enterovirus (HEV)-infected cells and anti-HEV 3C protease potency by fluorescence resonance energy transfer. Antimicrob. Agents Chemother. 2009, 53, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Blay, V.; Tolani, B.; Ho, S.P.; Arkin, M.R. High-Throughput Screening: Today’s biochemical and cell-based approaches. Drug Discov. Today 2020, 25, 1807–1821. [Google Scholar] [CrossRef]
- Salaun, C.; Takizawa, H.; Galindo, A.; Munro, K.R.; McLellan, J.; Sugimoto, I.; Okino, T.; Tomkinson, N.C.O.; Chamberlain, L.H. Development of a novel high-throughput screen for the identification of new inhibitors of protein S-acylation. J. Biol. Chem. 2022, 298, 102469. [Google Scholar] [CrossRef]
- Coquerel, Q.; Legendre, C.; Frangieh, J.; Waard, S.; Montnach, J.; Cmarko, L.; Khoury, J.; Hassane, C.S.; Bréard, D.; Siegler, B.; et al. Screening an In-House Isoquinoline Alkaloids Library for New Blockers of Voltage-Gated Na+ Channels Using Voltage Sensor Fluorescent Probes: Hits and Biases. Molecules 2022, 27, 4133. [Google Scholar] [CrossRef]
- Chen, S.; Chen, L.L.; Luo, H.B.; Sun, T.; Chen, J.; Ye, F.; Cai, J.H.; Shen, J.K.; Shen, X.; Jiang, H.L. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique. Acta Pharmacol. Sin. 2005, 26, 99–106. [Google Scholar] [CrossRef]
- Liu, Y.C.; Huang, V.; Chao, T.C.; Hsiao, C.D.; Lin, A.; Chang, M.F.; Chow, L.P. Screening of drugs by FRET analysis identifies inhibitors of SARS-CoV 3CL protease. Biochem. Biophys. Res. Commun. 2005, 333, 194–199. [Google Scholar] [CrossRef]
- Brown, A.S.; Ackerley, D.F.; Calcott, M.J. High-Throughput Screening for Inhibitors of the SARS-CoV-2 Protease Using a FRET-Biosensor. Molecules 2020, 25, 4666. [Google Scholar] [CrossRef]
- Takahashi, D.; Kim, Y.; Lovell, S.; Prakash, O.; Groutas, W.C.; Chang, K.O. Structural and inhibitor studies of norovirus 3C-like proteases. Virus Res. 2013, 178, 437–444. [Google Scholar] [CrossRef]
- Kilianski, A.; Mielech, A.M.; Deng, X.; Baker, S.C. Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 2013, 87, 11955–11962. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Cho, J.H.; Jeong, P.; Lee, Y.; Lim, J.J.; Park, K.R.; Eom, S.H.; Kim, Y.C. Benserazide, the first allosteric inhibitor of Coxsackievirus B3 3C protease. FEBS Lett. 2015, 589, 1795–1801. [Google Scholar] [CrossRef]
- van der Linden, L.; Ulferts, R.; Nabuurs, S.B.; Kusov, Y.; Liu, H.; George, S.; Lacroix, C.; Goris, N.; Lefebvre, D.; Lanke, K.H.; et al. Application of a cell-based protease assay for testing inhibitors of picornavirus 3C proteases. Antivir. Res. 2014, 103, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, J.; Fan, S.; Jin, Z.; Huang, C. Targeting stress granules: A novel therapeutic strategy for human diseases. Pharmacol. Res. 2020, 161, 105143. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Kedersha, N.; Anderson, P.; Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118876. [Google Scholar] [CrossRef]
- Zhang, Q.; Sharma, N.R.; Zheng, Z.M.; Chen, M. Viral Regulation of RNA Granules in Infected Cells. Virol. Sin. 2019, 34, 175–191. [Google Scholar] [CrossRef]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e28. [Google Scholar] [CrossRef]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e17. [Google Scholar] [CrossRef]
- McCormick, C.; Khaperskyy, D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017, 17, 647–660. [Google Scholar] [CrossRef]
- Eiermann, N.; Haneke, K.; Sun, Z.; Stoecklin, G.; Ruggieri, A. Dance with the Devil: Stress Granules and Signaling in Antiviral Responses. Viruses 2020, 12, 984. [Google Scholar] [CrossRef]
- Deater, M.; Tamhankar, M.; Lloyd, R.E. TDRD3 is an antiviral restriction factor that promotes IFN signaling with G3BP1. PLoS Pathog. 2022, 18, e1010249. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Lloyd, R.E. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J. Virol. 2015, 89, 2575–2589. [Google Scholar] [CrossRef]
- Tsai, W.C.; Lloyd, R.E. Cytoplasmic RNA Granules and Viral Infection. Annu. Rev. Virol. 2014, 1, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Brownsword, M.J.; Locker, N. A little less aggregation a little more replication: Viral manipulation of stress granules. Wiley Interdiscip. Rev. RNA 2023, 14, e1741. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Cardenas, A.M.; Marissen, W.E.; Lloyd, R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2007, 2, 295–305. [Google Scholar] [CrossRef]
- Ye, X.; Pan, T.; Wang, D.; Fang, L.; Ma, J.; Zhu, X.; Shi, Y.; Zhang, K.; Zheng, H.; Chen, H.; et al. Foot-and-Mouth Disease Virus Counteracts on Internal Ribosome Entry Site Suppression by G3BP1 and Inhibits G3BP1-Mediated Stress Granule Assembly via Post-Translational Mechanisms. Front. Immunol. 2018, 9, 1142. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.S.; Jogi, M.; Yoo, J.S.; Onomoto, K.; Koike, S.; Iwasaki, T.; Yoneyama, M.; Kato, H.; Fujita, T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 2013, 87, 9511–9522. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hu, Z.; Fan, S.; Zhang, Q.; Zhong, Y.; Guo, D.; Qin, Y.; Chen, M. Picornavirus 2A protease regulates stress granule formation to facilitate viral translation. PLoS Pathog. 2018, 14, e1006901. [Google Scholar] [CrossRef]
- Fung, G.; Ng, C.S.; Zhang, J.; Shi, J.; Wong, J.; Piesik, P.; Han, L.; Chu, F.; Jagdeo, J.; Jan, E.; et al. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS ONE 2013, 8, e79546. [Google Scholar] [CrossRef]
- Humoud, M.N.; Doyle, N.; Royall, E.; Willcocks, M.M.; Sorgeloos, F.; van Kuppeveld, F.; Roberts, L.O.; Goodfellow, I.G.; Langereis, M.A.; Locker, N. Feline Calicivirus Infection Disrupts Assembly of Cytoplasmic Stress Granules and Induces G3BP1 Cleavage. J. Virol. 2016, 90, 6489–6501. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, J.; Han, L.; Zhuang, M.W.; Xu, Y.; Zhang, J.; Nan, M.L.; Xiao, Y.; Zhan, P.; Liu, X.; et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther. 2022, 7, 22. [Google Scholar] [CrossRef]
- Zheng, Z.Q.; Wang, S.Y.; Xu, Z.S.; Fu, Y.Z.; Wang, Y.Y. SARS-CoV-2 nucleocapsid protein impairs stress granule formation to promote viral replication. Cell Discov. 2021, 7, 38. [Google Scholar] [CrossRef]
- Luo, L.; Li, Z.; Zhao, T.; Ju, X.; Ma, P.; Jin, B.; Zhou, Y.; He, S.; Huang, J.; Xu, X.; et al. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci. Bull. 2021, 66, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Ciccosanti, F.; Di Rienzo, M.; Romagnoli, A.; Colavita, F.; Refolo, G.; Castilletti, C.; Agrati, C.; Brai, A.; Manetti, F.; Botta, L.; et al. Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection. Antivir. Res. 2021, 190, 105064. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Bai, Y.; Zhang, X.; Gao, T.; Liu, Y.; Li, E.; Wang, X.; Cao, Z.; Zhu, L.; Dong, Q.; et al. SARS-CoV-2 N Protein Antagonizes Stress Granule Assembly and IFN Production by Interacting with G3BPs to Facilitate Viral Replication. J. Virol. 2022, 96, e0041222. [Google Scholar] [CrossRef]
- Desmet, C.J.; Ishii, K.J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012, 12, 479–491. [Google Scholar] [CrossRef]
- Fung, G.; Shi, J.; Deng, H.; Hou, J.; Wang, C.; Hong, A.; Zhang, J.; Jia, W.; Luo, H. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ. 2015, 22, 2087–2097. [Google Scholar] [CrossRef]
- Wo, X.; Yuan, Y.; Xu, Y.; Chen, Y.; Wang, Y.; Zhao, S.; Lin, L.; Zhong, X.; Wang, Y.; Zhong, Z.; et al. TAR DNA-Binding Protein 43 is Cleaved by the Protease 3C of Enterovirus A71. Virol. Sin. 2021, 36, 95–103. [Google Scholar] [CrossRef]
- Patick, A.K.; Binford, S.L.; Brothers, M.A.; Jackson, R.L.; Ford, C.E.; Diem, M.D.; Maldonado, F.; Dragovich, P.S.; Zhou, R.; Prins, T.J.; et al. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999, 43, 2444–2450. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Chow, W.A.; Jiang, C.; Guan, M. Anti-HIV drugs for cancer therapeutics: Back to the future? Lancet Oncol. 2009, 10, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, K.; Yazdanpanah, N.; Saghazadeh, A.; Rezaei, N. Computational drug discovery and repurposing for the treatment of COVID-19: A systematic review. Bioorg. Chem. 2021, 106, 104490. [Google Scholar] [CrossRef] [PubMed]
- Klibanov, O.M.; Williams, S.H.; Smith, L.S.; Olin, J.L.; Vickery, S.B. Telaprevir: A novel NS3/4 protease inhibitor for the treatment of hepatitis C. Pharmacotherapy 2011, 31, 951–974. [Google Scholar] [CrossRef] [PubMed]
- Carmine, A.A.; Brogden, R.N.; Heel, R.C.; Speight, T.M.; Avery, G.S. Trifluridine: A review of its antiviral activity and therapeutic use in the topical treatment of viral eye infections. Drugs 1982, 23, 329–353. [Google Scholar] [CrossRef] [PubMed]
- Pène, V.; Li, Q.; Sodroski, C.; Hsu, C.S.; Liang, T.J. Dynamic Interaction of Stress Granules, DDX3X, and IKK-α Mediates Multiple Functions in Hepatitis C Virus Infection. J. Virol. 2015, 89, 5462–5477. [Google Scholar] [CrossRef]
- Fernández-Carrillo, C.; Pérez-Vilaró, G.; Díez, J.; Pérez-Del-Pulgar, S. Hepatitis C virus plays with fire and yet avoids getting burned. A review for clinicians on processing bodies and stress granules. Liver Int. 2018, 38, 388–398. [Google Scholar] [CrossRef]
- Garaigorta, U.; Heim, M.H.; Boyd, B.; Wieland, S.; Chisari, F.V. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012, 86, 11043–11056. [Google Scholar] [CrossRef]
- Burgess, H.M.; Mohr, I. Defining the Role of Stress Granules in Innate Immune Suppression by the Herpes Simplex Virus 1 Endoribonuclease VHS. J. Virol. 2018, 92, e00829-18. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, J.; Yang, X.; Quan, K.; Wang, H.; Ying, L.; Xie, N.; Ou, M.; Wang, K. FRET Nanoflares for Intracellular mRNA Detection: Avoiding False Positive Signals and Minimizing Effects of System Fluctuations. J. Am. Chem. Soc. 2015, 137, 8340–8343. [Google Scholar] [CrossRef]
- Christopeit, T.; Øverbø, K.; Danielson, U.H.; Nilsen, I.W. Efficient screening of marine extracts for protease inhibitors by combining FRET based activity assays and surface plasmon resonance spectroscopy based binding assays. Mar. Drugs 2013, 11, 4279–4293. [Google Scholar] [CrossRef]
- Klickstein, J.A.; Mukkavalli, S.; Raman, M. AggreCount: An unbiased image analysis tool for identifying and quantifying cellular aggregates in a spatially defined manner. J. Biol. Chem. 2020, 295, 17672–17683. [Google Scholar] [CrossRef] [PubMed]
- Chusainow, J.; Ajuh, P.M.; Trinkle-Mulcahy, L.; Sleeman, J.E.; Ellenberg, J.; Lamond, A.I. FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35. RNA N. Y. 2005, 11, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhuang, M.W.; Han, L.; Zhang, J.; Nan, M.L.; Zhan, P.; Kang, D.; Liu, X.; Gao, C.; Wang, P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 2020, 5, 299. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]



| Number | Name | CAS | Description | FRET Screen | SGs Screen |
|---|---|---|---|---|---|
| 1 | Amprenavir | 161814-49-9 | HIV protease inhibitor | − | − |
| 2 | Ritonavir | 155213-67-5 | HIV protease inhibitor | − | − |
| 3 | Lopinavir | 192725-17-0 | HIV protease inhibitor | − | − |
| 4 | Atazanavir | 198904-31-3 | HIV protease inhibitor | + | − |
| 5 | Darunavir | 206361-99-1 | HIV protease inhibitor | − | − |
| 6 | Saquinavir mesylate | 149845-06-7 | HIV protease inhibitor | − | − |
| 7 | Nelfinavir | 159989-64-7 | HIV protease inhibitor | − | − |
| 8 | Asunaprevir | 630420-16-5 | HCV NS3pro inhibitor | + | − |
| 9 | Boceprevir | 394730-60-0 | HCV NS3pro inhibitor | − | − |
| 10 | Ledipasvir | 1256388-51-8 | HCV NS5Apro inhibitor | − | − |
| 11 | Simeprevir | 923604-59-5 | HCV NS3/4Apro inhibitor | − | − |
| 12 | Telaprevir | 402957-28-2 | HCV NS3/4Apro inhibitor | + | + |
| 13 | Daclatasvir | 1009119-64-5 | HCV NS5Apro inhibitor | − | − |
| 14 | Tenofovir Disoproxil Fumarate | 202138-50-9 | HIV-1 reverse transcriptase (RT) inhibitor | − | − |
| 15 | Entecavir Hydrate | 209216-23-9 | HBV replication inhibitor | − | − |
| 16 | Abacavir | 136470-78-5 | HIV RT inhibitor | − | − |
| 17 | MK-5172 | 1350514-68-9 | HCV NS3/4A pro inhibitor | − | − |
| 18 | Oseltamivir | 196618-13-0 | Influenza neuraminidase inhibitor | − | − |
| 19 | Oseltamivir acid | 187227-45-8 | Influenza neuraminidase inhibitor | − | − |
| 20 | Peramivir | 330600-85-6 | Influenza neuraminidase inhibitor | + | − |
| 21 | PSI-7977 | 1190307-88-0 | HCV NS5B polymerase inhibitor | − | − |
| 22 | Rilpivirine | 500287-72-9 | nonnucleoside RT inhibitor | − | − |
| 23 | Lomibuvir (VX-222) | 1026785-59-0 | HCV NS5B polymerase inhibitor | − | − |
| 24 | Elvitegravir | 697761-98-1 | HIV-1 integrase inhibitor | − | − |
| 25 | Raltegravir | 518048-05-0 | HIV-1 integrase inhibitor | + | − |
| 26 | S/GSK1349572 | 1051375-16-6 | HIV-1 integrase inhibitor | − | − |
| 27 | Fumagillin | 23110-15-8 | Methionine aminopeptidase-2inhibitor | − | − |
| 28 | Tenofovir | 147127-20-6 | HIV RT inhibitor | − | − |
| 29 | Cidofovir | 113852-37-2 | viral DNA synthesis inhibitor | − | − |
| 30 | Maraviroc | 376348-65-1 | CCR5 inhibitor | − | − |
| 31 | Arbidol HCl | 131707-23-8 | viral fusion inhibitor | − | − |
| 32 | Didanosine | 69655-05-6 | RT inhibitor | − | − |
| 33 | Emtricitabine | 143491-57-0 | RT inhibitor | − | − |
| 34 | Lamivudine | 134678-17-4 | Nucleoside analog RT inhibitor | − | − |
| 35 | Nevirapine | 129618-40-2 | Non-nucleoside RT inhibitor | − | − |
| 36 | Trifluridine | 70-00-8 | HSV replication inhibitor | + | + |
| 37 | Acyclovir | 59277-89-3 | viral replication inhibitor | − | − |
| 38 | Favipiravir (T 705) | 259793-96-9 | RNA-dependent RNA polymerase inhibitor | − | − |
| 39 | Efavirenz | 154598-52-4 | RT inhibitor | + | − |
| 40 | Idoxuridine | 54-42-2 | nucleoside analogues | − | − |
| 41 | Oseltamivir phosphate | 204255-11-8 | Neuraminidase inhibitor | + | − |
| 42 | Penciclovir | 39809-25-1 | neuraminidase inhibitor | − | − |
| 43 | Salicylanilide | 87-17-2 | antiviral | − | − |
| 44 | Valganciclovir | 175865-59-5 | viral DNA polymerase inhibitor | − | − |
| 45 | Famciclovir | 104227-87-4 | Hsv-2 polymerase inhibitor | − | − |
| 46 | Moroxydine HCl | 3160-91-6 | Virus Proliferation inhibitor | − | − |
| 47 | Valaciclovir | 124832-27-5 | virus DNA polymerase inhibitor | − | − |
| 48 | Vidarabine | 5536-17-4 | viral DNA synthesis inhibitor | − | − |
| 49 | Ganciclovir | 82410-32-0 | Viral replication inhibitor | − | − |
| 50 | Ribavirin | 36791-04-5 | antiviral | − | − |
| 51 | Zanamivir | 139110-80-8 | Influenza A/B virus neuraminidases inhibitor | − | − |
| 52 | Peramivir Trihydrate | 1041434-82-5 | Influenza viral neuraminidase inhibitor | − | − |
| 53 | Abacavir sulfate | 188062-50-2 | RT inhibitor | − | − |
| 54 | Adefovir Dipivoxil | 142340-99-6 | RT inhibitor | − | − |
| 55 | Zalcitabine | 7481-89-2 | RT inhibitor | − | − |
| 56 | Etravirine (TMC125) | 269055-15-4 | RT inhibitor | − | − |
| 57 | Stavudine (d4T) | 3056-17-5 | RT inhibitor | − | − |
| 58 | GSK1349572 | 1051375-19-9 | HIV integrase inhibitor | − | − |
| 59 | Rimantadine | 1501-84-4 | M2 proton channel inhibitor | − | − |
| 60 | GS-7340 | 379270-37-8 | HIV RT inhibitor | − | − |
| 61 | Rolipram | 61413-54-5 | PDE4 selective inhibitor | − | − |
| 62 | Telbivudine | 3424-98-4 | RT inhibitor | − | − |
| 63 | Artemisinine | 63968-64-9 | AKT signaling pathway inhibitor | − | − |
| 64 | Cepharanthine | 481-49-2 | viral proliferation inhibitor | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Jiang, Y.; Wu, C.; Zhou, D.; Gong, J.; Zhao, T.; Jin, Z. Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors. Molecules 2023, 28, 3020. https://doi.org/10.3390/molecules28073020
Zhang J, Jiang Y, Wu C, Zhou D, Gong J, Zhao T, Jin Z. Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors. Molecules. 2023; 28(7):3020. https://doi.org/10.3390/molecules28073020
Chicago/Turabian StyleZhang, Jingjing, Yingpei Jiang, Chunxiu Wu, Dan Zhou, Jufang Gong, Tiejun Zhao, and Zhigang Jin. 2023. "Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors" Molecules 28, no. 7: 3020. https://doi.org/10.3390/molecules28073020
APA StyleZhang, J., Jiang, Y., Wu, C., Zhou, D., Gong, J., Zhao, T., & Jin, Z. (2023). Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors. Molecules, 28(7), 3020. https://doi.org/10.3390/molecules28073020






