In Silico Exploration of Alternative Conformational States of VDAC
Abstract
1. Introduction
2. Results
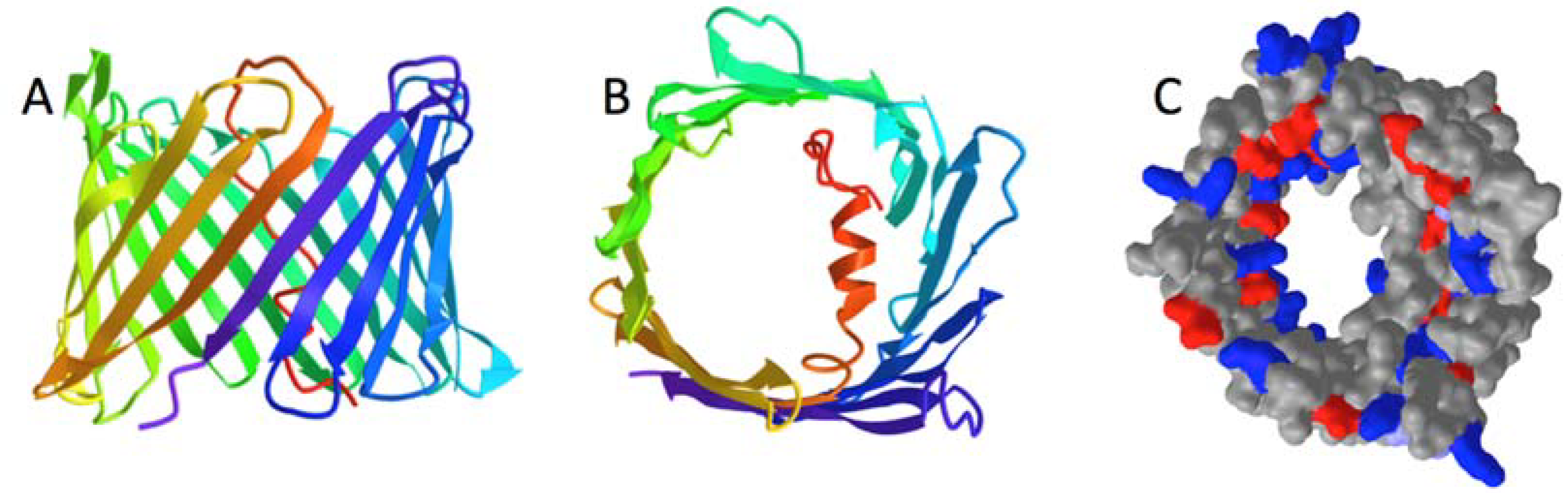
2.1. Folding Predictions for VDAC Monomer Constructs
- β-barrels with varying gaps (4.6–17.5 Å) between N- and C-terminal β-strands (“N-C gap”, orthogonal distance between adjacent β-strands measured at Cα atoms).
- “Partial” β-barrels with very wide “N-C gap” (28–31 Å) measured normal to cylinder axis.
- “Disrupted” β-barrels with short segments containing 4–6 β-strands tilted away from the main β-sheet having 9–12 β-strands.
- “Reverse” β-barrels, with left-hand rather than the normal right-hand twist observed in anti-parallel β-sheets in proteins.
- Collapsed β-sheet structures with no pore.
2.1.1. Predicted Topologies for hVDAC1
2.1.2. Predicted Topologies for hVDAC2 and ncVDAC
2.2. Predictions for VDAC Dimers and Domain Swapping
3. Discussion
3.1. Support for Basic Premise
- Deletion of the C-terminal β-strand from the sequence of ncVDAC yields a Class I β-barrel with or without (+/−) NTERM, with about equal N-C gaps (9.4 vs. 9.8 Å) (Table 1). The actual engineered construct yields a functional, voltage-gated pore with slightly smaller conductance than the wild-type 19-strand β-barrel in bilayers [40], consistent with the premise that Class I topologies with small N-C gaps can fold as functional β-barrels in a membrane environment.
- hVDAC2 constructs engineered with deletions of one to three β-strands at the C-terminal show pore-forming activity in bilayers, although only the 17-stranded variant display voltage-gating similar to the 19-stranded wild-type [41]. All three truncated sequences yield Class I folding predictions for hVDAC2 without NTERM (Table 1), but only the 17-stranded and wildtype 19-stranded sequences (corresponding to gatable pores) yield Class I topologies with NTERM present. The results are again consistent with the premise that Class I predictions represent pores in membranes, although the inverse is not necessarily true when NTERM is present. Instead, interactions (entanglement) between an extended N-terminal domain and the β-sheet appear to frustrate folding of the polypeptide into Class I β-barrels (evident in (p5) Figure 2) for truncation of β19–17 with both hVDAC2 and hVDAC1.
3.2. Structural Role of the N-Terminal Domain
3.3. Implications of the RoseTTAFold Predictions for VDAC Conformers
3.4. Implications for VDAC-VDAC Interactions
4. Materials and Methods
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Colombini, M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature 1979, 279, 643–645. [Google Scholar] [CrossRef]
- Mannella, C.A. Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications. J. Struct. Biol. 1998, 121, 207–218. [Google Scholar] [CrossRef]
- Mannella, C.A. VDAC-A Primal Perspective. Int. J. Mol. Sci. 2021, 22, 1685. [Google Scholar] [CrossRef] [PubMed]
- Colombini, M. Voltage gating in the mitochondrial channel, VDAC. J. Memb. Biol. 1989, 111, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, J.G.; Hoek, J.B. Regulation of hexokinase binding to VDAC. J. Bioenerg. Biomembr. 2008, 40, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Rostovtseva, T.K.; Sheldon, K.L.; Hassanzadeh, E.; Monge, C.; Saks, V.; Bezrukov, S.M.; Sackett, D.L. Tubulin binding blocks mitochondrial voltage-dependent anion channel and regulates respiration. Proc. Natl. Acad. Sci. USA 2008, 105, 18746. [Google Scholar] [CrossRef]
- Rostovtseva, T.; Colombini, M. VDAC channels mediate and gate the flow of ATP: Implications for the regulation of mitochondrial function. Biophys. J. 1997, 72, 1954–1962. [Google Scholar] [CrossRef]
- Colombini, M. The VDAC channel: Molecular basis for selectivity. Biochim. Biophys. Acta 2016, 1863, 2498–2502. [Google Scholar] [CrossRef]
- Tan, W.; Colombini, M. VDAC closure increases calcium ion flux. Biochim. Biophys. Acta 2007, 1768, 2510–2515. [Google Scholar] [CrossRef]
- Pastorino, J.G.; Shulga, N.; Hoek, J.B. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 2002, 277, 7610–7618. [Google Scholar] [CrossRef]
- De Stefani, D.; Bononi, A.; Romagnoli, A.; Messina, A.; De Pinto, V.; Pinton, P.; Rizzuto, R. VDAC1 selectively transfers apoptotic Ca2+ signals to mitochondria. Cell Death Differ. 2012, 19, 267–273. [Google Scholar] [CrossRef]
- Rosencrans, W.M.; Rajendran, M.; Bezrukov, S.M.; Rostovtseva, T.K. VDAC regulation of mitochondrial calcium flux: From channel biophysics to disease. Cell Calcium 2021, 94, 102356. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Mizrachi, D. VDAC1: From structure to cancer therapy. Front. Oncol. 2012, 2, 164. [Google Scholar] [CrossRef]
- Varughese, J.T.; Buchanan, S.K.; Pitt, A.S. The Role of Voltage-Dependent Anion Channel in Mitochondrial Dysfunction and Human Disease. Cells 2021, 10, 1737. [Google Scholar] [CrossRef] [PubMed]
- Hiller, S.; Garces, R.G.; Malia, T.J.; Orekhov, V.Y.; Colombini, M.; Wagner, G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 2008, 321, 1206–1210. [Google Scholar] [CrossRef]
- Bayrhuber, M.; Meins, T.; Habeck, M.; Becker, S.; Giller, K.; Villinger, S.; Vonrhein, C.; Griesinger, C.; Zweckstetter, M.; Zeth, K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA 2008, 105, 15370–15375. [Google Scholar] [CrossRef]
- Ujwal, R.; Cascio, D.; Colletier, J.-P.; Faham, S.; Zhang, J.; Toro, L.; Ping, P.; Abramson, J. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA 2008, 105, 17742–17747. [Google Scholar] [CrossRef]
- Hiller, S.; Abramson, J.; Mannella, C.; Wagner, G.; Zeth, K. The 3D structures of VDAC represent a native conformation. Trends Biochem. Sci. 2010, 35, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Zachariae, U.; Schneider, R.; Briones, R.; Gattin, Z.; Demers, J.P.; Giller, K.; Maier, E.; Zweckstetter, M.; Griesinger, C.; Becker, S.; et al. β-Barrel mobility underlies closure of the voltage-dependent anion channel. Structure 2012, 20, 1540–1549. [Google Scholar] [CrossRef]
- Krammer, E.-M.; Vu, G.H.T.; Homblé, F.; Prévost, M. Dual Mechanism of Ion Permeation through VDAC Revealed with Inorganic Phosphate Ions and Phosphate Metabolites. PLoS ONE 2015, 10, e0121746. [Google Scholar] [CrossRef] [PubMed]
- Bergdoll, L.; Grabe, M.; Abramson, J. An Assessment of How VDAC Structures Have Impacted Our Understanding of Their Function. In Molecular Basis for Mitochondrial Signaling; Rostovtseva, T.K., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 141–160. [Google Scholar]
- Haloi, N.; Wen, P.-C.; Cheng, Q.; Yang, M.; Natarajan, G.; Camara, A.K.S.; Kwok, W.-M.; Tajkhorshid, E. Structural basis of complex formation between mitochondrial anion channel VDAC1 and Hexokinase-II. Commun. Biol. 2021, 4, 667. [Google Scholar] [CrossRef]
- Shuvo, S.R.; Ferens, F.G.; Court, D.A. The N-terminus of VDAC: Structure, mutational analysis, and a potential role in regulating barrel shape. Biochim. Biophys. Acta 2016, 1858, 1350–1361. [Google Scholar] [CrossRef]
- De Pinto, V.; Prezioso, G.; Thinnes, F.; Link, T.A.; Palmieri, F. Peptide-specific antibodies and proteases as probes of the transmembrane topology of the bovine heart mitochondrial porin. Biochemistry 1991, 30, 10191–10200. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.; Dias, J.A.; D’Arcangelis, D.; Mannella, C.A. Peptide-specific antibodies as probes of the topography of the voltage-gated channel in the mitochondrial outer membrane of Neurospora crassa. J. Biol. Chem. 1995, 270, 16694–16700. [Google Scholar] [CrossRef]
- Konstantinova, S.A.; Mannella, C.A.; Skulachev, V.P.; Zorov, D.B. Immunoelectron microscopic study of the distribution of porin on outer membranes of rat heart mitochondria. J. Bioenerg. Biomembr. 1995, 27, 93–99. [Google Scholar] [CrossRef]
- Guo, X.W.; Smith, P.R.; Cognon, B.; D’Arcangelis, D.; Dolginova, E.; Mannella, C.A. Molecular design of the voltage-dependent, anion-selective channel in the mitochondrial outer membrane. J. Struct. Biol. 1995, 114, 41–59. [Google Scholar] [CrossRef]
- Peng, S.; Blachly-Dyson, E.; Forte, M.; Colombini, M. Large scale rearrangement of protein domains is associated with voltage gating of the VDAC channel. Biophys. J. 1992, 62, 123–131; discussion 131–135. [Google Scholar] [CrossRef]
- Zimmerberg, J.; Parsegian, V.A. Polymer inaccessible volume changes during opening and closing of a voltage-dependent ionic channel. Nature 1986, 323, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Kinnally, K.W.; Mannella, C.A. Circular dichroism studies of the mitochondrial channel, VDAC, from Neurospora crassa. Biophys. J. 1996, 71, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Lessen, H.J.; Fleming, P.J.; Fleming, K.G.; Sodt, A.J. Transmembrane Beta-Barrel Proteins Rigidify the Bacterial Outer Membrane. Biophys. J. 2019, 116, 327a. [Google Scholar] [CrossRef]
- Baker, D. What has de novo protein design taught us about protein folding and biophysics? Protein Sci. 2019, 28, 678–683. [Google Scholar] [CrossRef]
- Koga, N.; Koga, R.; Liu, G.; Castellanos, J.; Montelione, G.T.; Baker, D. Role of backbone strain in de novo design of complex α/β protein structures. Nat. Commun. 2021, 12, 3921. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Service, R.F. Protein structures for all. Science 2021, 374, 1426–1427. [Google Scholar] [CrossRef]
- White, S.H.; Wimley, W.C.; Ladokhin, A.S.; Hristova, K. Protein folding in membranes: Determining energetics of peptide-bilayer interactions. Meth. Enzymol. 1998, 295, 62–87. [Google Scholar]
- Schredelseker, J.; Paz, A.; López, C.J.; Altenbach, C.; Leung, C.S.; Drexler, M.K.; Chen, J.N.; Hubbell, W.L.; Abramson, J. High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J. Biol. Chem. 2014, 289, 12566–12577. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.R.; Zenezini Chiozzi, R.; Roelofs, M.C.; Hevler, J.F.; Ravi, R.T.; Maitan, P.; Zhang, M.; Henning, H.; Bromfield, E.G.; Howes, S.C.; et al. In-cell structures of conserved supramolecular protein arrays at the mitochondria-cytoskeleton interface in mammalian sperm. Proc. Natl. Acad. Sci. USA 2021, 118, e2110996118. [Google Scholar] [CrossRef] [PubMed]
- Popp, B.; Court, D.A.; Benz, R.; Neupert, W.; Lill, R. The Role of the N and C Termini of Recombinant Neurospora Mitochondrial Porin in Channel Formation and Voltage-dependent Gating. J. Biol. Chem. 1996, 271, 13593–13599. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.R.; Mahalakshmi, R. Evolutionary selection of a 19-stranded mitochondrial β-barrel scaffold bears structural and functional significance. J. Biol. Chem. 2020, 295, 14653–14665. [Google Scholar] [CrossRef]
- Yang, A.S.; Honig, B. Free energy determinants of secondary structure formation: II. Antiparallel beta-sheets. J. Mol. Biol. 1995, 252, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Koppel, D.A.; Kinnally, K.W.; Masters, P.; Forte, M.; Blachly-Dyson, E.; Mannella, C.A. Bacterial expression and characterization of the mitochondrial outer membrane channel. Effects of n-terminal modifications. J. Biol. Chem. 1998, 273, 13794–13800. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Gupta, R.; Blanco, L.P.; Yang, S.; Shteinfer-Kuzmine, A.; Wang, K.; Zhu, J.; Yoon, H.E.; Wang, X.; Kerkhofs, M.; et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 2019, 366, 1531–1536. [Google Scholar] [CrossRef]
- Najbauer, E.E.; Becker, S.; Giller, K.; Zweckstetter, M.; Lange, A.; Steinem, C.; de Groot, B.L.; Griesinger, C.; Andreas, L.B. Structure, gating and interactions of the voltage-dependent anion channel. Eur. Biophys. J. 2021, 50, 159–172. [Google Scholar] [CrossRef]
- Teijido, Ó.; Ujwal, R.; Hillerdal, C.-O.; Kullman, L.; Rostovtseva, T.K.; Abramson, J. Affixing N-terminal α-Helix to the Wall of the Voltage-dependent Anion Channel Does Not Prevent Its Voltage Gating. J. Biol. Chem. 2012, 287, 11437–11445. [Google Scholar] [CrossRef] [PubMed]
- Keinan, N.; Tyomkin, D.; Shoshan-Barmatz, V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol. Cell. Biol. 2010, 30, 5698–5709. [Google Scholar] [CrossRef]
- Höhr, A.I.C.; Lindau, C.; Wirth, C.; Qiu, J.; Stroud, D.A.; Kutik, S.; Guiard, B.; Hunte, C.; Becker, T.; Pfanner, N.; et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science 2018, 359, eaah6834. [Google Scholar] [CrossRef]
- Xu, X.; Colombini, M. Autodirected insertion: Preinserted VDAC channels greatly shorten the delay to the insertion of new channels. Biophys. J. 1997, 72, 2129–2136. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics 2020, 36, 131–135. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Lu, S.; Madej, T.; Marchler, G.H.; Cheng, T.; Chong, L.C.; et al. iCn3D: From Web-Based 3D Viewer to Structural Analysis Tool in Batch Mode. Front. Mol. Biosci. 2022, 9, 831740. [Google Scholar] [CrossRef] [PubMed]

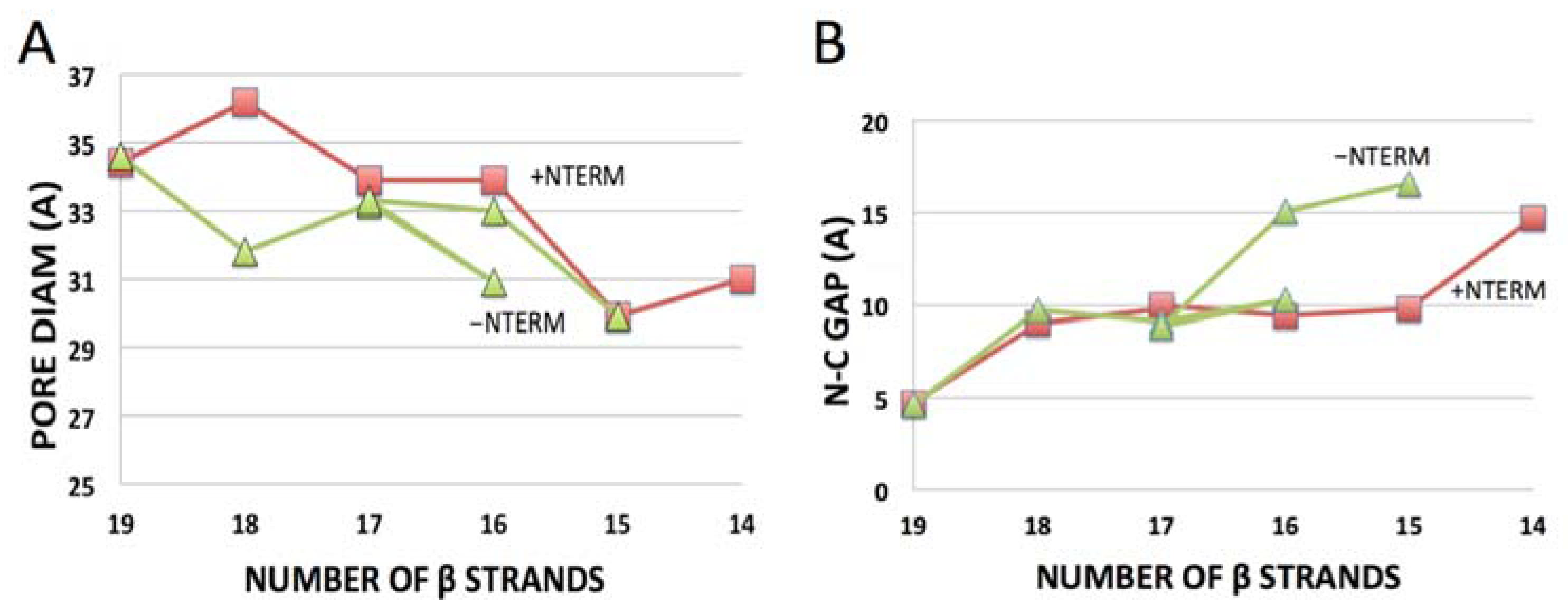

| CONSTRUCT + NTERM | CLASS | N-C GAP (Å) | lDDT | CONSTRUCT − NTERM | CLASS | N-C GAP (Å) | lDDT |
|---|---|---|---|---|---|---|---|
| HVDAC1 | I | 4.7 | 0.81 | HVDAC1 - N | I | 4.6 | 0.88 |
| HVDAC1 - β19 | I | 9.0 | 0.81 | HVDAC1 - N,β19 | I | 9.7 | 0.88 |
| HVDAC1 - β(18–19) | I | 9.8 | 0.81 | HVDAC1 - N,β(18–19) | I | 9.1 | 0.89 |
| HVDAC1 - β(17–19) | III | − | 0.81 | HVDAC1 - N,β(17–19) | I | 10.3 | 0.89 |
| HVDAC1 - β(13–14) | I | 10.0 | 0.80 | HVDAC1 - N,β(13–14) | I | 8.8 | 0.85 |
| HVDAC1 - β(13–14,19) | I | 9.4 | 0.80 | HVDAC1 - N,β(13–14,19) | I | 15.1 | 0.86 |
| HVDAC1 - β(13–14,18–19) | I | 9.8 | 0.80 | HVDAC1 - N,β(13–14,18–19) | I | 16.6 | 0.85 |
| HVDAC1 - β(13–14,17–19) | I | 14.7 | 0.78 | HVDAC1 - N,β(13–14,17–19) | II | 31.0 | 0.88 |
| HVDAC2 | I | 7.5 | 0.81 | HVDAC2 - N | I | 5.3 | 0.88 |
| HVDAC2 - β19 | IV | − | 0.78 | HVDAC2 - N,β19 | I | 9.9 | 0.85 |
| HVDAC2 - β(18–19) | I | 9.7 | 0.78 | HVDAC2 - N,β(18,19) | I | 9.1 | 0.88 |
| HVDAC2 - β(17–19) | III | − | 0.80 | HVDAC2 - N,β(17–19) | I | 16.9 | 0.87 |
| HVDAC2 - β(13–14) | III | − | 0.76 | HVDAC2 - N,β(13–14) | III | − | 0.82 |
| HVDAC2 - β(13–14,19) | III | − | 0.76 | HVDAC2 - N,β(13–14,19) | I | 9.7 | 0.83 |
| HVDAC2 - β(13–14,18–19) | III | − | 0.76 | HVDAC2 - N,β(13–14,18–19) | II | 28.5 | 0.85 |
| HVDAC2 - β(13–14,17–19) | I | 17.5 | 0.77 | HVDAC2 - N,β(13–14,17–19) | II | 29.7 | 0.86 |
| NCVDAC | I | 7.3 | 0.77 | NCVDAC - N | I | 5.6 | 0.84 |
| NCVDAC - β19 | I | 9.8 | 0.79 | NCVDAC - N,β19 | I | 9.4 | 0.83 |
| NCVDAC - β(18–19) | V | − | 0.78 | NCVDAC - N,β(18–19) | I | 9.4 | 0.85 |
| NCVDAC - β(17–19) | III, IV | − | 0.77 | NCVDAC - N,β(17–19) | II | 27.9 | 0.87 |
| NCVDAC - β(13–14) | IV | − | 0.77 | NCVDAC - N,β(13–14) | IV | − | 0.84 |
| NCVDAC - β(13–14,19) | V | − | 0.76 | NCVDAC - N,β(13–14,19) | I | 10.4 | 0.84 |
| NCVDAC - β(13–14,18–19) | I | 10.6 | 0.78 | NCVDAC - N,β(13–14,18–19) | I | 10.6 | 0.85 |
| NCVDAC - β(13–14,17–19) | III | − | 0.77 | NCVDAC - N,β(13–14,17–19) | I | 10.0 | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannella, C. In Silico Exploration of Alternative Conformational States of VDAC. Molecules 2023, 28, 3309. https://doi.org/10.3390/molecules28083309
Mannella C. In Silico Exploration of Alternative Conformational States of VDAC. Molecules. 2023; 28(8):3309. https://doi.org/10.3390/molecules28083309
Chicago/Turabian StyleMannella, Carmen. 2023. "In Silico Exploration of Alternative Conformational States of VDAC" Molecules 28, no. 8: 3309. https://doi.org/10.3390/molecules28083309
APA StyleMannella, C. (2023). In Silico Exploration of Alternative Conformational States of VDAC. Molecules, 28(8), 3309. https://doi.org/10.3390/molecules28083309





