Annotation and Identification of Phytochemicals from Eleusine indica Using High-Performance Liquid Chromatography Tandem Mass Spectrometry: Databases-Driven Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Tandem LCMS Analysis for Phytochemicals Annotation and Identification
2.1.1. Data Processing, Enrichment and Phytochemicals Annotation by MZmine 2.53
2.1.2. Phytochemicals Annotation by GNPS
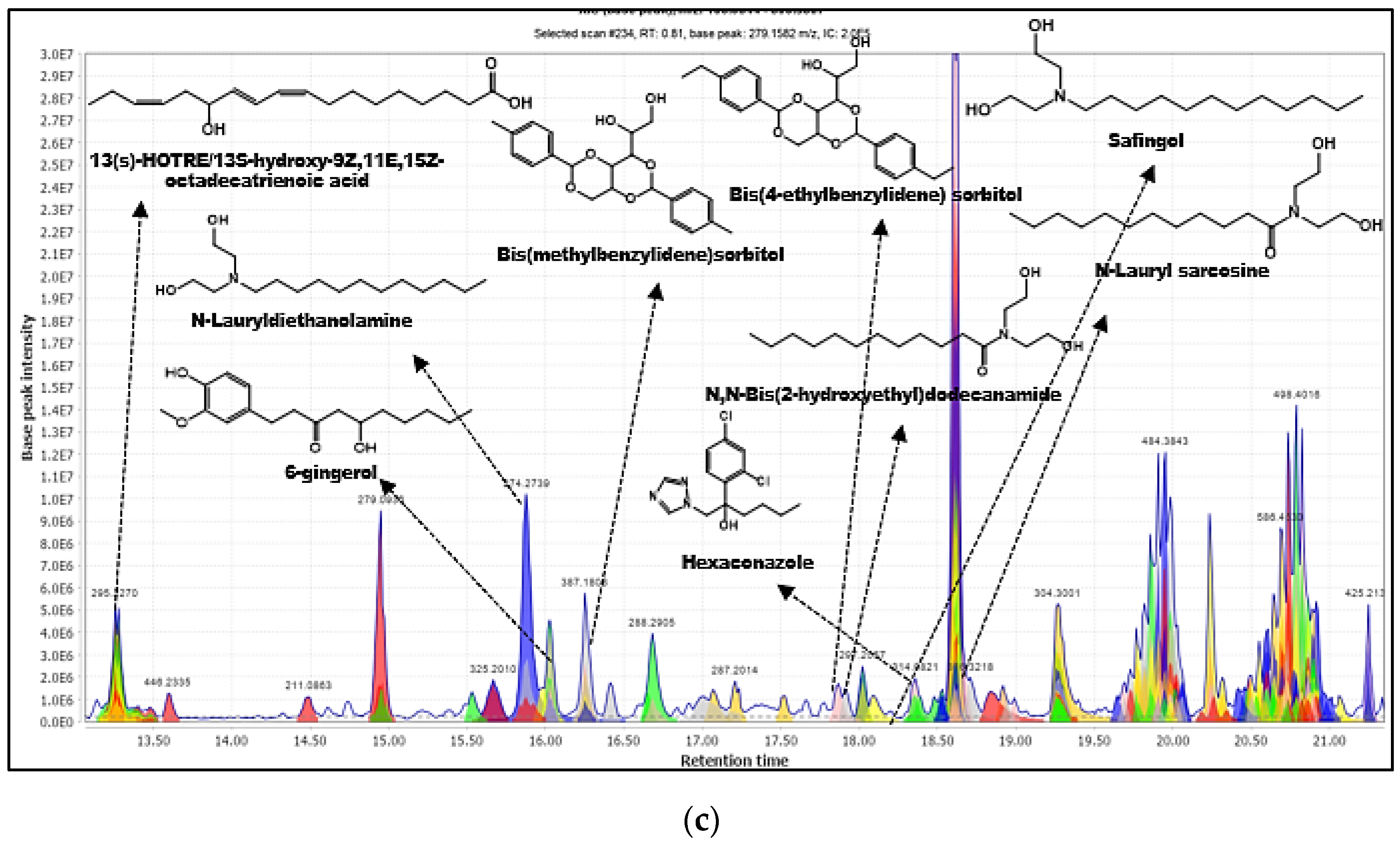
2.1.3. Phytochemicals Annotation by Compound Discoverer (CD) 3.0
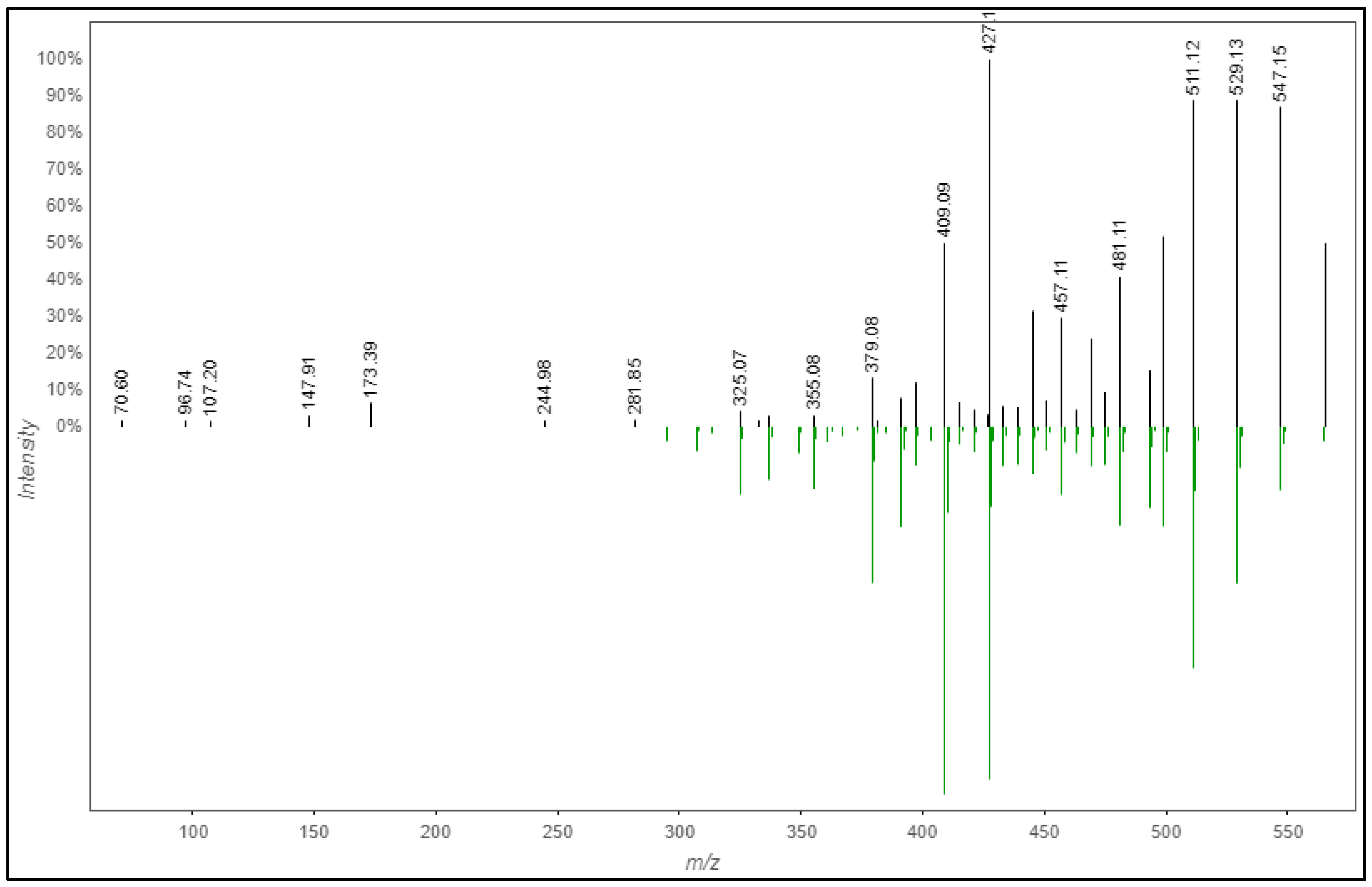
2.1.4. Phytochemicals Annotation by SIRIUS 4.0
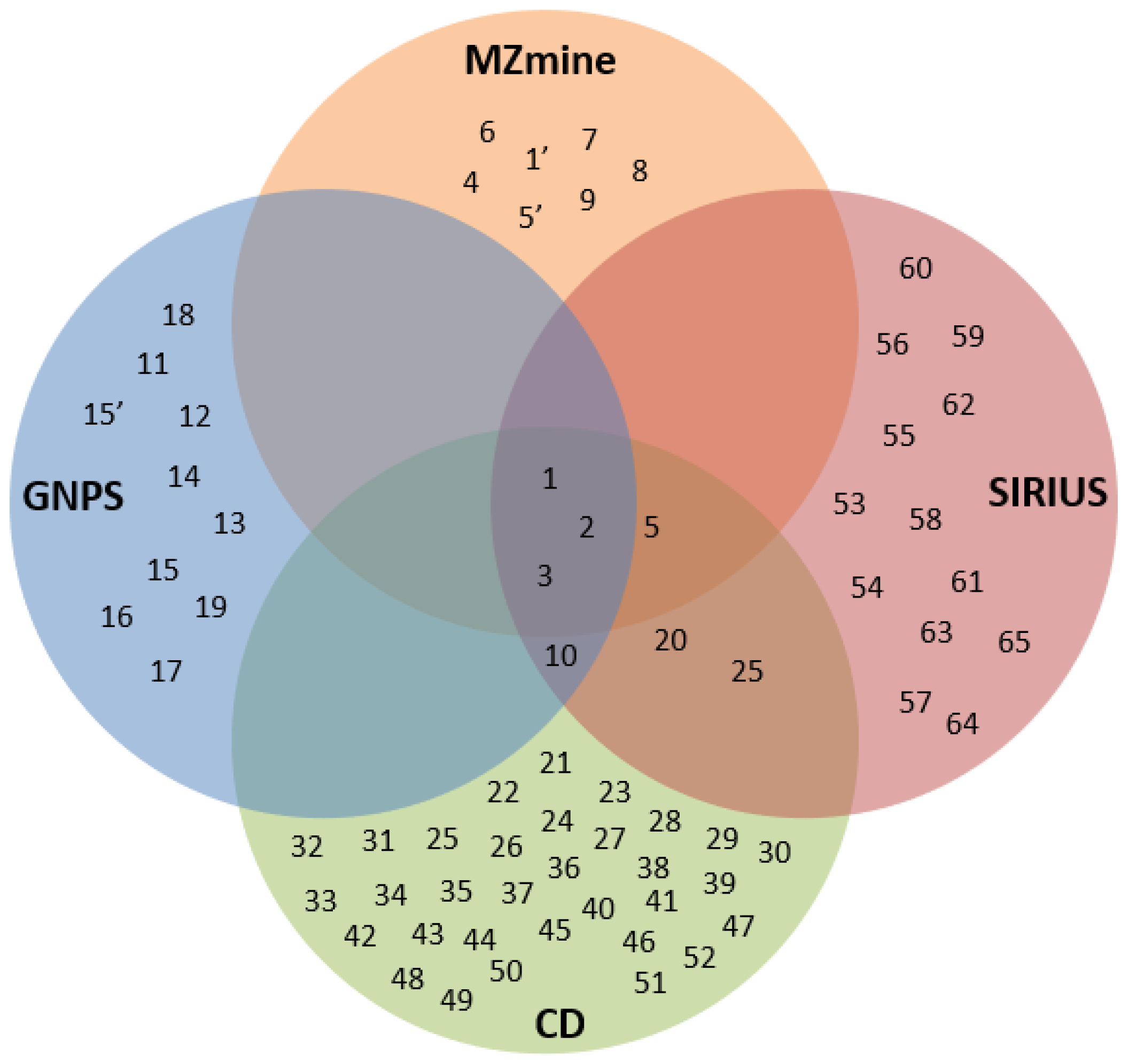
2.2. Verification of the Identified Phytochemicals
3. Materials and Methods
3.1. Materials and Reagents
3.1.1. Plant Materials and Extraction
3.1.2. Chemicals and Solvents
3.2. LCMS/MS Condition
3.2.1. Sample Preparation
3.2.2. LCMS Optimization
3.3. LCMS Data Analysis
3.3.1. MZmine 2.53 Data Pre-Treatment and Processing
3.3.2. Global Natural Social Molecular Networking (GNPS)
3.3.3. Compound Discoverer™ 3.0
3.3.4. SIRIUS 4.0
3.4. Isolation and Purification of Compound 1
3.4.1. General Chromatographic Procedure
3.4.2. Semi-Preparative and Recycling HPLC
3.4.3. Structural Elucidation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wolfender, J.L.; Rodriguez, S.; Hostettmann, K. Liquid chromatography coupled to mass spectrometry and nuclear magnetic resonance spectroscopy for the screening of plant constituents. J. Chromatogr. A 1998, 794, 299–316. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. BioSyst. 2012, 8, 470–481. [Google Scholar] [CrossRef]
- Covington, B.C.; McLean, J.A.; Bachmann, B.O. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 2017, 34, 6–24. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.32.31–30.32.24. [Google Scholar] [CrossRef]
- Ramos, A.E.F.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef]
- Alvarez-Rivera, G.; Ballesteros-Vivas, D.; Parada-Alfonso, F.; Ibañez, E.; Cifuentes, A. Recent applications of high resolution mass spectrometry for the characterization of plant natural products. TrAC Trends Anal. Chem. 2019, 112, 87–101. [Google Scholar] [CrossRef]
- Lim, T.K. Edible Medicinal and Non-Medicinal Plants; Springer: Cham, Switzerland, 2016; Volume 12. [Google Scholar]
- Ettebong, O.; Ubulom, P.M.E.; Obot, D.N. A Systematic review on Eleucine indica (L.) Gaertn.: From ethnomedicinal uses to pharmacological activities. J. Med. Plants Stud. 2020, 8, 262–274. [Google Scholar]
- Zakri, Z.H.M.; Suleiman, M.; Shean Yeaw, N.; Ngaini, Z.; Maili, S.; Salim, F. Eleusine Indica for Food and Medicine. J. Agrobiotechnol. 2021, 12, 68–87. [Google Scholar] [CrossRef]
- Burkill, I.H. A Dictionary of economic products of the Malay Peninsular. Minist. Agric. Co-Oper. Kuala Lumpur 1966, 137, 255. [Google Scholar]
- Hussain, A.G.; Noor, N.M.; Hussin, K. Nature’s Medicine: A Collection of Medicinal Plants from Malaysia’s Rainforest; Landskap Malaysia: Kuala Lumpur, Malaysia, 2015; Volume 1, Issue/No. a.; p. 374. [Google Scholar]
- Kulip, J.; Unchi, S.; Majawat, G. Research Contributions towards Optimization of Resources in Tropical Ecosystems. In Proceedings of the 4th Seminar on Tropical Ecosystem Research in Sabah, Sabah, 2000; Institute of Development Studies: Brighton, UK; Available online: https://books.google.co.th/books/about/4th_Seminar_on_Tropical_Ecosystem_Resear.html?id=sKwMAQAAMAAJ&redir_esc=y (accessed on 17 February 2023).
- Al-Zubairi, A.S.; Abdul, A.B.; Abdelwahab, S.I.; Peng, C.Y.; Mohan, S.; Elhassan, M.M. Eleucine indica Possesses Antioxidant, Antibacterial and Cytotoxic Properties. Evid. Based Complement. Altern. Med. 2011, 2011, 965370. [Google Scholar] [CrossRef]
- De Melo, G.O.; Muzitano, M.F.; Legora-Machado, A.; Almeida, T.A.; De Oliveira, D.B.; Kaiser, C.R.; Koatz, V.L.; Costa, S.S. C-glycosylflavones from the aerial parts of Eleusine indica inhibit LPS-induced mouse lung inflammation. Planta Med. 2005, 71, 362–363. [Google Scholar] [CrossRef] [PubMed]
- Sukor, S.; Zahari, Z.; Rahim, N.; Yusoff, J.; Salim, F. Chemical Constituents and Antiproliferative Activity of Eleusine indica. Sains Malays. 2022, 51, 873–882. [Google Scholar] [CrossRef]
- Desai, A.V. Phytochemical Investigation of Eleusine Indica for in-Vivo Diuretic and in-Vitro Anti-Urolithiatic Activity. World J. Pharm. Res. 2017, 6, 1292–1304. [Google Scholar] [CrossRef]
- Iberahim, R.; Yaacob, W.A.; Ibrahim, N. Phytochemistry, cytotoxicity and antiviral activity of Eleusine indica (sambau). In Proceedings of the 2015 UKM FST Postgraduate Colloquium, 1 September 2015; p. 030013. [Google Scholar]
- Iqbal, M.; Gnanaraj, C. Eleusine indica L. possesses antioxidant activity and precludes carbon tetrachloride (CCl₄)-mediated oxidative hepatic damage in rats. Environ. Health Prev. Med. 2012, 17, 307–315. [Google Scholar] [CrossRef]
- Phuong, N.M.; Sung, T.V.; Ripperger, H.; Adam, G. Sterol Glucosides from Eleucine indica. Planta Med. 1994, 60, 498. [Google Scholar] [CrossRef]
- Peñaloza, E.M.C.; Casanova, L.M.; Costa, S.S.; Leal, I.C.R.; Aguiar, P.F.d. Metabolite fingerprinting and profiling of the medicinal grass Eleusine indica based on HPLC-DAD, UPLC-DAD-MS/MS and NMR analyses. J. Braz. Chem. Soc. 2018, 29, 2522–2534. [Google Scholar] [CrossRef]
- Wang, D.; Fu, Z.; Xing, Y.; Tan, Y.; Han, L.; Yu, H.; Wang, T. Rapid identification of chemical composition and metabolites of Pingxiao Capsule in vivo using molecular networking and untargeted data-dependent tandem mass spectrometry. Biomed. Chromatogr. 2020, 34, e4882. [Google Scholar] [CrossRef]
- Sulaiman, N.A.S.; Yunus, Y.; Ahmad, R.; Salim, F. Diastereoseparation of C-7 Pentacyclic Oxindole Alkaloids and Their Vibrational Circular Dichroism Characteristics. Malays. J. Chem. 2021, 23, 34–41. [Google Scholar]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Hilu, K.W.; De Wet, J.M.J.; Seigler, D. Flavonoid patterns and systematics in Eleusine. Biochem. Syst. Ecol. 1978, 6, 247–249. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Simonato, M.; Fochi, I.; Vedovelli, L.; Giambelluca, S.; Carollo, C.; Padalino, M.; Carnielli, V.P.; Cogo, P. Urinary metabolomics reveals kynurenine pathway perturbation in newborns with transposition of great arteries after surgical repair. Metabolomics 2019, 15, 145. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Miralles, P.; López, A.; Dualde, P.; Coscollà, C.; Yusà, V. Liquid chromatography-Orbitrap Tribrid high-resolution mass spectrometry using data dependent-tandem mass spectrometry with triple stage fragmentation as a screening tool to perform identification and risk assessment of unknown substances in food contact epoxy resin. J. Sep. Sci. 2021, 44, 3020–3030. [Google Scholar] [CrossRef]
- Ohizumi, Y.; Kawada, M.; Kamada, M.; Nakajima, A.; Kajima, K.; Uozumi, N.; Hara, Y.; Guo, Y.; Ishibashi, M. Isolation of Adenosine and Cordysinin B from Anredera cordifolia that Stimulates CRE-Mediated Transcription in PC12 Cells. Planta Med. Int. Open 2021, 8, e19–e24. [Google Scholar] [CrossRef]
- Hoffmann, M.A.; Nothias, L.F.; Ludwig, M.; Fleischauer, M.; Gentry, E.C.; Witting, M.; Dorrestein, P.C.; Dührkop, K.; Böcker, S. High-confidence structural annotation of metabolites absent from spectral libraries. Nat. Biotechnol. 2022, 40, 411–421. [Google Scholar] [CrossRef]
- Huang, F.Y.; Yu, Y.; Wu, X.J. Characterization and Properties of Cellulose Oleate. Adv. Mater. Res. 2011, 197–198, 1306–1309. [Google Scholar] [CrossRef]
- Yang, X.; Kang, M.-C.; Lee, K.-W.; Kang, S.-M.; Lee, W.-W.; Jeon, Y.-J. Antioxidant activity and cell protective effect of loliolide isolated from Sargassum ringgoldianum subsp. coreanum. Algae 2011, 26, 201–208. [Google Scholar] [CrossRef]
- Yuan, Z.; Zheng, X.; Zhao, Y.; Liu, Y.; Zhou, S.; Wei, C.; Hu, Y.; Shao, H. Phytotoxic Compounds Isolated from Leaves of the Invasive Weed Xanthium spinosum. Molecules 2018, 23, 2840. [Google Scholar] [CrossRef]
- Grabarczyk, M.; Wińska, K.; Mączka, W.; Potaniec, B.; Anioł, M. Loliolide—The most ubiquitous lactone. Acta Univ. Lodziensis. Folia Biol. Oecolog. 2015, 11, 1–8. [Google Scholar] [CrossRef]
- Kasim, N.; Afzan, A.; Mediani, A.; Low, K.H.; Ali, A.M.; Mat, N.; Wolfender, J.-L.; Ismail, N.H. Correlation of chemical profiles obtained from 1H-NMR and LC–MS metabolomics with α-glucosidase inhibition activity for varietal selections of Ficus deltoidea. Phytochem. Anal. 2022, 33, 1235–1245. [Google Scholar] [CrossRef]
- Shaharuddin, N.H.; Ahmad, R.; Salim, F. Isolation of pteropodic acid from Malaysian Uncaria lanosa VAR. ferrea by using liquid chromatography mass spectrometry dereplication approach. Malays. J. Chem. 2021, 25, 959–965. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Dictionary of Natural Products (DNP) Database, v26.2. 2017. Available online: https://dmnp.chemnetbase.com/ (accessed on 25 March 2023).
- Cerrato, A.; Cannazza, G.; Capriotti, A.L.; Citti, C.; La Barbera, G.; Laganà, A.; Montone, C.M.; Piovesana, S.; Cavaliere, C. A new software-assisted analytical workflow based on high-resolution mass spectrometry for the systematic study of phenolic compounds in complex matrices. Talanta 2020, 209, 120573. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Rutz, A.; Dounoue-Kubo, M.; Ollivier, S.; Bisson, J.; Bagheri, M.; Saesong, T.; Ebrahimi, S.N.; Ingkaninan, K.; Wolfender, J.L.; Allard, P.M. Taxonomically Informed Scoring Enhances Confidence in Natural Products Annotation. Front. Plant Sci. 2019, 10, 1329. [Google Scholar] [CrossRef]

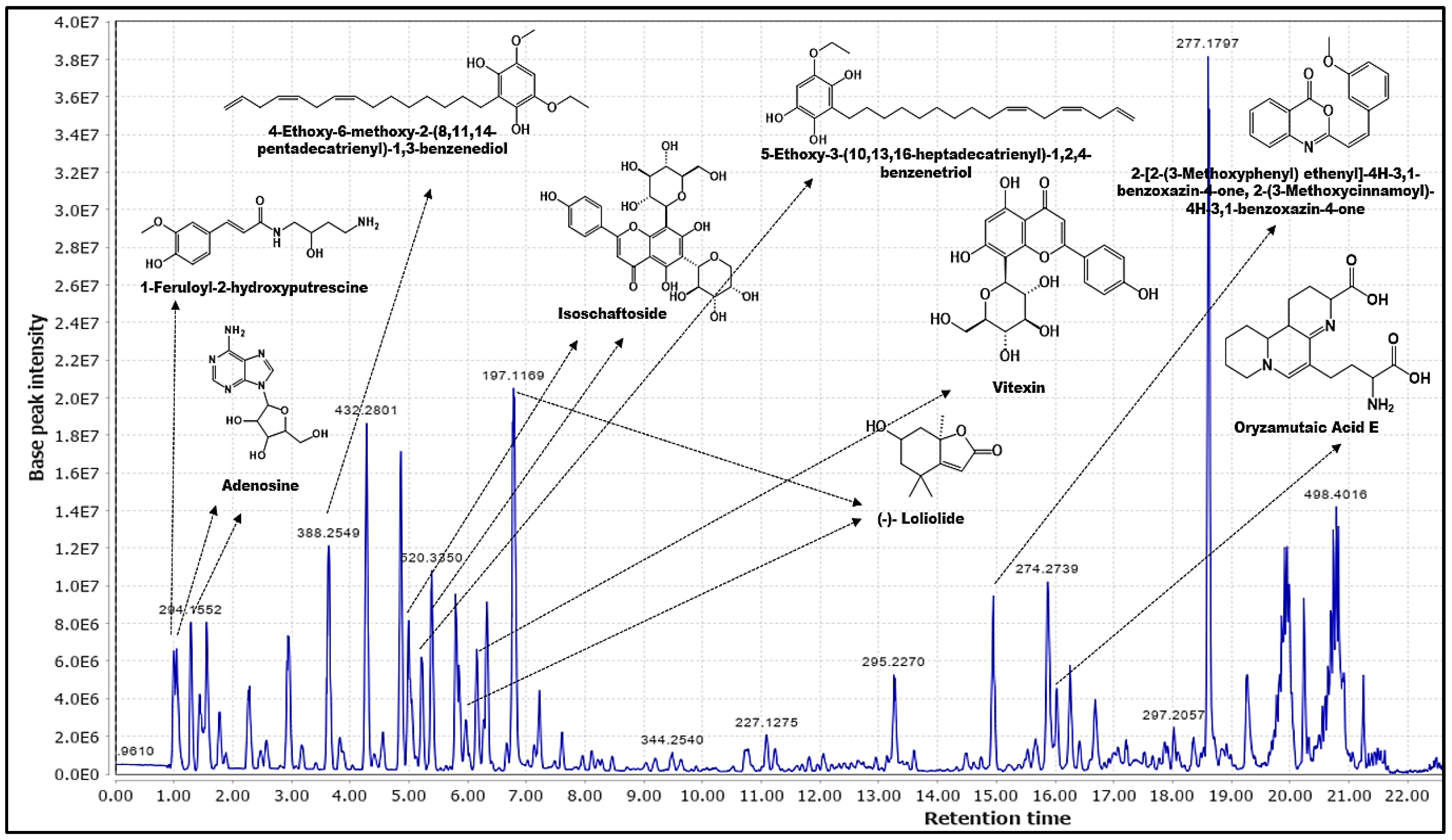




| Compound Name | [M + H]+ (m/z) | Rt (min) | Database Sourced |
|---|---|---|---|
| Loliolide (1) | 197.1174 | 6.78 | LR_positive.csv |
| Isoschaftoside (2) | 565.1553 | 5.00 | LR_positive.csv |
| Vitexin (3) | 433.1132 | 6.16 | LR_positive.csv |
| 4-Ethoxy-6-methoxy-2-(8,11,14-pentadecatrienyl)-1,3-benzenediol (4) | 388.2603 | 3.63 | DNP_Poaceae.csv |
| Adenosine (5) | 268.1044 | 1.28 | LR_positive.csv DNP_Poaceae.csv |
| Adenosine (5′) | 268.1044 | 1.05 | LR_positive.csv DNP_Poaceae.csv |
| 5-Ethoxy-3-(10,13,16-heptadecatrienyl)-1,2,4-benzenetriol (6) | 402.2763 | 5.22 | DNP_Poaceae.csv |
| Loliolide (1′) | 197.1174 | 5.97 | LR_positive.csv |
| Oryzamutaic Acid E (7) | 335.1830 | 16.02 | DNP_Poaceae.csv |
| 1-Feruloyl-2-hydroxyputrescine (8) | 280.1425 | 1.04 | DNP_Poaceae.csv |
| Isoschaftoside (2′) | 565.1552 | 5.38 | LR_positive.csv |
| 2-(3-Methoxycinnamoyl)-4H-3,1-benzoxazin-4-one (9) | 279.0906 | 14.95 | DNP_Poaceae.csv |
| No | Compound Name | Cluster Index | Library Class | Cosine | MZ Error ppm | Spectral m/z/ Library m/z | Instrument | Ion Mode | Data Source | Ion Source |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L-phenylalanine (10) | 1 | Bronze | 1.00 | 0 | 166.09/ 166.09 | Orbitrap | Positive | Trent Northen | LC-ESI |
| 2 | Vitexin (3) | 770 | Gold | 0.96 | 5 | 433.11/ 433.11 | Orbitrap | Positive | BMDMS-NP | ESI |
| 3 | Tris(2-butoxyethyl) phosphate (11) | 660 | Bronze | 0.94 | 0 | 399.25/ 399.25 | qTof | Positive | Nediljko Budisa | ESI |
| 4 | Loliolide (1) | 25 | Bronze | 0.94 | 1 | 197.12/ 197.12 | Orbitrap | Positive | Lihini Aluwihare | LC-ESI |
| 5 | Sulfamethazine (12) | 222 | Bronze | 0.92 | 9 | 279.09/ 279.09 | Hybrid FT | Positive | Massbank | ESI |
| 6 | Undecaethylene glycol (13) | 928 | Bronze | 0.92 | 2 | 503.31/ 503.31 | QQQ | Positive | Rob Knight | ESI |
| 7 | 13-Keto-9Z,11E-octadecadienoic acid (14) | 206 | Bronze | 0.85 | 1 | 277.22/ 277.22 | qTof | Positive | Wolfender/Litaudon | ESI |
| 8 | N-Fructosyl isoleucine (15) | 279 | Bronze | 0.84 | 4 | 294.15/ 294.15 | qTof | Positive | Massbank | ESI |
| 9 | Isoshaftoside (2) | 1053 | Gold | 0.82 | 0 | 565.16/ 565.16 | Orbitrap | Positive | BMDMS-NP | ESI |
| 10 | 9-Oxo-10E,12Z-octadecadienoic acid (16) | 295 | Bronze | 0.82 | 2 | 295.23/ 295.23 | IT-FT/ion trap with FTMS | Positive | Rob Knight | ESI |
| 11 | N-(1-Deoxy-1-fructosyl) phenylalanine (17) | 448 | Bronze | 0.77 | 1 | 328.14/ 328.14 | qTof | Positive | Claudia Maier | LC-ESI |
| 12 | N-Lauroylsarcosine (18) | 175 | Bronze | 0.76 | 1 | 272.22/ 272.22 | Hybrid FT | Positive | Massbank | ESI |
| 13 | N-Fructosyl isoleucine (15′) | 188 | Bronze | 0.72 | 2 | 276.14/ 276.14 | qTof | Positive | Massbank | ESI |
| 14 | 9,10-epoxy-12Z-octadecenoic acid (19) | 313 | Bronze | 0.72 | 2 | 297.24/ 297.24 | HCD | Positive | Rob Knight | ESI |
| No | Compound Name | Molecular Formula | [M + H]+ | Fragmentation Ions (MS/MS) | Rt (min) | ∆Mass [Da] | ∆Mass [ppm] | mzCloud Similarity | Reference | FISh Score | MIC * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 6-Gingerol (20) | C17H26O4 | 295.1905 | 277.1803, 173.4033 | 16.029 | 0.00012 | 0.42 | 70.0 | ChemSpider | 85.3 | 2 |
| 2 | -13S-hydroxy-9Z,11E,15Z-octadecatrienoic acid (21) | C18H30O3 | 295.2270 | 277.2160, 151.1113 | 13.268 | 0.00019 | 0.64 | 89.8 | DNP, ChemSpider | 89.71 | 2 |
| 3 | 3,5-Di-tert-butyl-4-hydroxybenzaldehyde (22) | C15H22O2 | 235.1692 | 219.1388, 179.1068 | 19.915 | 0.00007 | 0.30 | 95.9 | DNP, Chemspider | 80 | 2 |
| 4 | 5-Hydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy (23) | C23H24O12 | 493.1350 | 331.08015, 313.7641 | 7.132 | 0.00038 | 0.77 | 99.3 | ChemSpider | 73.91 | 2 |
| 5 | 7-Aminooctadecanoic acid (24) | C18H37NO3 | 316.2849 | 106.4888, 115.0000 | 20.015 | 0.00051 | 1.60 | - | DNP, ChemSpider | 100.00 | 2 |
| 6 | 9-Oxo-10E, 12E-octadecadienoic acid (25) | C18H30O3 | 295.2268 | 277.1609, 151.1115 | 20.489 | 0.00013 | 0.46 | 90.6 | ChemSpider, DNP | 77.78 | 2 |
| 7 | Adenosine (5) | C10H13N5O4 | 268.1041 | 136.0612, 112.7441 | 1.286 | 0.00028 | 1.04 | 99.3 | DNP ChemSpider | 83.33 | 2 |
| 8 | Alpha-linolenic acid (26) | C18H30O2 | 279.2319 | 279.2311, 261.2196 | 20.898 | 0.00008 | 0.28 | 89.5 | CD Database | 74.66 | 2 |
| 9 | Benazol-P (27) | C13H11N3O | 226.0975 | 126.0975, 120.0550 | 20.731 | 0.00003 | 0.13 | 95.8 | CD Database | 29.41 | 2 |
| 10 | Bis(4-ethylbenzylidene) sorbitol (28) | C24H39O6 | 415.2122 | 135.0799, 119.0851 | 17.867 | 0.00116 | 2.80 | 99.8 | CD Database | 75.00 | 2 |
| 11 | Bis(methylbenzylidene) sorbitol (29) | C22H26O6 | 387.1802 | 119.0491, 105.0692 | 16.258 | 0.00056 | 1.46 | 99.8 | CD Database | 44.44 | 2 |
| 12 | Citroflex A-4 (30) | C20H34O8 | 403.2331 | 185.0806, 129.0175 | 21.247 | 0.00046 | 1.46 | 95.3 | CD Database | 54.55 | 2 |
| 13 | Isoschaftoside (2) | C26H28O14 | 565.1564 | 427.1027, 409.0923 | 4.996 | 0.00113 | 2.00 | 97.1 | DNP | 45.16 | 2 |
| 14 | Hexaconazole (31) | C14H17C12N3O | 314.0824 | 184.9910, 158.9758 | 18.355 | 0.00056 | 1.78 | 98.1 | ChemSpider | 70.00 | 2 |
| 15 | L-phenylalanine (10) | C9H11NO2 | 166.0861 | 120.0802, 103.0537 | 1.557 | 0.00003 | −0.17 | 99.9 | ChemSpider | 87.50 | 2 |
| 16 | Mugineic acid (32) | C12H20N2O7 | 305.1344 | 227.1023, 191.0811 | 1.023 | 0.00035 | 1.14 | 90 | DNP | 71.43 | 2 |
| 17 | N-Lauryl sarcosine (33) | C15H29NO3 | 272.2946 | 230.4062, 90.0543 | 18.662 | 0.00018 | 0.66 | 77.8 | ChemSpider | 82.35 | 2 |
| 18 | Tricin 5-o-β-D-glucoside (34) | C23H24O12 | 493.1350 | 331.0818, 329.4911 | 8.108 | 0.00038 | 0.77 | 99.2 | ChemSpider | 66.67 | 2 |
| 19 | Vitexin (3) | C21H20O10 | 433.1136 | 397.0921, 313.0708 | 6.159 | 0.00051 | 1.18 | 97.8 | ChemSpider | 22.58 | 2 |
| 20 | 2,4,8,11-Dodecatetranoic acid (35) | C16H25NO | 248.2011 | 117.0693, 105.0693 | 20.684 | 0.00001 | 0.03 | - | DNP | 71.43 | 3 |
| 21 | Decaethylene glycol (36) | C20H42O11 | 459.2809 | 177.1119, 133.0855 | 4.864 | 0.00071 | 1.54 | 88 | ChemSpider | 85.71 | 2 |
| 22 | 3,6,9,12,15,18-Hexaoxaicosane-1,20-diol (37) | C14H30O8 | 327.2018 | 173.4045, 146.7485 | 2.949 | 0.00068 | 2.09 | 92.2 | CD Database | 75.00 | 2 |
| 23 | Octaethylene glycol (38) | C16H34O9 | 371.2282 | 329.1156, 133.0853, | 3.634 | 0.00050 | 1.34 | 97.2 | ChemSpider | 100 | 2 |
| 24 | Tris(2-butxyethyl) phosphate (39) | C18H39O7P | 399.2512 | 299.1618, 199.0726 | 20.240 | 0.00067 | 2.68 | 98.0 | CD Database | 77.8 | 3 |
| 25 | 3,6,9,12,15-Pentaoxapentacosan-1-ol (40) | C20H42O6 | 379.3095 | 173.3948, 89.0958 | 20.021 | 0.00056 | 1.48 | 84.6 | CD Database | 82.4 | 4 |
| 26 | 3,6,9,12,15-Tetraoxadocosan-1-ol (41) | C18H38O5 | 335.2802 | 133.0855, 89.059245 | 20.053 | 0.00046 | 1.37 | - | CD Database | 83.33 | 2 |
| 27 | 3-[Dodecyl(2-hydroxyethyl) amino]-1,2-propanediol (42) | C17H37NO3 | 304.2850 | 256.2627, 122.0806 | 17.006 | 0.00041 | 1.36 | - | ChemSpider | 80.00 | 3 |
| 28 | 3′-Geranyl-3,4,2′,4′-tetrahydroxy-6′-methoxydihydrochalcone (43) | C26H32O6 | 441.2253 | 173.4048, 159.6250 | 18.919 | −0.00185 | −4.21 | - | Arita Lab 6549 Flavonoid S | 100.00 | 2 |
| 29 | Loliolide (1) | C11H16O3 | 197.1173 | 179.1063, 133.1007 | 6.781 | 0.00007 | 0.37 | - | DNP | 46.15 | 3 |
| 30 | 6-(1,3-Benzodioxol-5-yl)-2-oxo-4-phenyl-1,2-dihydro-3-pyridinecarbonitrile (44) | C19H12N2O3 | 317.0925 | 289.0975, 271.0869 | 17.520 | 0.00049 | 1.55 | - | ChemSpider | 44.83 | 3 |
| 31 | Dibutyl phthalate (45) | C16H22O4 | 279.1592 | 233.0775, 149.0229 | 20.694 | 0.00012 | 0.45 | 86 | CD Database | 24.24 | 2 |
| 32 | Methyl 9,10-dihydroxystearate (46) | C19H3804 | 331.2845 | 285.2999, 173.3954 | 19.266 | 0.00040 | 1.21 | 82 | CD Database | 100.00 | 3 |
| 33 | Methyl 9H-b-carboline-3-carboxylate (47) | C13H10N2O2 | 227.0814 | 210.0726, 182.0786 | 3.826 | −0.00002 | −0.10 | 73 | ChemSpider | - | 3 |
| 34 | Hexaethylene glycol (48) | C12H26O7 | 283.1752 | 270.8245, 173.3921 | 2.277 | 0.00010 | 0.34 | 80 | ChemSpider | 100.00 | 3 |
| 35 | N,N-Bis(2-hydroxyethyl)dodecanamide (49) | C16H33NO3 | 288.2534 | 228.2041, 106.0856 | 17.915 | 0.00031 | 1.08 | 78 | CD Database | 50.00 | 2 |
| 36 | N-Lauryldiethanolamine (50) | C16H35NO2 | 274.2741 | 106.0862, 88.0751 | 15.873 | 0.00024 | 0.86 | - | CD Database | 70.00 | 2 |
| 37 | Nonaethylene glycol (51) | C18H38O10 | 415.2544 | 221.1385, 177.1166 | 4.279 | 0.00064 | 1.54 | 83 | ChemSpider | 100.00 | 2 |
| 38 | Safingol (52) | C18H39NO2 | 302.3055 | 102.2445 | 18.091 | 0.00009 | 0.31 | 93 | ChemSpider | 66.67 | 2 |
| No | Annotation (CSI: FingerID) | [M + H]+ | Fragment Ions | Rt (min) | Molecular Formula | COSMIC | Classification | Smiles | Links | MIC * |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2,6-Bis[2-[2-(2-aminoethoxy) ethoxy] ethylamino] pyridine (53) | 372.64 | 327.2019, 133.0853 | 2.94 | C17H33N5O4 | 0.8221 | Aminopyrimidines and derivatives | C1=CC(=NC(=C1)NCCOCCOCCN)NCCOCCOCCN | PubChem:(86235228) | 2 |
| 2 | N-[6-[3,5-diamino-2-[3-amino-6-(aminomethyl)-4,5-dihydroxyoxan-2-yl] oxy-6-hydroxycyclohexyl] oxyhexyl]-4-oxopentanamide (54) | 520.33 | 503.3067, 133.0851 | 5.39 | C23H45N5O8 | 0.722 | Dialkyl ether | CC(=O)CCC(=O)NCCCCCCOC1C(C(CC(C1OC2C(C(C(C(O2)CN)O)O)N)N)N)O | PubChem:(90164161) | 2 |
| 3 | Istamycin C1 (55) | 432.28 | 415.2545, 133.0854 | 4.28 | C19H37N5O6 | 0.611 | alkanolamine | CCNCC1CCC(C(O1)OC2C(CC(C(C2O)N(C)C(=O)CNC=O)OC)N)N | COCONUT, KEGG, Natural products, CHEBI, PubChem | 2 |
| 4 | Netilmicin (56) | 476.31 | 459.2808, 133.0854 | 4.87 | C21H41N5O7 | 0.595 | Dialkyl ether | CCNC1CC(C(C(C1OC2C(C(C(CO2)(C)O)NC)O)O)OC3C(CC=C(O3)CN)N)N | KEGG, COCONUT, Natural | 2 |
| 5 | Adenosine (5) | 268.10 | 136.0612 | 1.28 | C10H13N5O4 | 0.399 | Purine nucleosides | C1=NC(=C2C(=N1)N(C=N2)C3C(C(C(O3)CO)O)O)N | HMDB, SuperNatural HSDB, MeSH, Plantcyc, PubMed, NORMAN, COCONUT, KNApSAcK, Natural Products, PubChem, CHEBI, KEGG | 2 |
| 6 | N-{1-[(5-carbamimidamido-1-hydroxypentan-2-yl)-C-hydroxycarbonimidoyl]-2-methylpropyl-}2-[(1-hydroxyethylidene) amino]-4-methylpentanimidic acid (57) | 415.30 | 119.0851, 145.0637 | 20.87 | C19H38N6O4 | 0.371 | Dipeptides | CC(C)CC(C(=O)NC(C(C)C)C(=O)NC(CCCN=C(N)N)CO)NC(=O)C | COCONUT:(CNP0335012) | 2 |
| 7 | Plakoridine A (58) | 572.44 | 555.4106, 133.0851 | 19.85 | C35H57NO5 | 0.368 | Fatty amides | CCCCCCCCCCCCCCCCC(=O)C=C1C(C(C(N1CCC2=CC=C(C=C2)O)CCC)C(=O)OC)O | Natural product, COCONUT | 2 |
| 8 | Isoschaftoside (2) | 565.16 | 547.1456, 427.1027 | 5.00 | C26H28O14 | 0.350 | Flavanoid 8-C- Glycoside | C1C(C(C(C(O1)C2=C3C(=C(C(=C2O)C4C(C(C(C(O4)CO)O)O)O)O)C(=O)C=C(O3)C5=CC=C(C=C5)O)O)O)O | CHEBI, COCONUT, HMDB, KNApSAcK, MeSH, Natural Products, Pubchem, PubMed, SuperNatural, ZINC bio | 2 |
| 9 | Schaftoside (59) | 565.17 | 529.1347, 427.1027 | 5.36 | C26H28O14 | 0.350 | Flavanoid 8-C- Glycoside | C1C(C(C(C(O1)C2=C(C(=C3C(=C2O)C(=O)C=C(O3)C4=CC=C(C=C4)O)C5C(C(C(C(O5)CO)O)O)O)O)O)O)O | CHEBI, COCONUT, HMDB, KEGG, KNApSAcK, MeSH, Natural Products, Pubchem, PubMed, SuperNatural, ZINC bio | 2 |
| 10 | Broussonetine M1(60) | 348.28 | 173.1167, 155.1062 | 13.26 | C18H37NO5 | 0.295 | Long chain fatty acid | C(CCCCC(CCCCO)O)CCCC1C(C(C(N1)CO)O)O | COCONUT, Natural Product, PubChem, PubMed, KNapSAck, Supernatural | 2 |
| 11 | Vitexin (3) | 433.11 | 397.0921, 313.0708 | 6.11 | C21H20O10 | 0.283 | Phenolic glycoside | C1=CC(=CC=C1C2=CC(=O)C3=C(O2)C(=C(C=C3O)O)C4C(C(C(C(O4)CO)O)O)O)O | HMDB, SuperNatural, ZINC, MeSH, Plantcyc, PubMed, NORMAN, COCONUT, KNApSAcK, Natural Products, CHEBI, PubChem, KEGG, Biocyc | 2 |
| 12 | Fructose phenylalanine (61) | 328.14 | 292.118, 264.1228 | 1.45 | C15H21NO7 | 0.219 | Hexoses | C1=CC=C(C=C1)CC(C(=O)O)NCC(=O)C(C(C(CO)O)O)O | MeSH, PubMed | 2 |
| 13 | Loliolide (1) | 197.12 | 179.1063, 133.1007 | 6.78 | C11H16O3 | 0.171 | Dihydrofurans | O=C1OC2(C(=C1)C(C)(C)CC(O)C2)C | CHEBI, COCONUT, PubChem, PubMed, Natural Product, MeSH, KNapSAck, Supernatural | 2 |
| 14 | Justicidin B (62) | 365.11 | 203.0527, 185.0419 | 1.00 | C21H16O6 | 0.158 | Phenolic Glycoside | COC1=CC2=CC3=C(C(=C2C=C1OC)C4=CC5=C(C=C4)OCO5)C(=O)OC3 | CHEBI, COCONUT, PubChem, PubMed, Natural Product, MeSH, KNapSAck, Supernatural | 2 |
| 15 | Isoleucinopine (63) | 262.13 | 244.1180, 216.1230 | 1.05 | C11H19NO6 | 0.136 | Alpha Amino Acid | O=C(O)CCC(NC(C(=O)O)C(C)CC)C(=O)O | COCONUT, Natural Product, Pubchem | 2 |
| 16 | Gingerol (20) | 295.19 | 277.1803, 137.0592 | 16.03 | C17H26O4 | 0.093 | Carboxylic acids | O=C(CCC1=CC=C(O)C(OC)=C1)CC(O)CCCCC | CHEBI, COCONUT, PubChem, PubMed, Natural Product, MeSH, KNapSAck, Supernatural | 2 |
| 17 | 11-Oxooctadeca-9,12-dienoic acid (25) | 295.23 | 277.2160, 259.2054 | 13.26 | C18H30O3 | 0.071 | Lineolic acids and derivatives | O=C(C=CCCCCC)C=CCCCCCCCC(=O)O | COCONUT, PubChem, Natural Product, KNapSAck, Supernatural | 2 |
| 18 | L-phenylalanine (10) | 166.09 | 131.0486, 120.0802 | 1.55 | C9H11NO2 | 0.736 | Phenylalanine and derivatives | O=C(O)C(N)CC=1C=CC=CC1 | CHEBI, COCONUT, PubChem, PubMed, Natural Product, MeSH, KNapSAck, Supernatural, NORMAN, HMDB, Plantcyc | 2 |
| 19 | Watsonol B (64) | 277.18 | 231.1739, 137.0591 | 18.61 | C17H24O3 | 0.109 | Prenol lipids | O=C(OCC(=C)C1CCC(=C)C2CCC(=C)C2C1O)C | COCONUT, KNapSAcK, Natural Products, PubChem, SuperNatural | 2 |
| 20 | Embelin (65) | 295.19 | 203.1792, 137.0592f | 18.61 | C17H26O4 | 0.239 | Prenol lipids | CCCCCCCCCCCC1=C(C(=O)C=C(C1=O)O)O | COCONUT, KNapSAcK, Natural Products, PubChem, SuperNatural, CHEBI, KEGG, MeSH, ZINC bio, NORMAN | 2 |
| No | [M + H]+ | RT (min) | Molecular Formula | Class of Compound | COSMIC |
|---|---|---|---|---|---|
| 1 | 498.40 | 20.79 | C26H51N5O4 | Aminopyrimidines and derivatives | 0.730 |
| 2 | 454.38 | 20.82 | C24H47N5O3 | Azoles | 0.310 |
| 3 | 542.43 | 20.73 | C28H55N5O5 | Amino acids and derivatives | 0.378 |
| 4 | 388.25 | 3.64 | C17H33N5O5 | Heteroaromatic Compounds | 0.607 |
| 5 | 484.38 | 19.95 | C25H49N5O4 | Aminopyrimidines and derivatives | 0.735 |
| 6 | 528.41 | 19.91 | C27H53N5O5 | Benzenoids | 0.546 |
| 7 | 274.27 | 15.88 | C16H35NO2 | 1,2-aminoalcohols | 0.087 |
| 8 | 440.36 | 19.99 | C23H45N5O3 | Heteroaromatic Compounds | 0.652 |
| 9 | 279.09 | 14.95 | C12H11FN4O3 | Benzoyl derivatives | 0.314 |
| 10 | 399.25 | 20.23 | C18H39O7P | Trialkyl phosphates | 0.190 |
| 11 | 490.32 | 6.33 | C28H43NO6 | Benzenoids | 0.293 |
| 12 | 586.45 | 20.69 | C30H59N5O6 | Peptides | 0.372 |
| 13 | 572.24 | 19.85 | C29H57N5O6 | N-Acyl amines | 0.370 |
| 14 | 294.16 | 1.29 | C12H23NO7 | Alpha Amino Acids and derivatives | 0.111 |
| 15 | 415.25 | 4.28 | C18H38O10 | Polyethylene glycols | 0.303 |
| 16 | 454.38 | 20.82 | C24H47N5O3 | Azoles | 0.310 |
| 17 | 327.22 | 18.63 | C18H30O5 | Fatty acid methyl esters | 0.063 |
| 18 | 372.26 | 2.94 | C17H33N5O4 | Aminopyrimidines and derivatives | 0.855 |
| 19 | 276.14 | 1.28 | C8H17N7O4 | Primary amines | 0.096 |
| 20 | 371.23 | 3.64 | C16H34O9 | Polyethylene glycols | 0.329 |
| 21 | 534.35 | 6.80 | C24H47N5O8 | Dialkyl ethers | 0.138 |
| 22 | 484.39 | 19.95 | C25H49N5O4 | Aminopyrimidines and derivatives | 0.735 |
| 23 | 280.14 | 1.05 | C11H21NO7 | Alpha amino acids | 0.170 |
| 24 | 520.33 | 5.39 | C23H45N5O8 | Dialkyl ethers | 0.632 |
| 25 | 402.27 | 5.21 | C18H35N5O5 | Dialkyl ethers | 0.474 |
| 26 | 459.28 | 4.87 | C20H42O11 | Polyethylene glycols | 0.298 |
| 27 | 446.29 | 5.79 | C20H39N5O6 | Heteroaromatic Compounds | 0.102 |
| 28 | 388.25 | 3.64 | C17H33N5O5 | Heteroaromatic Compounds | 0.606 |
| 29 | 344.23 | 2.95 | C15H29N5O4 | Dialkyl ethers | 0.625 |
| 30 | 564.34 | 5.85 | C25H49N5O9 | Carboxylic acid amides | 0.103 |
| 31 | 387.18 | 16.25 | C29H20 | Phenols | 0.219 |
| 32 | 630.48 | 20.64 | C23H63N5O7 | Fatty amides | 0.372 |
| 33 | 410.35 | 20.85 | C22H43N502 | Aminopyrimidines and derivatives | 0.193 |
| 34 | 388.26 | 3.63 | C17H33N5O5 | Aryl thiothers | 0.605 |
| 35 | 304.30 | 19.27 | C21H37N | Phenylmethylamines | 0.278 |
| 36 | 440.36 | 19.99 | C23H45N5O3 | Heteroaromatic compounds | 0.463 |
| 37 | 268.10 | 1.05 | C15H13BO4 | Glycosyl compounds | 0.506 |
| 38 | 328.23 | 2.29 | C15H29N5O3 | Aminopyrimidines and derivatives | 0.416 |
| 39 | 277.18 | 16.03 | C17H24O3 | Prenol lipids | 0.135 |
| 40 | 371.23 | 3.64 | C16H34O9 | Polyethylene glycol | 0.329 |
| 41 | 310.13 | 1.44 | C15H19NO6 | Phenylalanine and derivatives | 0.145 |
| 42 | 372.26 | 2.94 | C17H33N5O4 | Aminopyrimidines and derivatives | 0.855 |
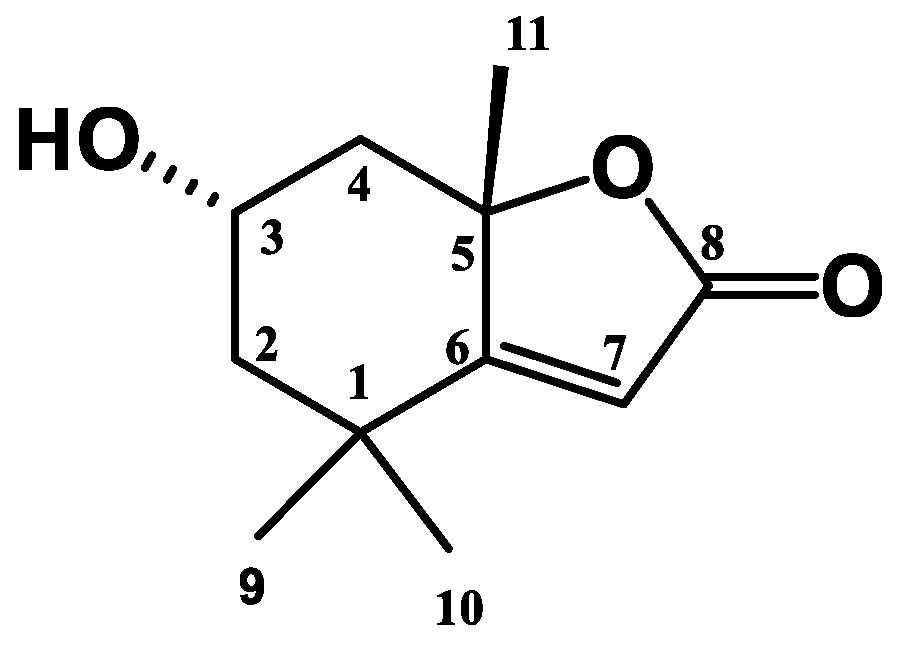
| Loliolide | ||||||||
|---|---|---|---|---|---|---|---|---|
| Position | δH (ppm) | δC (ppm) | δH (ppm) a (CD3OD) | δC (ppm) a (CD3OD) | HMBC | HMQC | COSY | NOESY |
| 1 | - | 35.73 | - | 35.75 | - | - | - | - |
| 2 | α: 2.01 (dt, J = 14.4, 3.1) β: 1.55 (dd, J = 14.4, 3.7,) | 49.53 | α: 1.50 (dt, J = 14.4, 3.1) β: 1.98 (dd, J = 4.0, 16.0) | 46.53 | C-1, C-4, C-3, C-8 C-1, C-10, C-11 | C-2 | H2β, H3 H2α, H3 | - |
| 3 | 4.33 (m) | 66.1 | 4.19 | 65.80 | C-1, C-5, C-11 | C-3 | H2α, H11 | H11, H2β, H9 |
| 4 | α: 1.78 (d, J = 4.0) β: 2.44 (dt, J = 13.6, 2.4) | 46.99 | α: 1.71 (d, J = 4.0) β: 2.46 (dt, J = 4.0, 16.0) | 44.99 | C-3, C-4, C-5, C-8, C-10 C-3, C-4, C-5, C-8, C-9 | C-4 | H4β, H3 H2α, H3 | - |
| 5 | - | 87.8 | - | 87.52 | - | - | - | - |
| 6 | - | 170.1 | - | 172.12 | - | - | - | - |
| 7 | 5.69 (s) | 112.2 | 5.80 | 111.88 | C-1, C-5, C-6, C-8, C-9 | C-7 | - | - |
| 8 | - | 181.5 | - | 184.13 | - | - | - | - |
| 9 | 1.47 | 25.8 | 1.45 | 25.52 | C-1, C-4, C-8, C-11 | C-9 | - | - |
| 10 | 1.27 | 26.3 | 1.26 | 29.57 | C-1, C-3, C-4, C-5, C-7, C-8, C-9 | C-10 | - | - |
| 11 | 1.78 | 29.8 | 1.74 | 25.99 | C-3, C-4, C-5, C-8, C-10 | C-11 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sukor, N.S.M.; Zakri, Z.H.M.; Rasol, N.E.; Salim, F. Annotation and Identification of Phytochemicals from Eleusine indica Using High-Performance Liquid Chromatography Tandem Mass Spectrometry: Databases-Driven Approach. Molecules 2023, 28, 3111. https://doi.org/10.3390/molecules28073111
Sukor NSM, Zakri ZHM, Rasol NE, Salim F. Annotation and Identification of Phytochemicals from Eleusine indica Using High-Performance Liquid Chromatography Tandem Mass Spectrometry: Databases-Driven Approach. Molecules. 2023; 28(7):3111. https://doi.org/10.3390/molecules28073111
Chicago/Turabian StyleSukor, Nur Syahirah Mad, Zikry Hamizan Md Zakri, Nurulfazlina Edayah Rasol, and Fatimah Salim. 2023. "Annotation and Identification of Phytochemicals from Eleusine indica Using High-Performance Liquid Chromatography Tandem Mass Spectrometry: Databases-Driven Approach" Molecules 28, no. 7: 3111. https://doi.org/10.3390/molecules28073111
APA StyleSukor, N. S. M., Zakri, Z. H. M., Rasol, N. E., & Salim, F. (2023). Annotation and Identification of Phytochemicals from Eleusine indica Using High-Performance Liquid Chromatography Tandem Mass Spectrometry: Databases-Driven Approach. Molecules, 28(7), 3111. https://doi.org/10.3390/molecules28073111





