Novel PD-L1-Targeted Phenyl-Pyrazolone Derivatives with Antioxidant Properties
Abstract
:1. Introduction
2. Results
2.1. Drug Synthesis
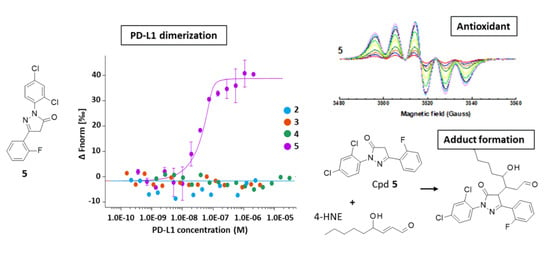
2.2. Drug Binding to PD-L1 and Inhibition of PD-1 Signaling in Cells
2.3. Antioxidant Activity
2.4. Aldehyde Reactivity
3. Discussion
4. Materials and Methods
4.1. Synthesis of the Compounds
- 1-(2,4-Dichlorophenyl)-3-(2-methoxyphenyl)-1H-pyrazol-5(4H)-one (3): The oil obtained was purified by chromatography on silica gel with cyclohexane/ethyl acetate (7:3) as eluent and recrystallized with ethanol. White solid. Yield: 51%. mp: 188 °C. Rf (cyclohexane/ethyl acetate 1:1): 0.6. 1H NMR (DMSO): 3.86 (s, 3H); 6.02 (s, 1H); 6.95 (t, 1H, J = 7.4 Hz,); 7.08 (d, 1H, J = 8.2 Hz); 7.29 (t, 1H, J = 7.2 Hz); 7.56 (d, 2H, J = 0.8 Hz); 7.81 (dd, 1H, J = 7.9 Hz, J’ = 1.4 Hz); 7.84 (s, 1H); 11,35 (s, 1H). LC-MS (ESI+) m/z 335.0 (MH+); tr = 2.92 min. 13C NMR (DMSO) δ 157, 154, 148, 135, 134, 133, 132, 130, 129, 128, 127, 122, 121, 112, 88, 56.
- 1-(2,4-dichlorophenyl)-3-ethyl-1H-pyrazol-5(4H)-one (4): Compound was purified by flash chromatography using with cyclohexane/ethyl acetate (1:1). Orange solid. Yield 29%. mp: 160 °C. Rf (cyclohexane/ethyl acetate 1:1): 0.3. 1H NMR (CDCl3) δ 7.51–7.50 (d, J = 2.0 Hz, 1H); 7.37–7.34 (d, J = 9.2 Hz, 1H); 7.32–7.29 (Dd, J = 9.2 Hz, J’ = 2.0 Hz, 1H); 3.39 (s, 2H); 2.53–2.46 (d, J = 7.7 Hz, 1H); 1.25–1.20 (t, J = 7.7 Hz, 3H). LC-MS (ESI) m/z 258 (MH+), tr 2.36 min. 13C NMR (CDCl3) δ 171, 161, 135, 133, 132, 130, 129, 128, 40, 25, 11.
- 1-(2,4-Dichlorophenyl)-3-(2-fluorophenyl)-1H-pyrazol-5-ol (5): The product was recrystallized with methanol. White solid. Yield: 34%. mp: 229 °C. Rf (cyclohexane/ethyl acetate 7:3): 0.7. 1H NMR (DMSO): 5.93 (d, 1H, J = 4,2 Hz); 7.21–7.39 (m, 3H); 7,60 (m, 2H); 7.88–7.94 (m, 2H); 11.65 (s, 1H). LC-MS (ESI+) m/z 323.3 (MH+); tr = 2.97 min. 13C NMR (DMSO) δ 161, 158, 155, 146, 135, 134, 132, 131, 130, 129, 128, 125, 121, 116, 87.
4.2. Microscale Thermophoresis (MST)
4.3. MST Dimerization Binding Assay
4.4. Fluorescence Resonance Energy Transfer (FRET) Assay
4.5. Cell Proliferation Assay
4.6. EPR Experiments
4.7. Aldehyde (4-HNE) Reactivity Measurements
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Offringa, R.; Kötzner, L.; Huck, B.; Urbahns, K. The expanding role for small molecules in immuno-oncology. Nat. Rev. Drug Discov. 2022, 21, 821–840. [Google Scholar] [CrossRef]
- Zak, K.M.; Grudnik, P.; Magiera, K.; Dömling, A.; Dubin, G.; Holak, T.A. Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2. Structure 2017, 25, 1163–1174. [Google Scholar] [CrossRef]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget 2017, 8, 72167–72181. [Google Scholar] [CrossRef] [Green Version]
- Sasmal, P.; Kumar Babasahib, S.; Prashantha Kumar, B.R.; Manjunathaiah Raghavendra, N. Biphenyl-based small molecule inhibitors: Novel cancer immunotherapeutic agents targeting PD-1/PD-L1 interaction. Bioorg. Med. Chem. 2022, 73, 117001. [Google Scholar] [CrossRef]
- Shaabani, S.; Gadina, L.; Surmiak, E.; Wang, Z.; Zhang, B.; Butera, R.; Zarganes-Tzitzikas, T.; Rodriguez, I.; Kocik-Krol, J.; Magiera-Mularz, K.; et al. Biphenyl Ether Analogs Containing Pomalidomide as Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Cell Death-Ligand 1 Interaction. Molecules 2022, 27, 3454. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Qi, Z.; Wang, T.; Zhang, X.; Zhang, K.; Wang, K.; Cheng, Y.; Xiao, Y.; Li, Z.; Jiang, S. Design, Synthesis, and Evaluation of PD-1/PD-L1 Antagonists Bearing a Benzamide Scaffold. ACS Med. Chem. Lett. 2022, 13, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, H.; Shen, L.; Xu, H.; Deng, M.; Cheng, M.; Wang, J. Discovery of benzo[d]isothiazole derivatives as novel scaffold inhibitors targeting the programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) interaction through “ring fusion” strategy. Bioorg. Chem. 2022, 123, 105769. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Cheng, Y.; Liu, X.; Wang, G.; Min, W.; Wang, X.; Yuan, K.; Hou, Y.; Li, J.; Zhang, H.; et al. Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents. Acta Pharm. Sin. B 2022, 12, 4446–4457. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Elumalai, S.; Murugesan, V.; Kunjiappan, S.; Pavadai, P.; Theivendren, P. Computational design of PD-L1 small molecule inhibitors for cancer therapy. Mol. Divers. 2022; ahead of print. [Google Scholar] [CrossRef]
- Wang, F.; Ye, W.; He, Y.; Zhong, H.; Zhu, Y.; Han, J.; Gong, X.; Tian, Y.; Wang, Y.; Wang, S.; et al. Identification of CBPA as a New Inhibitor of PD-1/PD-L1 Interaction. Int. J. Mol. Sci. 2023, 24, 3971. [Google Scholar] [CrossRef]
- DiFrancesco, M.; Hofer, J.; Aradhya, A.; Rufinus, J.; Stoddart, J.; Finocchiaro, S.; Mani, J.; Tevis, S.; Visconti, M.; Walawender, G.; et al. Discovery of small-molecule PD-1/PD-L1 antagonists through combined virtual screening and experimental validation. Comput. Biol. Chem. 2023, 102, 107804. [Google Scholar] [CrossRef]
- Hao, X.; Chen, Z.; Li, H.; Wei, M.; Zuo, Z.; Su, Q. Small-Molecule Drugs in Immunotherapy. Mini Rev. Med. Chem. 2022; ahead of print. [Google Scholar] [CrossRef]
- Sasikumar, P.G.; Ramachandra, M. Small Molecule Agents Targeting PD-1 Checkpoint Pathway for Cancer Immunotherapy: Mechanisms of Action and Other Considerations for Their Advanced Development. Front. Immunol. 2022, 13, 752065. [Google Scholar] [CrossRef]
- Le Biannic, R.; Magnez, R.; Klupsch, F.; Leleu-Chavain, N.; Thiroux, B.; Tardy, M.; El Bouazzati, H.; Dezitter, X.; Renault, N.; Vergoten, G.; et al. Pyrazolones as inhibitors of immune checkpoint blocking the PD-1/PD-L1 interaction. Eur. J. Med. Chem. 2022, 236, 114343. [Google Scholar] [CrossRef]
- Bailly, C. Potential use of edaravone to reduce specific side effects of chemo-, radio- and immuno-therapy of cancers. Int. Immunopharmacol. 2019, 77, 105967. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Hecquet, P.E.; Kouach, M.; Thuru, X.; Goossens, J.F. Chemical reactivity and uses of 1-phenyl-3-methyl-5-pyrazolone (PMP), also known as edaravone. Bioorg. Med. Chem. 2020, 28, 115463. [Google Scholar] [CrossRef]
- Leleu-Chavain, N.; Regnault, R.; Ahouari, H.; Le Biannic, R.; Kouach, M.; Klupsch, F.; Magnez, R.; Vezin, H.; Thuru, X.; Bailly, C.; et al. Antioxidant Properties and Aldehyde Reactivity of PD-L1 Targeted Aryl-Pyrazolone Anticancer Agents. Molecules 2022, 27, 3316. [Google Scholar] [CrossRef]
- Kamogawa, E.; Sueishi, Y. A multiple free-radical scavenging (MULTIS) study on the antioxidant capacity of a neuroprotective drug, edaravone as compared with uric acid, glutathione, and trolox. Bioorg. Med. Chem. Lett. 2014, 24, 1376–1379. [Google Scholar] [CrossRef]
- Goossens, J.F.; Thuru, X.; Bailly, C. Properties and reactivity of the folic acid and folate photoproduct 6-formylpterin. Free Radic. Biol. Med. 2021, 171, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Regnault, R.; Kouach, M.; Goossens, L.; Thuru, X.; Bailly, C.; Goossens, J.F. Mono- and bis-edaravone adducts formed in the presence of vanillin in an aqueous solution. Sep. Sci. Plus 2022, 5, 285–295. [Google Scholar] [CrossRef]
- Jaganjac, M.; Zarkovic, N. Lipid Peroxidation Linking Diabetes and Cancer: The Importance of 4-Hydroxynonenal. Antioxid. Redox Signal. 2022, 37, 1222–1233. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, T.; Li, J.; Xia, M.; Li, Y.; Wang, X.; Liu, C.; Zheng, T.; Chen, R.; Kan, D.; et al. Oxidative Stress and 4-hydroxy-2-nonenal (4-HNE): Implications in the Pathogenesis and Treatment of Aging-related Diseases. J. Immunol. Res. 2022, 2022, 2233906. [Google Scholar] [CrossRef]
- Vazdar, K.; Škulj, S.; Bakarić, D.; Margetić, D.; Vazdar, M. Chemistry and Reactivity of 4-hydroxy-2-nonenal (HNE) in Model Biological Systems. Mini Rev. Med. Chem. 2021, 21, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharma, P.; Bailey, T.; Bhattarai, S.; Subedi, U.; Miller, C.; Ara, H.; Kidambi, S.; Sun, H.; Panchatcharam, M.; et al. Electrophilic Aldehyde 4-Hydroxy-2-Nonenal Mediated Signaling and Mitochondrial Dysfunction. Biomolecules 2022, 12, 1555. [Google Scholar] [CrossRef] [PubMed]
- Aldini, G.; Vistoli, G.; Regazzoni, L.; Benfatto, M.C.; Bettinelli, I.; Carini, M. Edaravone inhibits protein carbonylation by a direct carbonyl-scavenging mechanism: Focus on reactivity, selectivity, and reaction mechanisms. Antioxid Redox Signal. 2010, 12, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Cai, Y.; Zhu, X.; Wang, J.; Gao, F.; Yang, M.; Mao, L.; Zhang, Z.; Sun, B. Edaravone Dexborneol Treatment Attenuates Neuronal Apoptosis and Improves Neurological Function by Suppressing 4-HNE-Associated Oxidative Stress After Subarachnoid Hemorrhage. Front. Pharmacol. 2022, 13, 848529. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hu, L. Immunomodulators targeting the PD-1/PD-L1 protein-protein interaction: From antibodies to small molecules. Med. Res. Rev. 2019, 39, 265–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Zhang, Y.; Guo, Y.; Pan, Z.; Zhong, S.; Jin, X.; Zhuang, W.; Chen, S.; Gao, J.; Huang, W.; et al. Discovery of phenyl-linked symmetric small molecules as inhibitors of the programmed cell death-1/programmed cell death-ligand 1 interaction. Eur. J. Med. Chem. 2021, 223, 113637. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, K.; Chen, H.; Feng, Z. Design, synthesis, evaluation, and SAR of 4-phenylindoline derivatives, a novel class of small-molecule inhibitors of the programmed cell death-1/ programmed cell death-ligand 1 (PD-1/PD-L1) interaction. Eur. J. Med. Chem. 2021, 211, 113001. [Google Scholar] [CrossRef] [PubMed]
- Russomanno, P.; Assoni, G.; Amato, J.; D’Amorev, V.M.; Scaglia, R.; Brancaccio, D.; Pedrini, M.; Polcaro, G.; La Pietra, V.; Orlando, P.; et al. Interfering with the Tumor-Immune Interface: Making Way for Triazine-Based Small Molecules as Novel PD-L1 Inhibitors. J. Med. Chem. 2021, 64, 16020–16045. [Google Scholar] [CrossRef]
- Deng, J.; Cheng, Z.; Long, J.; Dömling, A.; Tortorella, M.; Wang, Y. Small Molecule Inhibitors of Programmed Cell Death Ligand 1 (PD-L1): A Patent Review (2019–2021). Expert Opin. Ther. Pat. 2022, 32, 575–589. [Google Scholar] [CrossRef]
- Zhu, P.; Zhang, J.; Yang, Y.; Wang, L.; Zhou, J.; Zhang, H. Design, synthesis and biological evaluation of isoxazole-containing biphenyl derivatives as small-molecule inhibitors targeting the programmed cell death-1/ programmed cell death-ligand 1 immune checkpoint. Mol. Divers. 2022, 26, 245–264. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, K.; Gao, Y.; Yuan, D.; Ling, L.; Liu, J.; Wu, S.; Chen, R.; Li, H.; Xiong, Y.; et al. Discovery of quinazoline derivatives as novel small-molecule inhibitors targeting the programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) interaction. Eur. J. Med. Chem. 2022, 229, 113998. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Halim, S.A.; Alsalman, A.; Khan, A.; Elkord, E.; Al-Harrasi, A. Structure-based small inhibitors search combined with molecular dynamics driven energies for human programmed cell death-1 (PD-1) protein. J. Biomol. Struct. Dyn. 2023, 1–15. [Google Scholar] [CrossRef]
- Sun, C.; Yin, M.; Cheng, Y.; Kuang, Z.; Liu, X.; Wang, G.; Wang, X.; Yuan, K.; Min, W.; Dong, J.; et al. Novel Small-Molecule PD-L1 Inhibitor Induces PD-L1 Internalization and Optimizes the Immune Microenvironment. J. Med. Chem. 2023, 66, 2064–2083. [Google Scholar] [CrossRef]
- Li, Y.; Cui, R.; Fan, F.; Lu, Y.; Ai, Y.; Liu, H.; Liu, S.; Du, Y.; Qin, Z.; Sun, W.; et al. The Efficacy and Safety of Ischemic Stroke Therapies: An Umbrella Review. Front. Pharmacol. 2022, 13, 924747. [Google Scholar] [CrossRef] [PubMed]
- Neupane, P.; Thada, P.K.; Singh, P.; Faisal, A.R.; Rai, N.; Poude, P.; Waleed, M.S.; Quinonez, J.; Ruxmohan, S.; Jain, E. Investigating Edaravone Use for Management of Amyotrophic Lateral Sclerosis (ALS): A Narrative Review. Cureus 2023, 15, e33746. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Liu, S.; Yang, M. Antioxidant and anti-inflammatory agents in chronic liver diseases: Molecular mechanisms and therapy. World J. Hepatol. 2023, 15, 180–200. [Google Scholar] [CrossRef]
- Zeng, W.; Long, X.; Liu, P.S.; Xie, X. The interplay of oncogenic signaling, oxidative stress and ferroptosis in cancer. Int. J. Cancer. 2023; ahead of print. [Google Scholar] [CrossRef]
- Huot, J.R.; Baumfalk, D.; Resendiz, A.; Bonetto, A.; Smuder, A.J.; Penna, F. Targeting Mitochondria and Oxidative Stress in Cancer- and Chemotherapy-Induced Muscle Wasting. Antioxid. Redox Signal. 2023, 38, 352–370. [Google Scholar]
- Reyes-Jiménez, E.; Ramírez-Hernández, A.A.; Santos-Álvarez, J.C.; Velázquez-Enríquez, J.M.; Pina-Canseco, S.; Baltiérrez-Hoyos, R.; Vásquez-Garzón, V.R. Involvement of 4-hydroxy-2-nonenal in the pathogenesis of pulmonary fibrosis. Mol. Cell. Biochem. 2021, 476, 4405–4419. [Google Scholar] [CrossRef] [PubMed]
- Jaganjac, M.; Tirosh, O.; Cohen, G.; Sasson, S.; Zarkovic, N. Reactive aldehydes--second messengers of free radicals in diabetes mellitus. Free Radic. Res. 2013, 47, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Moreira, M.T.G.; Pereira, P.R.; Aquino, A.; Conte-Junior, C.A.; Paschoalin, V.M.F. Aldehyde Accumulation in Aged Alcoholic Beer: Addressing Acetaldehyde Impacts on Upper Aerodigestive Tract Cancer Risks. Int. J. Mol. Sci. 2022, 23, 14147. [Google Scholar] [CrossRef]
- Sutaria, S.R.; Gori, S.S.; Morris, J.D.; Xie, Z.; Fu, X.A.; Nantz, M.H. Lipid Peroxidation Produces a Diverse Mixture of Saturated and Unsaturated Aldehydes in Exhaled Breath That Can Serve as Biomarkers of Lung Cancer-A Review. Metabolites 2022, 12, 561. [Google Scholar] [CrossRef]
- Sruthi, C.R.; Raghu, K.G. Methylglyoxal induces ambience for cancer promotion in HepG2 cells via Warburg effect and promotes glycation. J. Cell. Biochem. 2022, 123, 1532–1543. [Google Scholar] [CrossRef]
- Liang, J.; Wang, B.; Yang, Y.; Liu, B.; Jin, Y. Approaching the Dimerization Mechanism of Small Molecule Inhibittors Targeting PD-L1 with Molecular Simulation. Int. J. Mol. Sci. 2023, 24, 1280. [Google Scholar] [CrossRef]
- Guzik, K.; Zak, K.M.; Grudnik, P.; Magiera, K.; Musielak, B.; Törner, R.; Skalniak, L.; Dömling, A.; Dubin, G.; Holak, T.A. Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1. J. Med. Chem. 2017, 60, 5857–5867. [Google Scholar] [CrossRef] [PubMed]
- Chai, I.; Kornyeyev, D.; Hsieh, E.; Magombedze, G.; Stapleton, L.; Hung, M.; Kwon, H.J.; Stefanutti, E.; Belzile, J.; Czerwieniec, G.; et al. Effects of small molecule-induced dimerization on the programmed death ligand 1 protein life cycle. Sci. Rep. 2022, 12, 21286. [Google Scholar] [CrossRef]
- Wang, T.; Cai, S.; Cheng, Y.; Zhang, W.; Wang, M.; Sun, H.; Guo, B.; Li, Z.; Xiao, Y.; Jiang, S. Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation. J. Med. Chem. 2022, 65, 3879–3893. [Google Scholar] [CrossRef]
- Kelliny, S.; Xiong, J.; Bobrovskaya, L.; Zhou, X.F. Preclinical validation of a novel oral Edaravone formulation for treatment of frontotemporal dementia. Neurotox. Res. 2021, 39, 1689–1707. [Google Scholar] [CrossRef] [PubMed]
- Song, M.M.; Chen, J.; Ye, S.M.; Lu, D.P.; Zhang, G.Y.; Liu, R.; Shen, Y.X. Targeted delivery of edaravone by liposomes for the treatment of ischemic stroke. Nanomedicine 2022, 17, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Tuo, W.; Bollier, M.; Leleu-Chavain, N.; Lemaire, L.; Barczyk, A.; Dezitter, X.; Klupsch, F.; Szczepanski, F.; Spencer, J.; Chavatte, P.; et al. Development of novel oxazolo[5,4-d]pyrimidines as competitive CB2 neutral antagonists based on scaffold hopping. Eur. J. Med. Chem. 2018, 146, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Magnez, R.; Thiroux, B.; Taront, S.; Segaoula, Z.; Quesnel, B.; Thuru, X. PD-1/PD-L1 binding studies using microscale thermophoresis. Sci. Rep. 2017, 7, 17623. [Google Scholar] [CrossRef] [Green Version]
- Romain, M.; Thiroux, B.; Tardy, M.; Quesnel, B.; Thuru, X. Measurement of Protein-Protein Interactions through Microscale Thermophoresis (MST). Bio. Protoc. 2020, 10, e3574. [Google Scholar] [CrossRef] [PubMed]
- Shimozono, S.; Miyawaki, A. Engineering FRET constructs using CFP and YFP. Methods Cell. Biol. 2008, 85, 381–393. [Google Scholar] [PubMed]
- Marasco, M.; Berteotti, A.; Weyershaeuser, J.; Thorausch, N.; Sikorska, J.; Krausze, J.; Brandt, H.J.; Kirkpatrick, J.; Rios, P.; Schamel, W.W.; et al. Molecular mechanism of SHP2 activation by PD-1 stimulation. Sci. Adv. 2020, 6, 4458. [Google Scholar] [CrossRef] [Green Version]
- Abe, S.; Kirima, K.; Tsuchiya, K.; Okamoto, M.; Hasegawa, T.; Houchi, H.; Yoshizumi, M.; Tamaki, T. The reaction rate of edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one (MCI-186)) with hydroxyl radical. Chem. Pharm. Bull. 2004, 52, 186–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Compounds | PD-L1 Binding (KD, nM) a | Bioactivity (IC50, nM) b | Proliferation (IC50, nM) c |
|---|---|---|---|
| 2 | 34 ± 3 | 23 ± 3 | 53 ± 11 |
| 3 | 45 ± 7 | 44 ± 9 | 79 ± 7 |
| 4 | 19 ± 3 | 12 ± 5 | nd |
| 5 | 27 ± 4 | 74 ± 7 | 43 ± 9 |
| BMS-202 | nd | 124 ± 12 | 53 ± 17 |
| Compounds | DMPO | DPPH EC50 (μM) | |
|---|---|---|---|
| kr (1010 M−1 s−1) | kr/ka | ||
| 1 | 25.9 | 60 | 35.81 |
| 2 | 19.3 | 45 | 36.46 |
| 3 | 18.8 | 44 | 34.02 |
| 4 | 4.40 | 10 | 40.94 |
| 5 | 9.74 | 23 | 38.18 |
| Products [M–H]- | Formula | Ion m/z Values (Observed) | Intensities | Errors (Δppm) | rdb |
|---|---|---|---|---|---|
Cpd 1 (edaravone)
| C9H15O2 C10H9N2O2 C19H25N2O3 C29H33N4O3 C39H43N6O4 | 155.10692 173.0712 329.18719 485.25602 659.33535 | 1.63 × 105 3.98 × 106 4.88 × 105 6.60 × 105 8.37 × 104 | 1.8 1.5 1.7 2.7 | 2.5 7.5 8.5 15.5 21.5 |
Cpd 3
| C9H15O2 C16H11N2O2Cl2 C25H27N2O4Cl2 C41H37N4O5Cl4 C57H49N6O7Cl6 | 155.10692 333.02066 489.13606 805.15335 1139.18175 | 1.92 × 105 3.84 × 106 3.23 × 106 1.56 × 106 2.53 × 105 | 1.6 4.0 3.7 2.6 2.5 | 2.5 11.5 12.5 23.5 33.5 |
Cpd 4
| C9H15O2 C11H9N2OCl2 C20H25N2O3Cl2 C31H33N4O3Cl4 C42H43N6O4Cl6 | 155.10689 255.00989 411.12532 649.13249 905.14908 | 1.01 × 105 3.45 × 106 9.70 × 105 2.05 × 106 3.76 × 105 | 1.5 4.9 4.0 3.6 2.1 | 2.5 7.5 8.5 15.5 21.5 |
Cpd 5
| C9H15O2 C15H8N2OCl2F C24H24N2O3Cl2F C39H31N4O3Cl4F2 C54H40N6O4Cl6F2 | 155.10691 321.00062 477.11605 781.11314 1103.12119 | 2.11 × 105 3.92 × 106 1.98 × 106 1.43 × 106 3.51 × 105 | 1.3 4.5 3.6 2.8 2.9 | 2.5 11.5 12.5 23.5 33.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regnault, R.; Klupsch, F.; El-Bouazzati, H.; Magnez, R.; Le Biannic, R.; Leleu-Chavain, N.; Ahouari, H.; Vezin, H.; Millet, R.; Goossens, J.-F.; et al. Novel PD-L1-Targeted Phenyl-Pyrazolone Derivatives with Antioxidant Properties. Molecules 2023, 28, 3491. https://doi.org/10.3390/molecules28083491
Regnault R, Klupsch F, El-Bouazzati H, Magnez R, Le Biannic R, Leleu-Chavain N, Ahouari H, Vezin H, Millet R, Goossens J-F, et al. Novel PD-L1-Targeted Phenyl-Pyrazolone Derivatives with Antioxidant Properties. Molecules. 2023; 28(8):3491. https://doi.org/10.3390/molecules28083491
Chicago/Turabian StyleRegnault, Romain, Frédérique Klupsch, Hassiba El-Bouazzati, Romain Magnez, Raphaël Le Biannic, Natascha Leleu-Chavain, Hania Ahouari, Hervé Vezin, Régis Millet, Jean-François Goossens, and et al. 2023. "Novel PD-L1-Targeted Phenyl-Pyrazolone Derivatives with Antioxidant Properties" Molecules 28, no. 8: 3491. https://doi.org/10.3390/molecules28083491