Targeted MRM Quantification of Urinary Proteins in Chronic Kidney Disease Caused by Glomerulopathies
Abstract
1. Introduction
2. Results
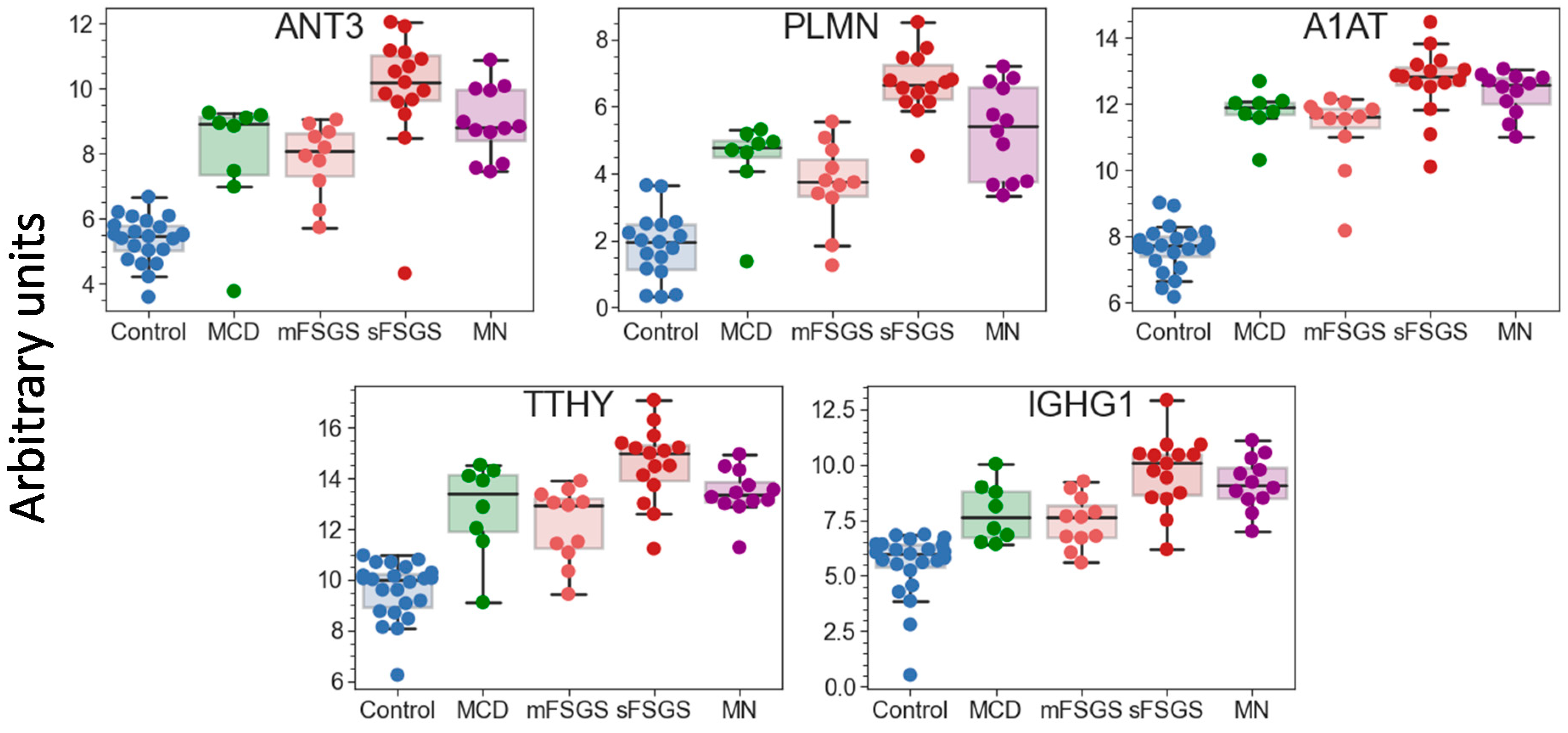
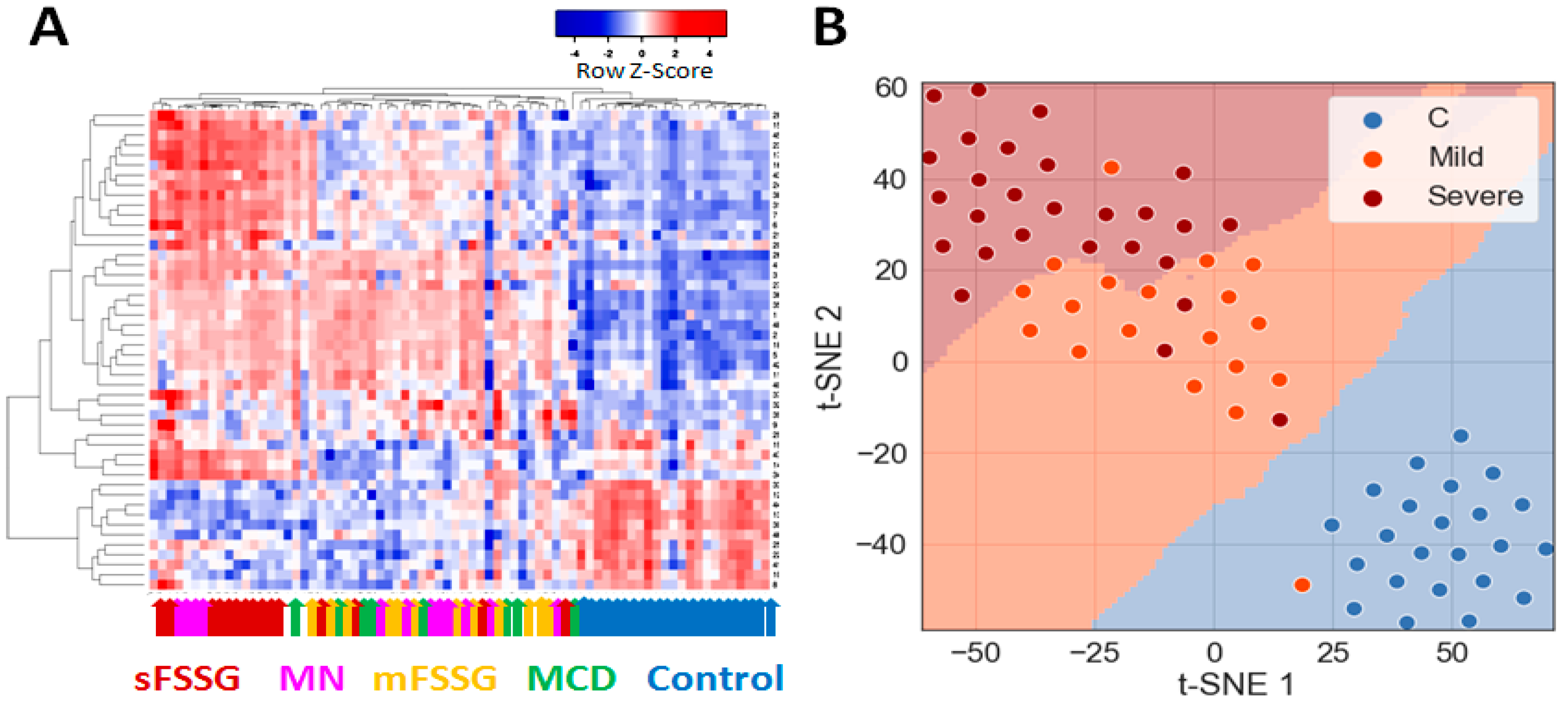
2.1. Identification of Significantly Different Urine Proteins
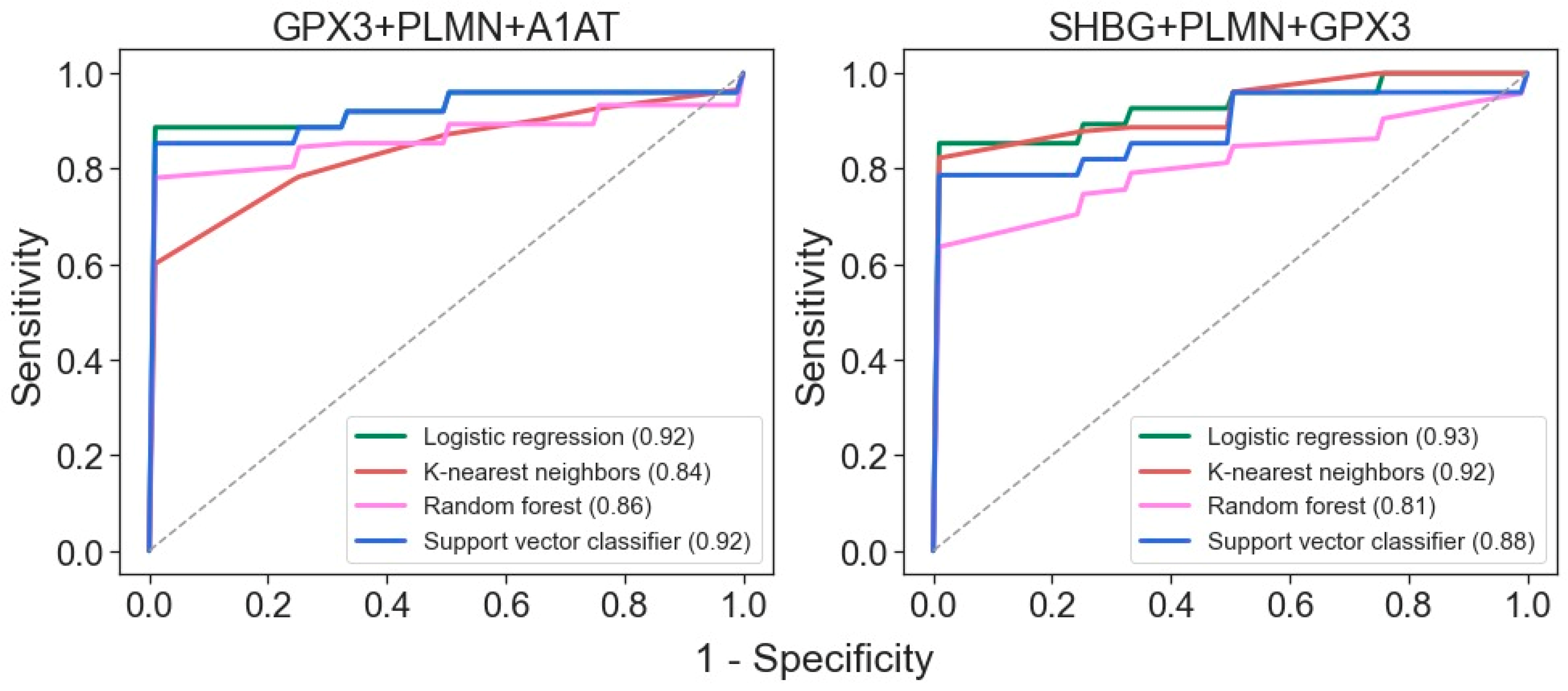
2.2. Building of a Binary Classifier for Distinguishing Mild and Severe Nephropathies
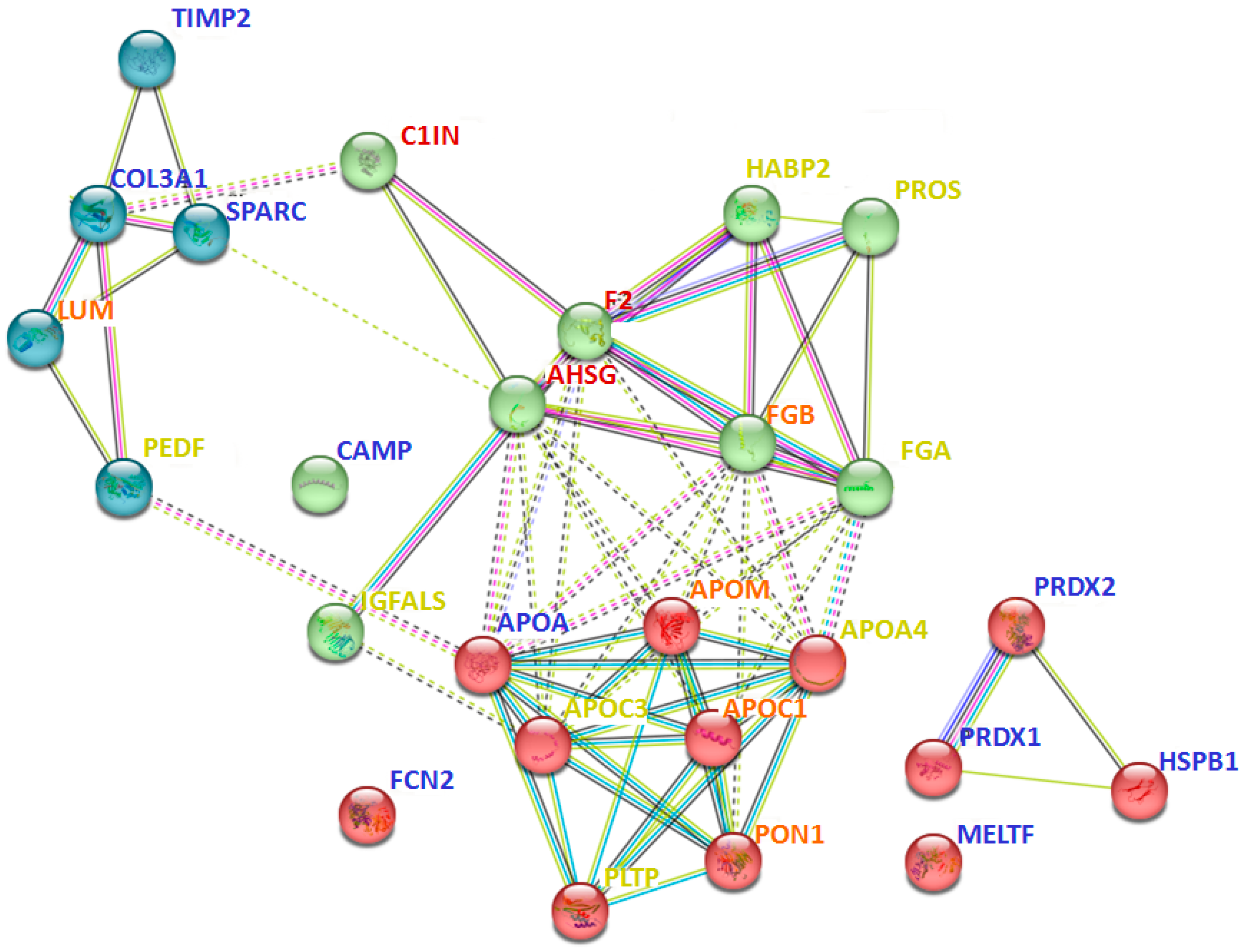
2.3. Perspective Potential Protein Markers Specific for CKD Glomerulopathies
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Urine Sample Preparation for LC-MS
4.3. Targeted Quantitative LC-MS/MS Using Multiple-Reaction Monitoring (MRM) with Stable-Isotope-Labeled Peptide Standards (SISs)
4.4. Data Analysis
4.5. Machine Learning for Binary Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Allen, C.; Arora, M.; Barber, R.M.; Bhutta, Z.A.; Brown, A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2015, 388, 1545–1602. [Google Scholar] [CrossRef]
- Hill, N.R.; Fatoba, S.T.; Oke, J.L.; Hirst, J.; O’Callaghan, C.A.; Lasserson, D.; Hobbs, R. Global Prevalence of Chronic Kidney Disease—A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0158765. [Google Scholar] [CrossRef]
- Schieppati, A.; Remuzzi, G. Chronic renal diseases as a public health problem: Epidemiology, social, and economic implications. Kidney Int. 2005, 68, S7–S10. [Google Scholar] [CrossRef] [PubMed]
- Bommer, J. Prevalence and socio-economic aspects of chronic kidney disease. Nephrol. Dial. Transplant. 2002, 17, 8–12. [Google Scholar] [CrossRef]
- Dhaun, N.; Bellamy, C.O.; Cattran, D.C.; Kluth, D.C. Utility of renal biopsy in the clinical management of renal disease. Kidney Int. 2014, 85, 1039–1048. [Google Scholar] [CrossRef]
- Sim, J.J.; Batech, M.; Hever, A.; Harrison, T.N.; Avelar, T.; Kanter, M.H.; Jacobsen, S.J. Distribution of biopsy-proven presumed primary glomerulonephropathies in 2000-2011 among a racially and ethnically diverse US population. Am. J. Kidney Dis. 2006, 68, 533–544. [Google Scholar] [CrossRef]
- Kitiyakara, C.; Eggers, P.; Kopp, J.B. Twenty-one-year trend in ESRD due to focal segmental glomerulosclerosis in the United States. Am. J. Kidney Dis. 2004, 44, 815–825. [Google Scholar] [CrossRef]
- Nagarai, N.; Mann, M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J. Proteome Res. 2011, 10, 637–645. [Google Scholar] [CrossRef]
- Coon, J.J.; Zürbig, P.; Dakna, M.; Dominiczak, A.F.; Decramer, S.; Fliser, D.; Frommberger, M.; Golovko, I.; Good, D.M.; Hergert-Rosenthal, S.; et al. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteom. Clin. Appl. 2008, 2, 964–973. [Google Scholar] [CrossRef]
- Chebotareva, N.; Vinogradov, A.; McDonnell, V.; Zakharova, N.V.; Indeykina, M.I.; Moiseev, S.; Nikolaev, T.N.; Kononikhin, A.S. Urinary Protein and Peptide Markers in Chronic Kidney Disease. Int. J. Mol. Sci. 2021, 22, 12123. [Google Scholar] [CrossRef]
- Good, D.M.; Zürbig, P.; Argilés, A.; Bauer, H.W.; Behrens, G.; Coon, J.J.; Dakna, M.; Decramer, S.; Delles, C.; Dominiczak, A.F.; et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol. Cell. Proteom. 2010, 9, 2424–2437. [Google Scholar] [CrossRef]
- Catanese, L.; Siwy, J.; Mavrogeorgis, E.; Amann, K.; Mischak, H.; Beige, J.; Rupprecht, H. A Novel Urinary Proteomics Classifier for Non-Invasive Evaluation of Interstitial Fibrosis and Tubular Atrophy in Chronic Kidney Disease. Proteomes 2021, 9, 32. [Google Scholar] [CrossRef]
- Pérez, V.; López, D.; Boixadera, E.; Ibernón, M.; Espinal, A.; Bonet, J.; Romero, R. Comparative differential proteomic analysis of minimal change disease and focal segmental glomerulosclerosis. BMC Nephrol. 2017, 18, 49. [Google Scholar] [CrossRef]
- Samavat, S.; Kalantari, S.; Nafar, M.; Rutishauser, D.; Rezaei-Tavirani, M.; Parvin, M.; Zubarev, R.A. Diagnostic Urinary Pro-teome Profile for Immunoglobulin A Nephropathy. Iran. J. Kid. Dis. 2015, 9, 239–248. [Google Scholar]
- Siwy, J.; Zürbig, P.; Argiles, A.; Beige, J.; Haubitz, M.; Jankowski, J.; Julian, B.A.; Linde, P.B.; Marx, D.; Mishkac, H.; et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol. Dial. Transplant. 2017, 32, 2079–2089. [Google Scholar]
- Fan, G.; Gong, T.; Lin, Y.; Wang, J.; Sun, L.; Wei, H.; Yang, X.; Liu, Z.; Li, X.; Zhao, L.; et al. Urine proteomics identifies biomarkers for diabetic kidney disease at different stages. Clin. Proteom. 2021, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Araumi, A.; Osaki, T.; Ichikawa, K.; Kudo, K.; Suzuki, N.; Watanabe, S.; Watanabe, M.; Konta, T. Urinary and plasma proteomics to discover biomarkers for diagnosing between diabetic nephropathy and minimal change nephrotic syndrome or mem-branous nephropathy. Biochem. Biophys. Rep. 2021, 27, 101102. [Google Scholar] [CrossRef]
- Pang, L.; Li, Q.; Li, Y.; Liu, Y.; Duan, N.; Li, H. Urine proteomics of primary membranous nephropathy using nanoscale liquid chromatography tandem mass spectrometry analysis. Clin. Proteom. 2018, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Dieplinger, H.; Dieplinger, B. Afamin—A pleiotropic glycoprotein involved in various disease states. Clin. Chim. Acta 2015, 446, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, Z.; Lu, C.; Yang, S.; Sun, H.; Guo, R.Y.; Sun, W.; Yue, H. Analysis of the differential urinary protein profile in IgA nephropathy patients of Uygur ethnicity. BMC Nephrol. 2018, 19, 358. [Google Scholar] [CrossRef]
- Mucha, K.; Bakun, M.; Jaźwiec, R.; Dadlez, M.; Florczak, M.; Bajor, M.; Gala, K.; Pączek, L. Complement components, proteolysis-related, and cell communication related proteins detected in urine proteomics are associated with IgA nephropathy. Pol. Arch. Med. Wewn. 2014, 124, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Gupta, R.; Negi, V.S.; Rajasekhar, L.; Misra, R.; Singh, P.; Chaturvedi, V.; Sinha, S. Urinary haptoglobin, alpha-1 anti-chymotrypsin and retinol binding protein identified by proteomics as potential biomarkers for lupus nephritis. Clin. Exp. Immunol. 2017, 188, 254–262. [Google Scholar] [CrossRef]
- Turnier, J.L.; Brunner, H.I.; Bennett, M.; Aleed, A.; Gulati, G.; Haffey, W.D.; Thornton, S.; Wagner, M.; Devarajan, P.; Witte, D.; et al. Discovery of SERPINA3 as a candidate urinary biomarker of lupus nephritis activity. Rheumatology 2018, 58, 321–330. [Google Scholar] [CrossRef]
- Rao, P.V.; Lu, X.; Standley, M.; Pattee, P.; Neelima, G.; Girisesh, G.; Dakshinamurthy, K.V.; Roberts, C.T., Jr.; Nagalla, S.S. Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care 2007, 30, 629–637. [Google Scholar] [CrossRef]
- Patel, D.N.; Kalia, K. Characterization of low molecular weight urinary proteins at varying time intervals in type 2 diabetes mellitus and diabetic nephropathy patients. Diabetol. Metab. Syndr. 2019, 11, 39. [Google Scholar] [CrossRef]
- Liao, W.-L.; Chang, C.-T.; Chen, C.-C.; Lee, W.-J.; Lin, S.-Y.; Liao, H.-Y.; Wu, C.-M.; Chang, Y.-W.; Chen, C.-J.; Tsai, F.-J. Urinary Proteomics for the Early Diagnosis of Diabetic Nephropathy in Taiwanese Patients. J. Clin. Med. 2018, 7, 483. [Google Scholar] [CrossRef] [PubMed]
- Addona, T.A.; Abbatiello, S.E.; Schilling, B.; Skates, S.J.; Mani, D.R.; Bunk, D.M.; Spiegelman, C.H.; Zimmerman, L.J.; Ham, A.-J.L.; Keshishian, H.; et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009, 27, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Gaither, C.; Popp, R.; Mohammed, Y.; Bochers, C.H. Determination of the concentration range for 267 proteins from 21 lots of commercial human plasma using highly multiplexed multiple reaction monitoring mass spectrometry. Analyst 2020, 145, 3634–3644. [Google Scholar] [CrossRef] [PubMed]
- Percy, A.J.; Yang, J.; Hardie, D.B.; Chambers, A.G.; Tamura-Wells, J.; Borchers, C.H. Precise quantitation of 136 urinary proteins by LC/MRM-MS using stable isotope labeled peptides as internal standards for biomarker discovery and/or verification studies. Methods 2015, 81, 24–33. [Google Scholar] [CrossRef]
- Cantley, L.G.; Colangelo, C.M.; Stone, K.L.; Chung, L.; Belcher, J.; Abbott, T.; Cantley, J.L.; Williams, K.R.; Parikh, C.R. Development of a targeted urine proteome assay for kidney diseases. Proteom. Clin. Appl. 2016, 10, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Makridakis, M.; Kontostathi, G.; Petra, E.; Stroggilos, R.; Lugirou, V.; Filip, S.; Duranton, F.; Mischak, H.; Argiles, A.; Zoidakis, J.; et al. Multiplexed MRM-based protein quantification of putative prognostic biomarkers for chronik kidney disease in plasma. Sci. Rep. 2020, 10, 4815. [Google Scholar] [CrossRef]
- Chebotareva, N.V.; Vinogradov, A.; Brzhozovskiy, A.G.; Kashirina, D.N.; Indeykina, M.I.; Bugrova, A.E.; Lebedeva, V.; Moiseev, S.; Nikolaev, E.N.; Kononikhin, A.S. Potential Urine Proteomic Biomarkers for Focal Segmental Glomerulosclerosis and Minimal Change Disease. Int. J. Mol. Sci. 2022, 23, 12607. [Google Scholar] [CrossRef] [PubMed]
- D’Agati, V.D.; Fogo, A.B.; Bruijn, J.A.; Jennette, J. Pathologic classification of focal segmental glomerulosclerosis: A working proposal. Am. J. Kidney Dis. 2004, 43, 368–382. [Google Scholar] [CrossRef]
- Choi, Y.W.; Kim, Y.G.; Song, M.-Y.; Moon, J.-Y.; Jeong, K.-H.; Lee, T.-W.; Ihm, C.-G.; Park, K.-S.; Lee, S.-H. Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin. Proteom. 2017, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.M.; Geyer, P.E.; Müller, J.B.; Strauss, M.T.; Koch, M.; Leypoldt, F.; Koertvelyessy, P.; Bittner, D.; Schipke, C.G.; Incesoy, E.I.; et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol. Syst. Biol. 2020, 16, e9356. [Google Scholar] [CrossRef]
- Kononikhin, A.S.; Zakharova, N.V.; Semenov, S.D.; Bugrova, A.E.; Brzhozovskiy, A.G.; Indeykina, M.I.; Fedorova, Y.; Kolykhalov, I.V.; Strelnikova, P.; Ikonnikova, A.Y.; et al. Prognosis of Alzheimer’s Disease Using Quantitative Mass Spectrometry of Human Blood Plasma Proteins and Machine Learning. Int. J. Mol. Sci. 2022, 23, 7907. [Google Scholar] [CrossRef]
- Guan, J.; Wang, M.; Zhao, M.; Ni, W.; Zhang, M. Discovery of fibrinogen γ-chain as a potential urinary biomarker for renal interstitial fibrosis in IgA nephropathy. BMC Nephrol. 2023, 24, 60. [Google Scholar] [CrossRef] [PubMed]
- Gaither, C.; Popp, R.; Bochers, S.P.; Skarphedinsson, K.; Eiriksson, F.F.; Thorsteinsdottir, M.; Mohammed, Y.; Borchers, C.H. Performance assessment of a 125 human plasma peptide mixture stored at room temperature for multiple reaction monitoring-mass spectrometry. J. Proteome Res. 2021, 20, 4292–4302. [Google Scholar] [CrossRef]
- Percy, A.J.; Borchers, C.H. Detailed Method for Performing the ExSTA Approach in Quantitative Bottom-Up Plasma Proteomics. Methods Mol. Biol. 2021, 2228, 353–384. [Google Scholar] [CrossRef]
- Mohammed, Y.; Pan, J.; Zhang, S.; Han, J.; Borchers, C.H. ExSTA: External standard addition method for accurate high-throughput quantitation in targeted proteomics experiments. Proteom. Clin. Appl. 2018, 12, 1600180. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- MacLean, B.X.; Pratt, B.S.; Egertson, J.D.; MacCoss, M.J.; Smith, R.D.; Baker, E.S. Using skyline to analyze data-containing liquid chromatography, ion mobility spectrometry, and mass spectrometry dimensions. J. Am. Soc. Mass Spectrom. 2018, 29, 2182–2188. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; Volume 445, pp. 51–56. [Google Scholar]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Machine Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
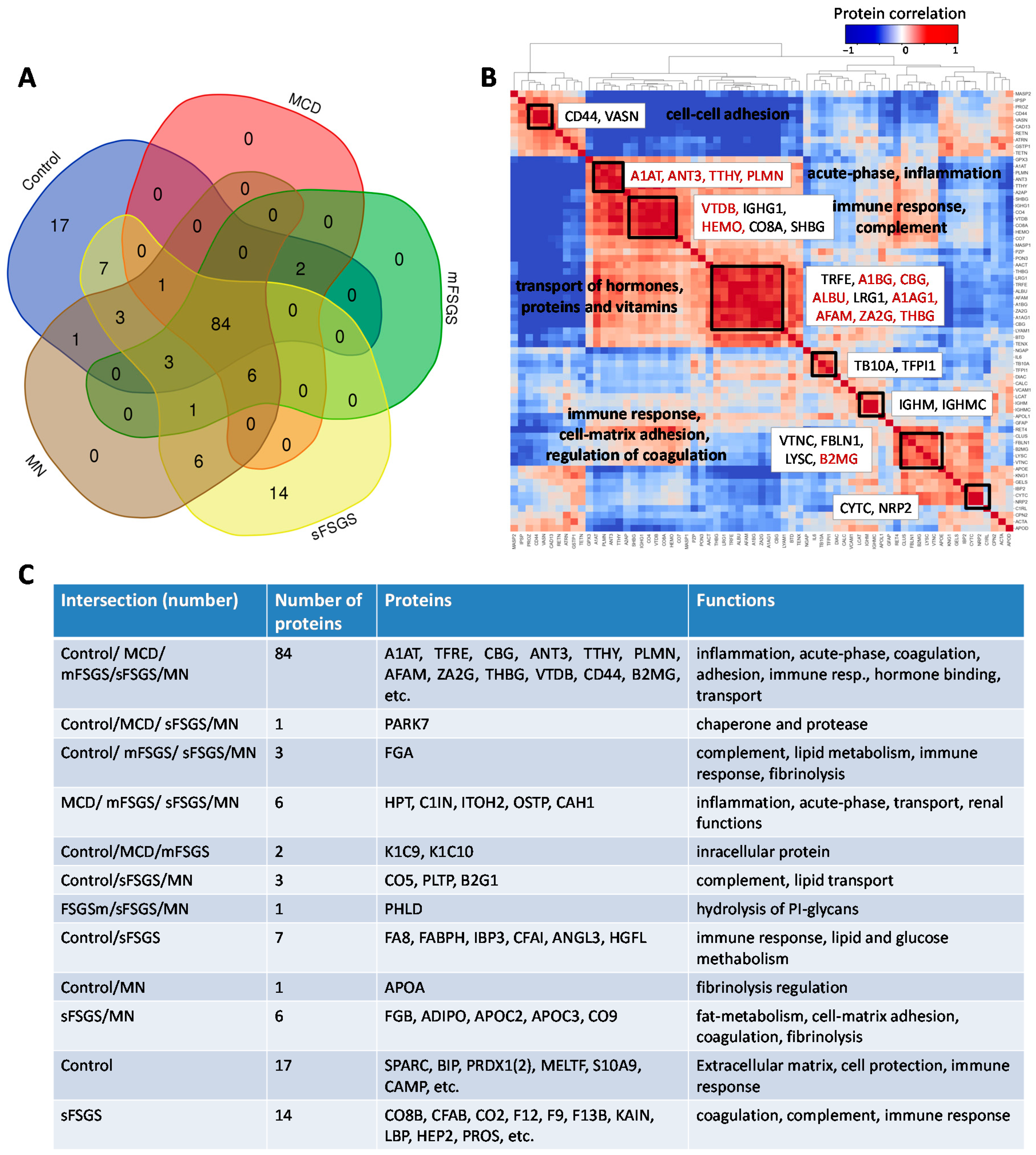
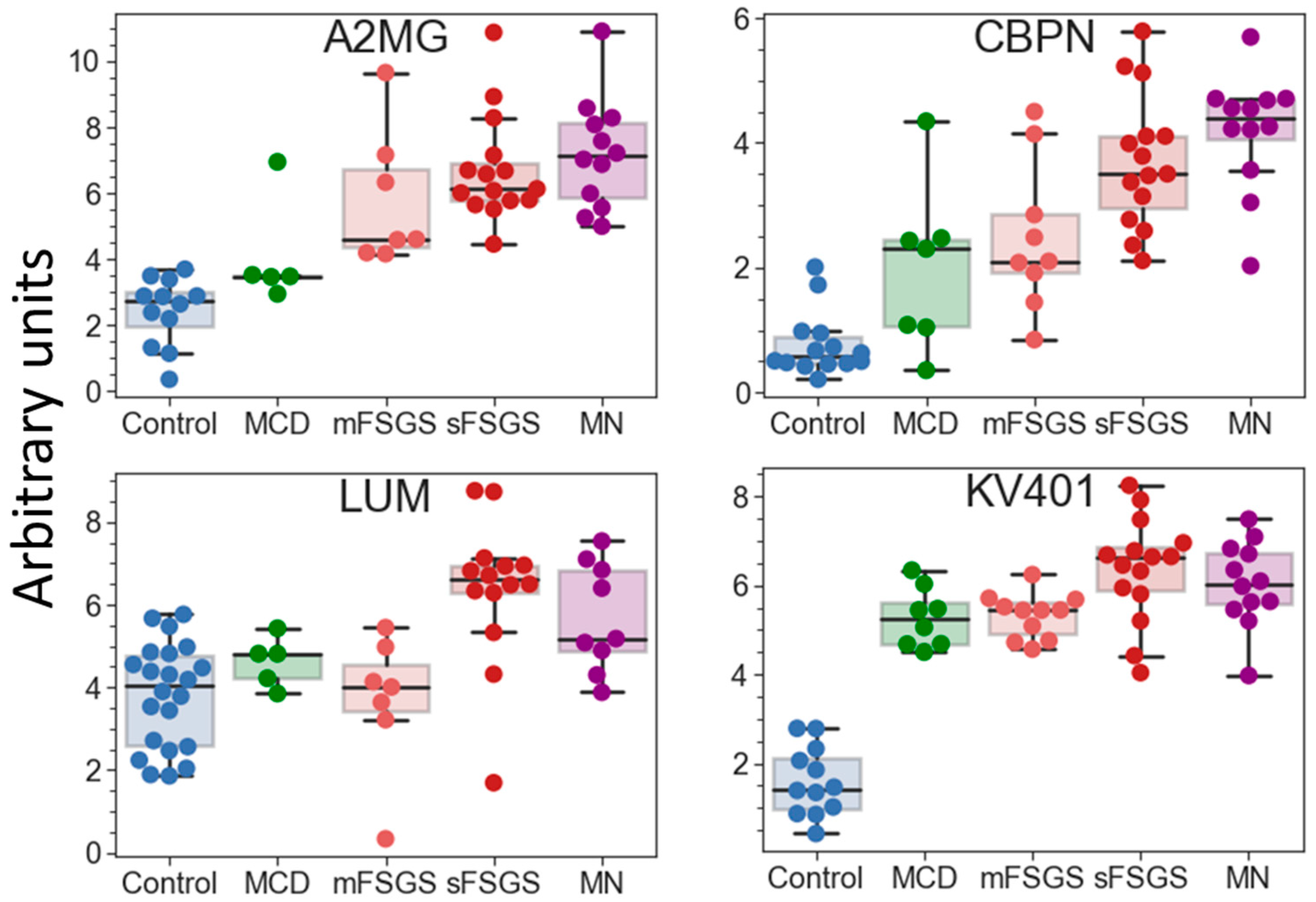
| Proteins | Control vs. | sFSGS vs. | mFSGS vs. MN | |||||
|---|---|---|---|---|---|---|---|---|
| MCD | mFSGS | sFSGS | MN | MCD | mFSGS | MN | ||
| ANT3 a, PLMN a | + | + | + | + | + | + | n.d. | n.d. |
| PZP | + | + | + | + | n.d. | n.d. | + | + |
| A1AT a, IGHG1 | + | + | + | + | n.d. | + | n.d. | n.d. |
| IPSP | + | + | + | + | n.d. | n.d. | + | n.d. |
| A1AG1 a, A1BG a, ALBU a, CBG a, CD44 a, LRG1, TRFE, TTHY a, VASN a, ZA2G a, AFAM a, THBG, AACT a, CAD13, PROZ, RETN b | + | + | + | + | n.d. | n.d. | n.d. | n.d. |
| PON3 | + | n.d. | + | + | n.d. | n.d. | n.d. | + |
| HEMO a | n.d. | n.d. | + | + | + | + | n.d. | n.d. |
| ATRN b | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. |
| BTD b | + | n.d. | + | + | n.d. | n.d. | n.d. | n.d. |
| GSTP1 b | n.d. | + | + | + | n.d. | n.d. | n.d. | n.d. |
| KNG1 b | + | + | n.d. | + | n.d. | n.d. | n.d. | n.d. |
| A2AP | + | n.d. | + | n.d. | n.d. | + | n.d. | n.d. |
| GPX3 | n.d. | n.d. | + | n.d. | + | + | n.d. | n.d. |
| SHBG | n.d. | n.d. | + | + | n.d. | + | n.d. | n.d. |
| APOE b | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| APOL1, TB10 A b | + | n.d. | n.d. | + | n.d. | n.d. | n.d. | n.d. |
| VTDB a, CO8A, MASP1, LYAM1 b | n.d. | n.d. | + | + | n.d. | n.d. | n.d. | n.d. |
| VTNC b | n.d. | + | n.d. | n.d. | n.d. | + | n.d. | n.d. |
| RET4 a,b | n.d. | n.d. | + | n.d. | n.d. | n.d. | n.d. | n.d. |
| Proteins | Group, % of Samples (Log2 Median) | ||||
|---|---|---|---|---|---|
| Control | MCD | mFSGS | sFSGS | MN | |
| Alpha-2-HS-glycoprotein (AHSG) a | 56.5 (4.86) | 100 (8.75) | 90.9 (7.48) | 100 (11.5) | 100 (9.49) |
| Carboxypeptidase N catalytic chain (CBPN) | 60.9 (−0.80) | 87.5 (2.45) | 81.8 (2.81) | 100 (5.01) | 100 (6.35) |
| Haptoglobin (HPT) a | 13 (Nan) | 62.5 (14.3) | 81.8 (15.4) | 93.3 (18.5) | 91.7 (17.9) |
| Immunoglobulin kappa variable 4-1 (KV401) | 52.3 (−0.92) | 100 (7.60) | 100 (7.85) | 100 (9.58) | 100 (8.71) |
| Osteopontin (OSTP) a | 4.34 (Nan) | 100 (5.83) | 100 (5.73) | 100 (6.32) | 100 (5.40) |
| Plasma protease C1 inhibitor (C1IN) | 39.1 (Nan) | 100 (6.53) | 100 (6.59) | 86.7 (9.75) | 91.7 (7.58) |
| Prothrombin (F2) a | 69.6 (5.90) | 87.5 (10.8) | 100 (9.28) | 93.3 (14.0) | 100 (11.5) |
| Alpha-2-macroglobulin (A2MG) a | 52.3 (−1.18) | 62.5 (4.62) | 63.6 (6.04) | 100 (8.85) | 100 (10.3) |
| Adiponectin (ADIPO) a | 21.7 (Nan) | 25 (Nan) | 27.3 (Nan) | 73.3 (4.72) | 66.7 (3.99) |
| Apolipoprotein C-I (APOC1) | 43.5 (Nan) | 25 (Nan) | 54.5 (3.80) | 80 (10.3) | 66.7 (9.60) |
| Apolipoprotein M (APOM) | 82.6 (3.07) | 50 (−0.21) | 63.6 (0.47) | 86.7 (5.53) | 66.7 (3.93) |
| Carbonic anhydrase 1 (CAH1) | 13 (Nan) | 50 (0.39) | 63.6 (1.55) | 100 (3.90) | 75 (3.60) |
| Complement C3 (CO3) a | 60.9 (2.30) | 37.5 (Nan) | 63.6 (4.52) | 86.7 (13.0) | 91.7 (10.3) |
| Complement component C9 (CO9) a | 0 (Nan) | 37.5 (Nan) | 45.5 (Nan) | 80 (10.3) | 75 (6.37) |
| Fibrinogen beta chain (FGB) | 13 (Nan) | 25 (Nan) | 27.3 (Nan) | 73.3 (11.6) | 75 (9.93) |
| Inter-alpha-trypsin inhibitor heavy chain H2 (ITOH2) | 13 (Nan) | 50 (2.86) | 63.6 (1.25) | 80 (9.14) | 75 (7.11) |
| Lumican (LUM) | 95.7 (5.60) | 62.5 (5.82) | 72.7 (4.58) | 86.7 (9.38) | 75 (7.20) |
| Phosphatidylinositol-glycan-specific phospholipase D (PHLD) | 0 (Nan) | 37.5 (Nan) | 45.5 (Nan) | 86.7 (3.42) | 75 (3.57) |
| Apolipoprotein(a) (APOA) | 100 (7.63) | 37.5 (Nan) | 45.5 (Nan) | 40 (Nan) | 58.3 (3.35) |
| Cathelicidin antimicrobial peptide (CAMP) | 100 (4.59) | 12.5 (Nan) | 18.2 (Nan) | 13.3 (Nan) | 8.3 (Nan) |
| Creatine kinase B-type (KCRB) | 82.6 (2.74) | 0 (Nan) | 0 (Nan) | 6.67 (Nan) | 0 (Nan) |
| Ficolin-2 (FCN2) | 87 (5.46) | 12.5 (Nan) | 9.1 (Nan) | 46.7 (Nan) | 16.7 (Nan) |
| Heat shock protein beta-1 (HSPB1) | 78.3 (5.02) | 12.5 (Nan) | 45.5 (Nan) | 13.3 (Nan) | 25 (Nan) |
| Melanotransferrin (MELTF) | 82.6 (2.94) | 0 (Nan) | 0 (Nan) | 0 (Nan) | 0 (Nan) |
| Metalloproteinase inhibitor 2 (TIMP2) | 91.3 (2.29) | 0 (Nan) | 9.1 (Nan) | 26.7 (Nan) | 8.3 (Nan) |
| Peroxiredoxin-1 (PRDX1) | 78.3 (3.48) | 0 (Nan) | 18.2 (Nan) | 6.7 (Nan) | 8.3 (Nan) |
| Peroxiredoxin-2 (PRDX2) | 69.6 (4.13) | 25 (Nan) | 27.3 (Nan) | 46.7 (Nan) | 33.3 (Nan) |
| SPARC (SPARC) | 73.9 (1.46) | 0 (Nan) | 0 (Nan) | 6.7 (Nan) | 8.3 (Nan) |
| Keratin type I cytoskeletal 10 (K1C10) a | 69.6 (8.09) | 62.5 (9.08) | 63.6 (7.07) | 6.7 (Nan) | 25 (Nan) |
| Keratin type I cytoskeletal 9 (K1C9) | 65.2 (4.30) | 62.5 (6.66) | 72.7 (4.21) | 26.7 (Nan) | 16.7 (Nan) |
| Apolipoprotein A-IV (APOA4) | 60.9 (1.51) | 62.5 (4.92) | 54.5 (5.22) | 86.7 (11.3) | 58.3 (6.50) |
| Apolipoprotein C-III (APOC3) | 17.4 (Nan) | 37.5 (Nan) | 45.5 (Nan) | 80 (9.18) | 58.3 (7.09) |
| Cartilage acidic protein 1 (CRAC1) | 0 (Nan) | 0 (Nan) | 9.1 (Nan) | 66.7 (1.53) | 25 (Nan) |
| Coagulation factor XII (FA12) | 0 (Nan) | 0 (Nan) | 18.2 (Nan) | 66.7 (4.61) | 25 (Nan) |
| Complement C5 (CO5) | 56.5 (−3.96) | 25 (Nan) | 45.5 (Nan) | 80 (5.49) | 58.3 (1.71) |
| Fibrinogen alpha chain (FGA) a | 69.6 (3.08) | 25 (Nan) | 54.5 (4.13) | 86.7 (8.69) | 58.3 (5.82) |
| Heparin cofactor 2 (HEP2) | 0 (Nan) | 12.5 (Nan) | 18.2 (Nan) | 73.3 (7.79) | 33.3 (Nan) |
| Hyaluronan-binding protein 2 (HABP2) | 0 (Nan) | 12.5 (Nan) | 18.2 (Nan) | 73.3 (4.83) | 33.3 (Nan) |
| Insulin-like growth-factor-binding protein complex acid labile subunit (IGFALS) | 17.4 (Nan) | 0 (Nan) | 9.1 (Nan) | 73.3 (4.08) | 33.3 (Nan) |
| Phospholipid transfer protein (PLTP) | 82.6 (−0.78) | 12.5 (Nan) | 18.2 (Nan) | 73.3 (2.14) | 50 (−1.34) |
| Pigment epithelium-derived factor (PEDF) a | 8.7 (Nan) | 25 (Nan) | 18.2 (Nan) | 73.3 (10.2) | 33.3 (Nan) |
| Vitamin-K-dependent protein S (PROS) | 0 (Nan) | 12.5 (Nan) | 18.2 (Nan) | 73.3 (5.18) | 33.3 (Nan) |
| MCD | mFSGS | sFSGS | MN | |
|---|---|---|---|---|
| n | 8 | 11 | 15 | 12 |
| Age (years) | 38 (27; 59) | 33.5 (27; 50) | 50.5 (39; 61) | 52.5 (49; 62) |
| Sex (%, F) | 75.0 | 50.0 | 30.8 | 25.0 |
| Proteinuria (g/24 h) | 3.24 (1.84; 3.65) | 2.3 (1.28; 3.37) | 5.0 (4.0; 8.64) | 3.25 (2.66; 4.05) |
| Nephrotic syndrome (%) | 37.5 | 36.4 | 93.3 | 41.7 |
| eGFR CKD-EPI a | 93 (70.8; 106) | 96.9 (71.3; 115.7) | 42.0 (37.0; 62.2) | 77.3 (44.9; 87.9) |
| Steroid-resistance (%) | 0 | 30.0 | 84.6 | N.d. |
| Failure response to other immunosuppressants (%) | 0 | 0 | 46.2 | 16.7 |
| Time to remission, months | 1 (0.0; 19.3) | 1 (0.26; 5.13) | 31 (8.8; 58.5) | 8 (1.25; 22.3) |
| Serum tests: | ||||
| Serum albumin (mg/mL) | 29.3 (19.7; 35.0) | 34.1 (29.1; 37.4) | 22.6 (17.1; 26.5) | 26.5 (21.2; 33.2) |
| Creatinine (μM) | 80.9 (71.4; 100.0) | 68.9 (58.2; 95.6) | 148 (122; 163) | 93.7 (79.0; 124) |
| Total cholesterol (mM) | 7.89 (7.04; 11.0) | 6.86 (6.23; 11.5) | 9.69 (8.3; 11.9) | 7.91 (7.05; 9.48) |
| Therapy: | ||||
| Corticosteroids (%) | 100 | 70 | 84.6 | 41.7 |
| Cyclosporin (%) | 62.5 | 50 | 69.2 | 25.0 |
| Cyclophosphamide (%) | 50 | 40 | 61.5 | 41.7 |
| Micophenolate mofetil (%) | 0 | 10 | 30.8 | 0 |
| Parameters | Score |
|---|---|
| eGFR CKD-EPI, mL/min/1.73 m2 | |
| >60 | 0 |
| 45–59 | 1 |
| 35–45 | 2 |
| <35 | 3 |
| Proteinuria, g/24 h | |
| >2 | 0 |
| 2–3 | 0.5 |
| 3–4 | 1 |
| 4–5 | 1.5 |
| 5–6 | 2 |
| 6–7 | 2.5 |
| >7 | 3 |
| Steroid resistance | |
| Absent | 0 |
| Present | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kononikhin, A.S.; Brzhozovskiy, A.G.; Bugrova, A.E.; Chebotareva, N.V.; Zakharova, N.V.; Semenov, S.; Vinogradov, A.; Indeykina, M.I.; Moiseev, S.; Larina, I.M.; et al. Targeted MRM Quantification of Urinary Proteins in Chronic Kidney Disease Caused by Glomerulopathies. Molecules 2023, 28, 3323. https://doi.org/10.3390/molecules28083323
Kononikhin AS, Brzhozovskiy AG, Bugrova AE, Chebotareva NV, Zakharova NV, Semenov S, Vinogradov A, Indeykina MI, Moiseev S, Larina IM, et al. Targeted MRM Quantification of Urinary Proteins in Chronic Kidney Disease Caused by Glomerulopathies. Molecules. 2023; 28(8):3323. https://doi.org/10.3390/molecules28083323
Chicago/Turabian StyleKononikhin, Alexey S., Alexander G. Brzhozovskiy, Anna E. Bugrova, Natalia V. Chebotareva, Natalia V. Zakharova, Savva Semenov, Anatoliy Vinogradov, Maria I. Indeykina, Sergey Moiseev, Irina M. Larina, and et al. 2023. "Targeted MRM Quantification of Urinary Proteins in Chronic Kidney Disease Caused by Glomerulopathies" Molecules 28, no. 8: 3323. https://doi.org/10.3390/molecules28083323
APA StyleKononikhin, A. S., Brzhozovskiy, A. G., Bugrova, A. E., Chebotareva, N. V., Zakharova, N. V., Semenov, S., Vinogradov, A., Indeykina, M. I., Moiseev, S., Larina, I. M., & Nikolaev, E. N. (2023). Targeted MRM Quantification of Urinary Proteins in Chronic Kidney Disease Caused by Glomerulopathies. Molecules, 28(8), 3323. https://doi.org/10.3390/molecules28083323








