Thiocholine-Mediated One-Pot Peptide Ligation and Desulfurization
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis of Thiocholine Peptide Thioester
2.2. Hydrolytic Stability and NCL Reactivity of Thiocholine Thioester
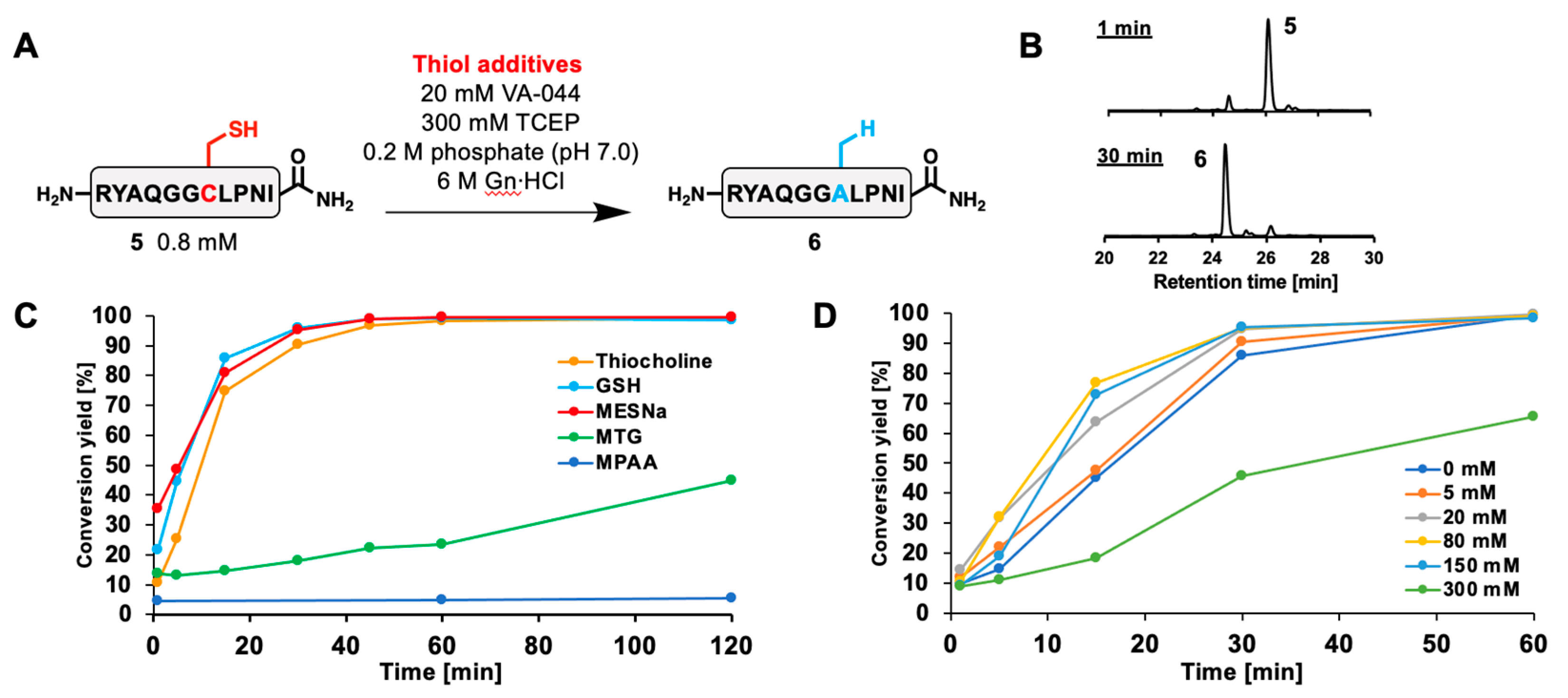
2.3. Desulfurization with Thiocholine
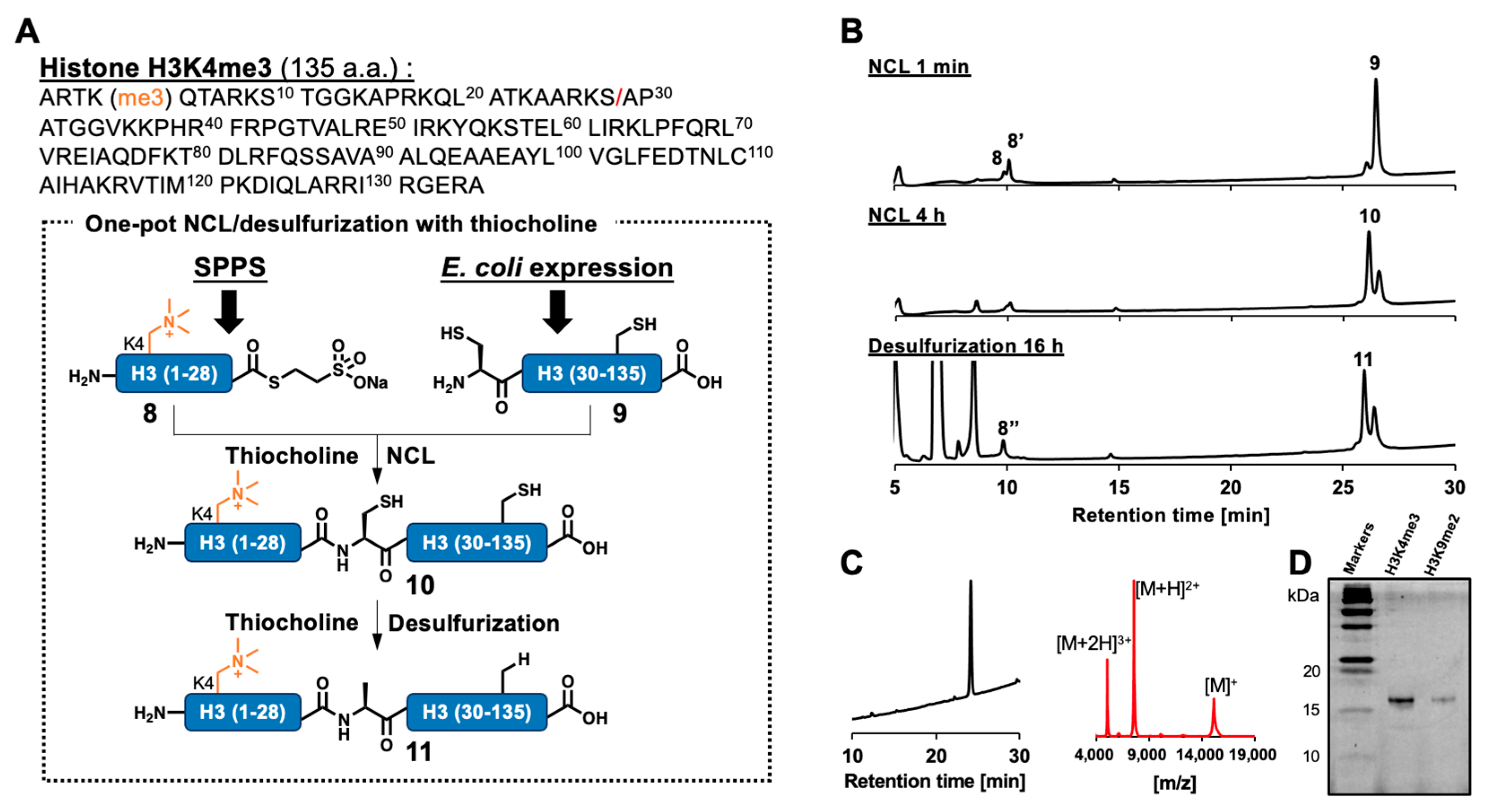
2.4. One-Pot NCL/Desulfurization for Histone H3K4me3 Semisynthesis
3. Materials and Methods
3.1. General
3.2. Synthesis of Thiocholine Chloride
3.3. Synthesis of Model Peptides
3.3.1. General Method of Peptide Synthesis
3.3.2. Synthesis of Model Peptide Thioesters
3.3.3. Synthesis of Other Model Peptides
3.4. Evaluation of Peptide Thioesters
3.4.1. Determination of Calibration Curves for Peptides 2 and 4
3.4.2. Hydrolysis Kinetics of Peptide Thioesters
3.4.3. NCL Kinetics of Peptide Thioesters
3.5. Desulfurization with Model Peptide
3.5.1. Desulfurization with Different Thiol Additives
3.5.2. Desulfurization with Different Concentrations of Thiocholine
3.6. Semisynthesis of Histone H3K4me3 and H3K9me2
3.6.1. Synthesis of Histone H3 N-Terminal Peptide Segments
3.6.2. Preparation of Histone H3 C-Terminal Peptide Segment by E. coli Expression
3.6.3. One-Pot NCL/Desulfurization for Full-Length H3K4me3 and H3K9me2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B.H. Synthesis of Proteins by Native Chemical Ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef]
- Agouridas, V.; Mahdi, O.E.; Diemer, V.; Cargoet, M.; Monbaliu, J.C.M.; Melnyk, O. Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations. Chem. Rev. 2019, 119, 7328–7443. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Sayers, J.; Premdjee, B.; Payne, R.J. Rapid and efficient protein synthesis through expansion of the native chemical ligation concept. Nat. Rev. Chem. 2018, 2, 0122. [Google Scholar] [CrossRef]
- Conibear, A.C.; Watson, E.E.; Payne, R.J.; Becker, C.F.W. Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc. Rev. 2018, 47, 9046–9068. [Google Scholar] [CrossRef]
- Diemer, V.; Firstova, O.; Agouridas, V.; Melnyk, O. Pedal to the Metal: The Homogeneous Catalysis of the Native Chemical Ligation Reaction. Chem. Eur. J. 2022, 28, e202104229. [Google Scholar]
- Hupe, D.J.; Jencks, W.P. Nonlinear structure-reactivity correlations. Acyl transfer between sulfur and oxygen nucleophiles. J. Am. Chem. Soc. 1977, 99, 451–464. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Kent, S.B.H. Insights into the Mechanism and Catalysis of the Native Chemical Ligation Reaction. J. Am. Chem. Soc. 2006, 128, 6640–6646. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Danishefsky, S.J. Free-Radical-Based, Specific Desulfurization of Cysteine: A Powerful Advance in the Synthesis of Polypeptides and Glycopolypeptides. Angew. Chem. Int. Ed. 2007, 46, 9248–9252. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Li, X. Advances in Native Chemical Ligation–Desulfurization: A Powerful Strategy for Peptide and Protein Synthesis. Chem. Eur. J. 2018, 24, 17397–17404. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.C., Jr.; Benner, J.; Xu, M.Q. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998, 7, 2256–2264. [Google Scholar] [CrossRef]
- Siman, P.; Blatt, O.; Moyal, T.; Danieli, T.; Lebendiker, M.; Lashuel, H.A.; Friedler, A.; Brik, A. Chemical Synthesis and Expression of the HIV-1 Rev Protein. ChemBioChem 2011, 12, 1097–1104. [Google Scholar] [CrossRef]
- Shimko, J.C.; North, J.A.; Bruns, A.N.; Poirier, M.G.; Ottesen, J.J. Preparation of Fully Synthetic Histone H3 Reveals That Acetyl-Lysine 56 Facilitates Protein Binding Within Nucleosomes. J. Mol. Biol. 2011, 408, 187–204. [Google Scholar] [CrossRef]
- Hayashi, G.; Sueoka, T.; Okamoto, A. In vitro and in cell analysis of chemically synthesized histone H2A with multiple modifications. Chem. Commun. 2016, 52, 4999. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Chen, C.C.; Gao, S.; Wang, Y.H.; Xiao, H.; Wang, F.; Tian, C.L.; Li, Y.M. Synthesis of L- and D-Ubiquitin by One-Pot Ligation and Metal-Free Desulfurization. Chem. Eur. J. 2016, 22, 7623–7628. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.E.; Liu, X.; Alonso-García, N.; Pereira, P.J.B.; Jolliffe, K.A.; Payne, R.J. Trifluoroethanethiol: An Additive for Efficient One-Pot Peptide Ligation-Desulfurization Chemistry. J. Am. Chem. Soc. 2014, 136, 8161–8164. [Google Scholar] [CrossRef] [PubMed]
- Ohkawachi, K.; Kobayashi, D.; Morimoto, K.; Shigenaga, A.; Denda, M.; Yamatsugu, K.; Kanai, M.; Otaka, A. Sulfanylmethyldimethylaminopyridine as a Useful Thiol Additive for Ligation Chemistry in Peptide/Protein Synthesis. Org. Lett. 2020, 22, 5289–5293. [Google Scholar] [CrossRef]
- Bagiyan, G.A.; Grachev, S.A.; Koroleva, I.K.; Soroka, N.V. The kinetics and mechanism of the reaction of the oxidation of aminothiols by hydrogen peroxide in aqueous solutions. Bull. Acad. Sci. USSR Div. Chem. Sci. 1976, 25, 961–966. [Google Scholar] [CrossRef]
- Chalker, J.M.; Lercher, L.; Rose, N.R.; Schofield, C.J.; Davis, B.G. Conversion of Cysteine into Dehydroalanine Enables Access to Synthetic Histones Bearing Diverse Post-Translational Modifications. Angew. Chem. Int. Ed. 2012, 51, 1835–1839. [Google Scholar] [CrossRef]
- Fang, G.M.; Li, Y.M.; Shen, F.; Huang, Y.C.; Li, J.B.; Lin, Y.; Cui, H.K.; Liu, L. Protein Chemical Synthesis by Ligation of Peptide Hydrazides. Angew. Chem. Int. Ed. 2011, 50, 7645–7649. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.S.; Tang, S.; Qi, Y.K.; Wang, Z.P.; Liu, L. Chemical Synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protcol. 2013, 8, 2483–2495. [Google Scholar] [CrossRef]
- Hackeng, T.M.; Griffin, J.H.; Dawson, P.E. Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. USA 1999, 96, 10068–10073. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.F.; Du, J.J.; Liu, Z.; Guo, J. Visible-Light-Induced Specific Desulfurization of Cysteinyl Peptide and Glycopeptide in Aqueous Solution. Org. Lett. 2016, 18, 1166–1169. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Neukirchen, S.; Cabrele, C.; Reiser, O. Visible-light photoredox-catalyzed desulfurization of thiol- and disulfide-containing amino acids and small peptides. J. Pept. Sci. 2017, 23, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, T.S.; Clayton, D.; Dowman, L.J.; Sayers, J.; Payne, R.J. Native Chemical Ligation–Photodesulfurization in Flow. J. Am. Chem. Soc. 2018, 140, 9020–9024. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Qu, Q.; Guan, C.J.; Chu, G.C.; Shi, J.; Li, Y.M. Efficient chemical synthesis for the analogue of ubiquitin-based probe Ub–AMC with native bioactivity. RSC Adv. 2016, 6, 47926. [Google Scholar] [CrossRef]
- Erickson, P.W.; Fulcher, J.M.; Spaltenstein, P.; Kay, M.S. Traceless Click-Assisted Native Chemical Ligation Enabled by Protecting Dibenzocyclooctyne from Acid-Mediated Rearrangement with Copper(I). Bioconjug. Chem. 2021, 32, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Park, J.H.; Ramlall, T.; Eliezer, D.; Rhoades, E.; Petersson, E.J. Chemoenzymatic Semi-synthesis Enables Efficient Production of Isotopically Labeled α-Synuclein with Site-Specific Tyrosine Phosphorylation. ChemBioChem 2021, 22, 1440–1447. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, L.; Shi, J.; Yu, H.Z. Desulfurization Mechanism of Cysteine in Synthesis of Polypeptides. Chin. J. Chem. Phys. 2015, 28, 269. [Google Scholar] [CrossRef]
- Qiu, W.; Shi, S.; Li, R.; Lin, X.; Rao, L.; Sun, Z. A Mild, General, Metal-Free Method for Desulfurization of Thiols and Disulfides Induced by Visible-Light. Chin. J. Chem. 2021, 39, 1255–1258. [Google Scholar] [CrossRef]
- Venneti, N.M.; Samala, G.; Morsy, R.M.I.; Mendoza, L.G.; Isidro-Llobet, A.; Tom, J.K.; Mukherjee, S.; Kopach, M.E.; Stockdill, J.L. Phosphine-Dependent Photoinitiation of Alkyl Thiols under Near-UV Light Facilitates User-Friendly Peptide Desulfurization. J. Am. Chem. Soc. 2023, 145, 1053–1061. [Google Scholar] [CrossRef]
- Sun, Z.; Ma, W.; Cao, Y.; Wei, T.; Mo, X.; Chow, H.T.; Tan, Y.; Cheung, C.H.P.; Liu, J.; Lee, H.K.; et al. Superfast desulfurization for protein chemical synthesis and modification. Chem 2022, 8, 2542–2557. [Google Scholar] [CrossRef]
- Roudier, F.; Ahmed, I.; Bérard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Park, S.; van Nocker, S. Genic and Global Functions for Paf1C in Chromatin Modification and Gene Expression in Arabidopsis. PLoS Genet. 2008, 4, e1000077. [Google Scholar] [CrossRef]
- Houden, A.; Demidov, D.; Gernand, D.; Meister, A.; Leach, C.R.; Schubert, I. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 2003, 33, 967–973. [Google Scholar]
- Jiang, D.; Kong, N.C.; Gu, X.; Li, Z.; He, Y. Arabidopsis COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development. PLoS Genet. 2011, 7, e1001330. [Google Scholar] [CrossRef]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef]
- Bang, D.; Kent, S.B.H. A One-Pot Total Synthesis of Crambin. Angew. Chem. Int. Ed. 2004, 43, 2534–2538. [Google Scholar] [CrossRef]
- Xu, L.; Jiang, H. Writing and Reading Histone H3 lysine 9 Methylation in Arabidopsis. Front. Plant Sci. 2020, 11, 452. [Google Scholar] [CrossRef]
- Jackson, J.P.; Johnson, L.; Jasencakova, Z.; Zhang, X.; PerezBurgos, L.; Singh, P.B.; Cheng, X.; Schubert, I.; Jenuwein, T.; Jacobsen, S.E. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 2004, 112, 308–315. [Google Scholar] [CrossRef]
- Soppe, W.J.J.; Jasencakova, Z.; Houben, A.; Kakutani, T.; Meister, A.; Huang, M.S.; Jacobsen, S.E.; Schubert, I.; Fransz, P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002, 21, 6549–6559. [Google Scholar] [CrossRef]
- Jasencakova, Z.; Soppe, W.J.J.; Meister, A.; Gernand, D.; Turner, B.M.; Schubert, I. Histone modifications in Arabidopsis—High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003, 33, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Naumann, K.; Fischer, A.; Hofmann, I.; Krauss, V.; Phalke, S.; Irmler, K.; Hause, G.; Aurich, A.C.; Dorn, R.; Jenuwein, T.; et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005, 24, 1418–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Du, X.; Tang, K.; Yang, Z.; Pan, L.; Zhu, P.; Luo, J.; Jiang, Y.; Zhang, H.; Wan, H.; et al. Arabidopsis AGDP1 links H3K9me2 to DNA methylation in heterochromatin. Nat. Commun. 2018, 9, 4547. [Google Scholar] [CrossRef]
- Walker, B.J.; Nair, G.P.; Marshall, L.F.; Bulović, V.; Bawendi, M.G. Narrow-Band Absorption-Enhanced Quantum Dot/J-Aggregate Conjugates. J. Am. Chem. Soc. 2009, 131, 9624–9625. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Kagawa, W.; Osakabe, A.; Kurumizaka, H. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl. Acad. Sci. USA 2010, 107, 10454–10459. [Google Scholar] [CrossRef] [PubMed]
- Kujirai, T.; Arimura, Y.; Fujita, R.; Horikoshi, N.; Machida, S.; Kurumizaka, H. Methods for Preparing Nucleosomes Containing Histone Variants; Part of the Methods in Molecular Biology Book Series; Orsi, G., Almouzni, G., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1832. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, S.; Nakajima, Y.; Kamo, N.; Osakabe, A.; Okamoto, A.; Hayashi, G.; Murakami, H. Thiocholine-Mediated One-Pot Peptide Ligation and Desulfurization. Molecules 2023, 28, 3655. https://doi.org/10.3390/molecules28093655
Suzuki S, Nakajima Y, Kamo N, Osakabe A, Okamoto A, Hayashi G, Murakami H. Thiocholine-Mediated One-Pot Peptide Ligation and Desulfurization. Molecules. 2023; 28(9):3655. https://doi.org/10.3390/molecules28093655
Chicago/Turabian StyleSuzuki, Sae, Yuya Nakajima, Naoki Kamo, Akihisa Osakabe, Akimitsu Okamoto, Gosuke Hayashi, and Hiroshi Murakami. 2023. "Thiocholine-Mediated One-Pot Peptide Ligation and Desulfurization" Molecules 28, no. 9: 3655. https://doi.org/10.3390/molecules28093655
APA StyleSuzuki, S., Nakajima, Y., Kamo, N., Osakabe, A., Okamoto, A., Hayashi, G., & Murakami, H. (2023). Thiocholine-Mediated One-Pot Peptide Ligation and Desulfurization. Molecules, 28(9), 3655. https://doi.org/10.3390/molecules28093655





