Molecular Profiling of Peanut under Raw, Roasting, and Autoclaving Conditions Using High-Resolution Magic Angle Spinning and Solution 1H NMR Spectroscopy
Abstract
:1. Introduction
2. Results
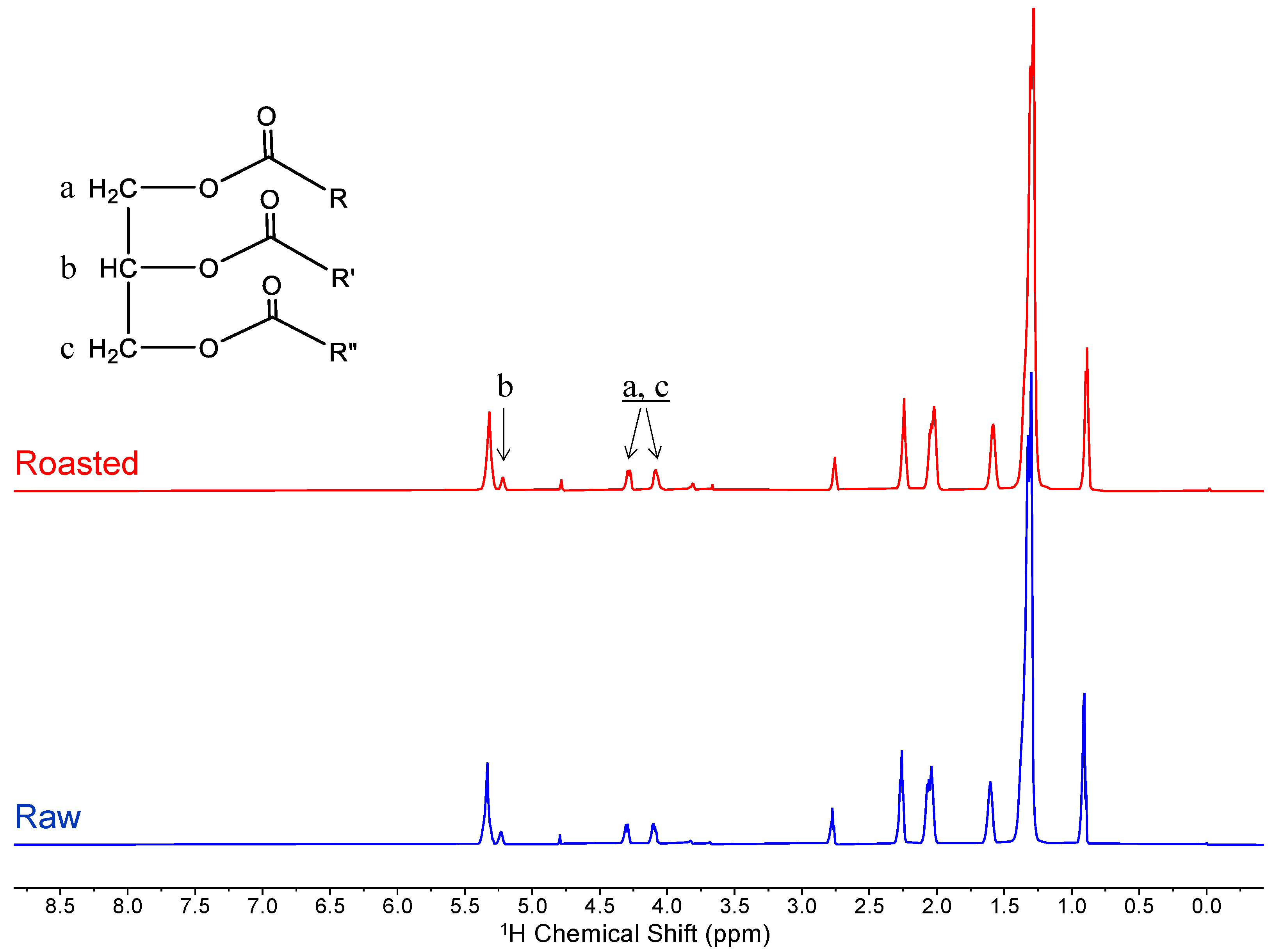
2.1. Predominance of Lipids Observed through Intact Peanut Analysis
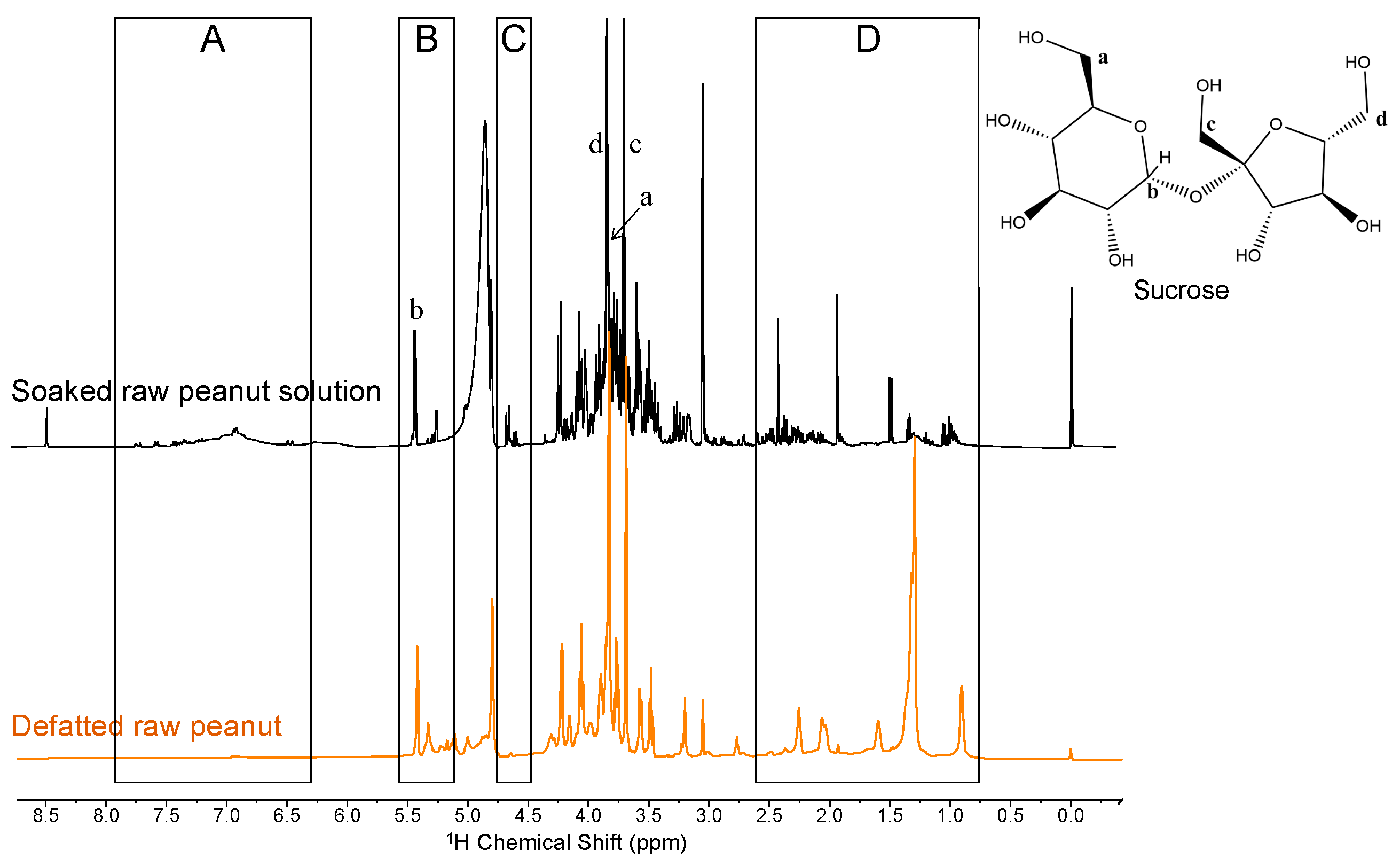
2.2. Sugars Revealed through the Analysis of Peanut Flour
2.3. Detecting Proteins through the Analysis of Soaked Peanut Solutions
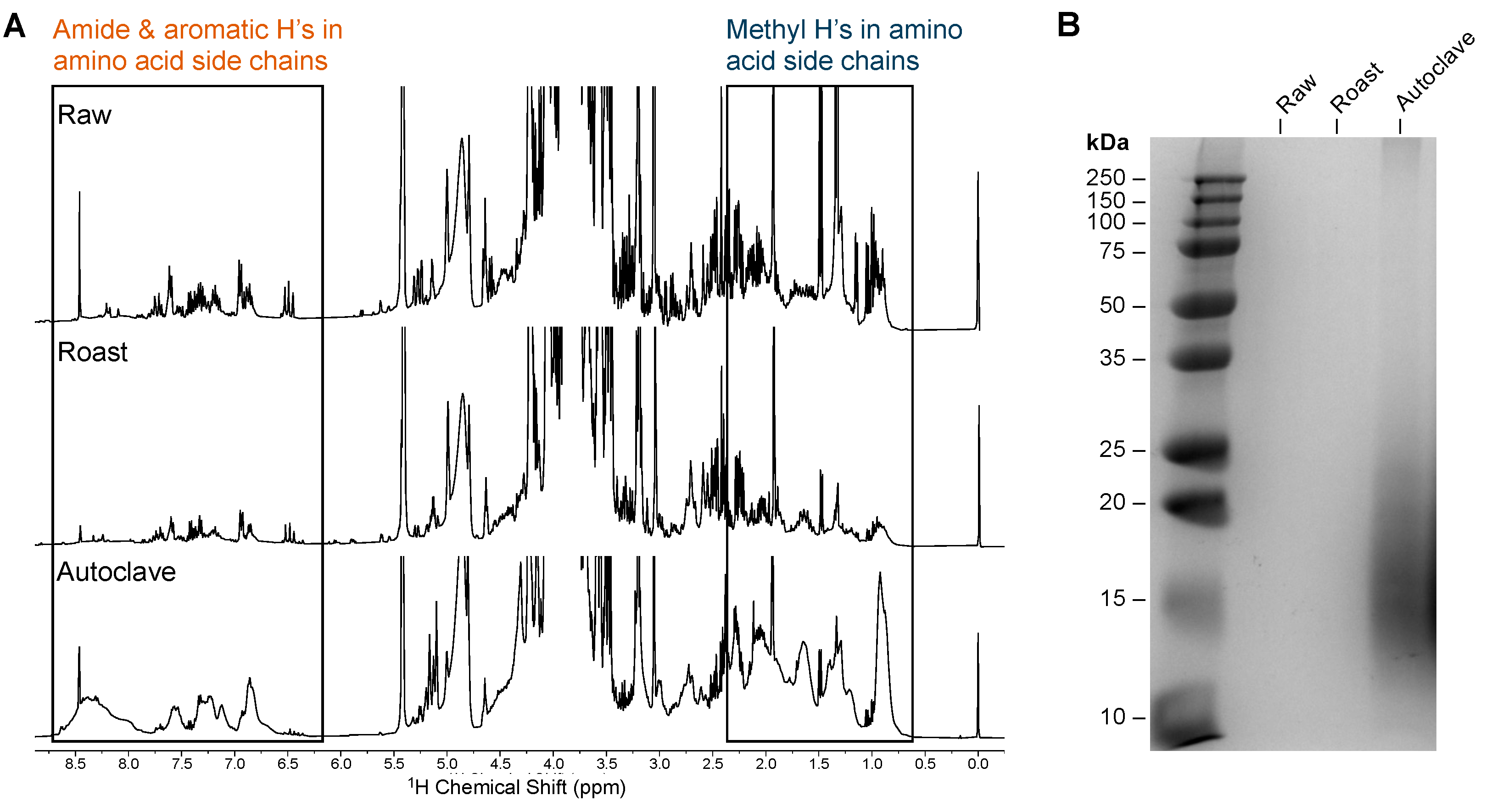
2.4. Differential Signature for Autoclaved Peanut
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.1.1. Physical Processing
4.1.2. Defatting Peanuts
4.2. NMR Sample Preparation
4.2.1. Solid Preparation
4.2.2. Solution Preparation
4.3. HR-MAS and Solution 1H NMR Spectroscopy
4.3.1. HR-MAS 1H NMR Analysis of Solid Peanut
4.3.2. Solution 1H NMR Analysis of Peanut-Soaked Solutions
4.4. SDS PAGE Analysis
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
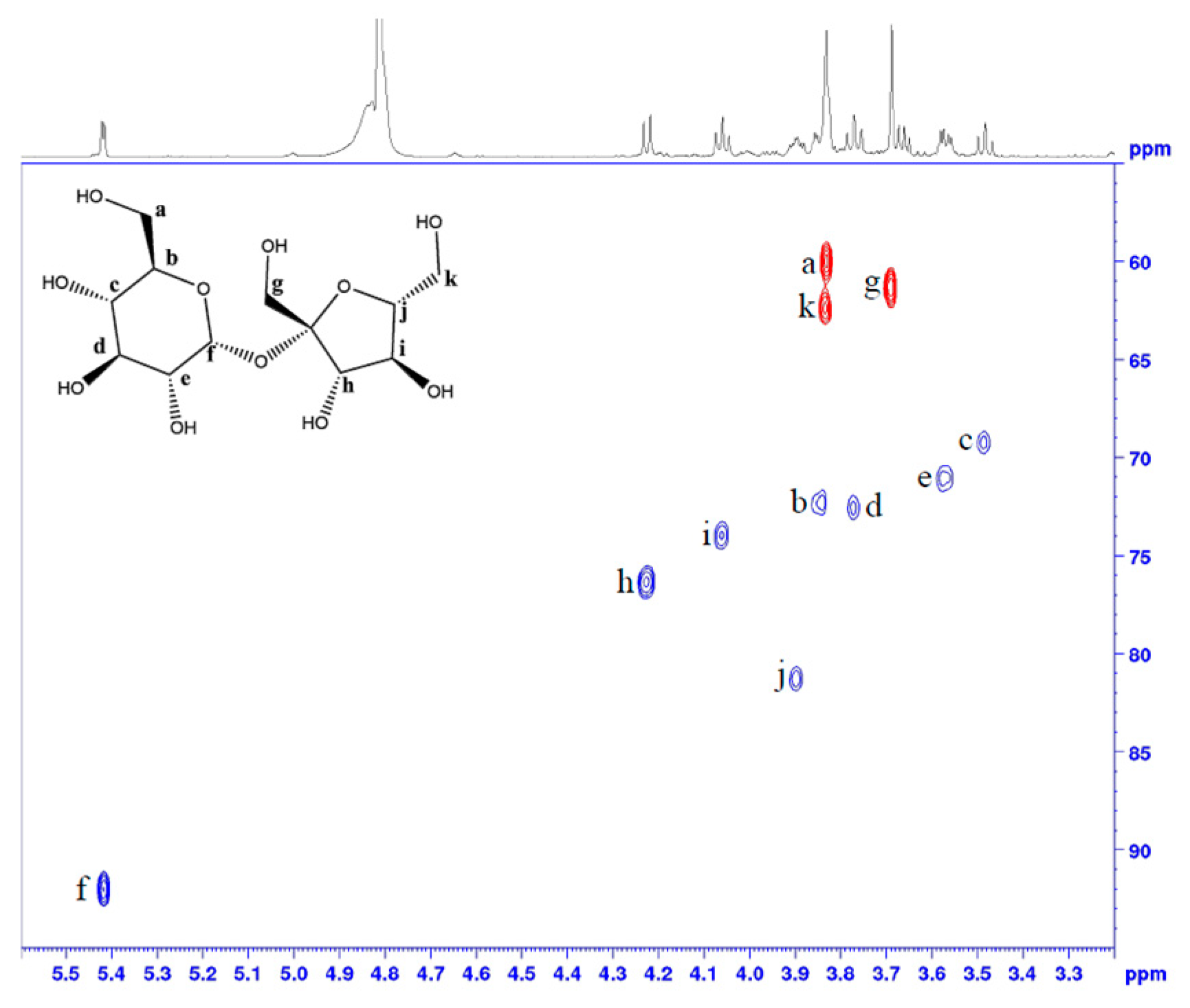
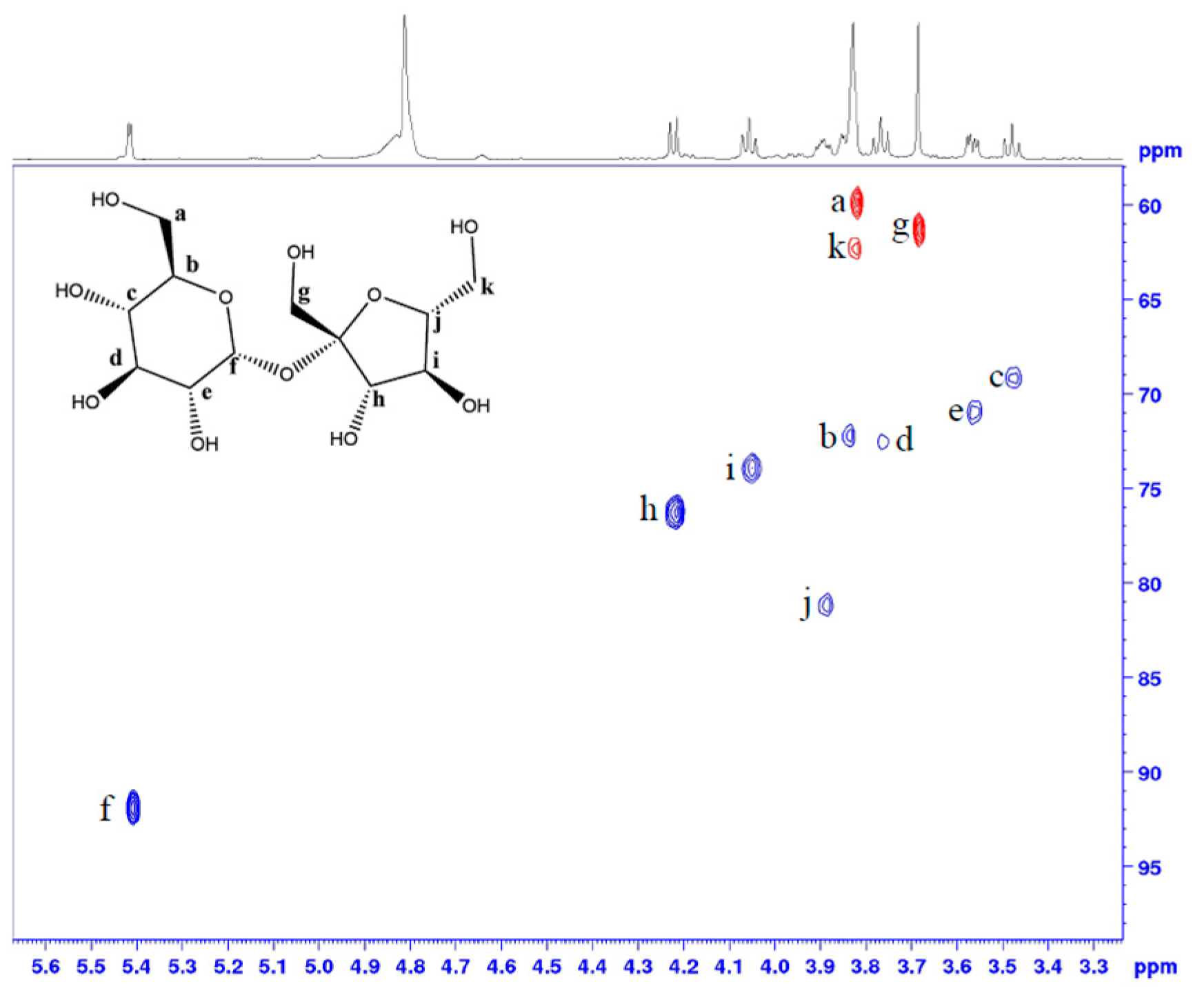
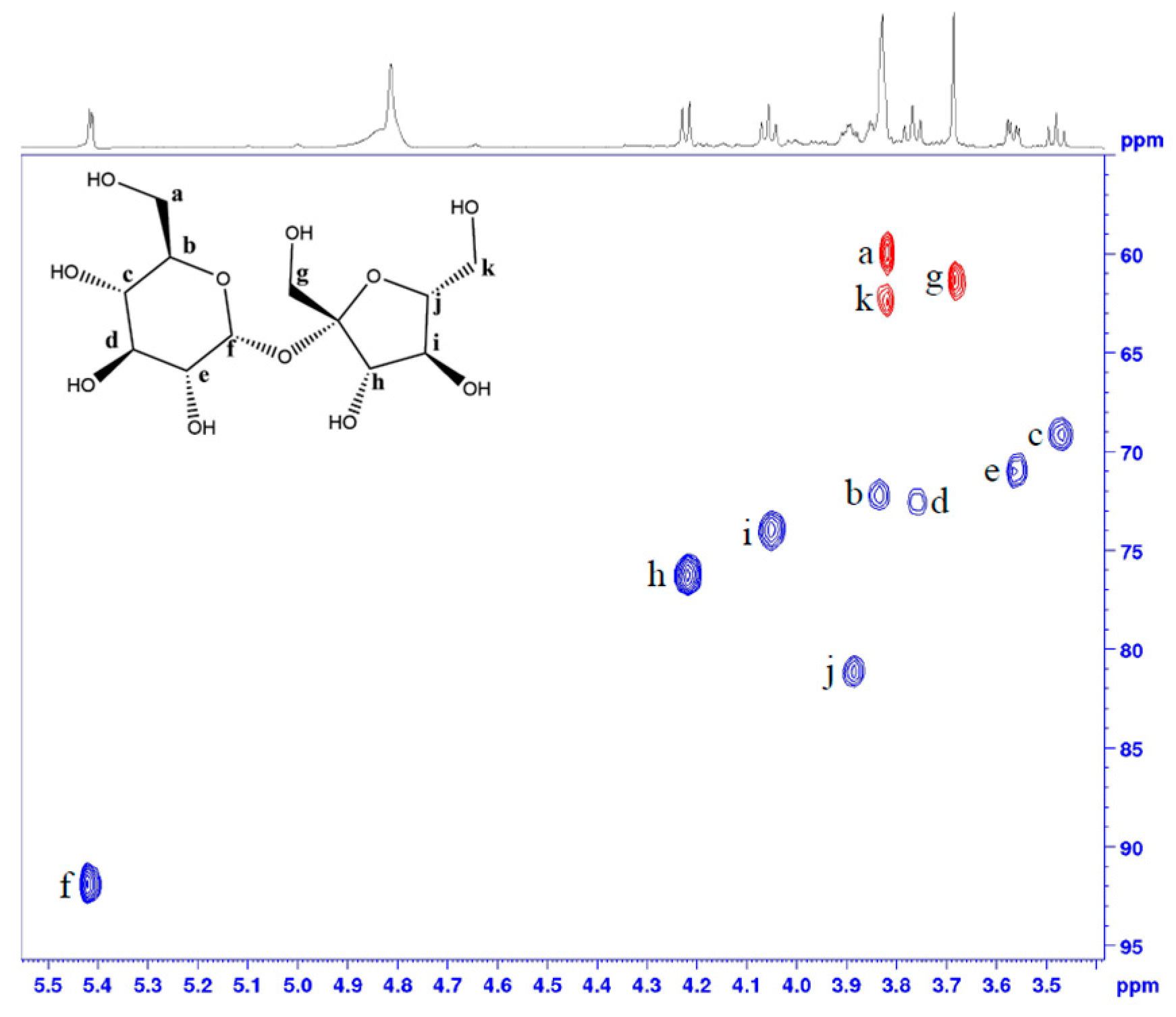
References
- Bonku, R.; Yu, J. Health aspects of peanuts as an outcome of its chemical composition. Food Sci. Hum. Wellness 2020, 9, 21–30. [Google Scholar] [CrossRef]
- Shah, F.; Shi, A.; Ashley, J.; Kronfel, C.; Wang, Q.; Maleki, S.J.; Adhikari, B.; Zhang, J. Peanut Allergy: Characteristics and Approaches for Mitigation. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1361–1387. [Google Scholar] [CrossRef] [PubMed]
- Mueller, G.A.; Maleki, S.J.; Pedersen, L.C. The molecular basis of peanut allergy. Curr. Allergy Asthma Rep. 2014, 14, 429. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Lin, J.; Bardina, L.; Grishina, G.A.; Lee, C.; Seo, W.H.; Sampson, H.A. What Characteristics Confer Proteins the Ability to Induce Allergic Responses? IgE Epitope Mapping and Comparison of the Structure of Soybean 2S Albumins and Ara h 2. Molecules 2016, 21, 622. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.M.; Jappe, U. Peanut allergens. Chem. Immunol. Allergy 2014, 100, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Young, C.T.; Schadel, W.E. Microstructure of Peanut Seed: A Review. Food Struct. 1990, 9, 317–328. [Google Scholar]
- Tian, Y.; Rao, H.; Zhang, K.; Tao, S.; Xue, W.-T. Effects of different thermal processing methods on the structure and allergenicity of peanut allergen Ara h 1. Food Sci. Nutr. 2018, 6, 1706–1714. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, Q.; Zhang, T.; Cai, Q.; Chen, Q. Thermal processing effects on peanut allergen Ara h 2 allergenicity in mice and its antigenic epitope structure. Food Chem. 2016, 212, 657–662. [Google Scholar] [CrossRef]
- Verhoeckx, K.C.; Vissers, Y.M.; Baumert, J.L.; Faludi, R.; Feys, M.; Flanagan, S.; Herouet-Guicheney, C.; Holzhauser, T.; Shimojo, R.; van der Bolt, N.; et al. Food processing and allergenicity. Food Chem. Toxicol. 2015, 80, 223–240. [Google Scholar] [CrossRef]
- Toda, M.; Hellwig, M.; Henle, T.; Vieths, S. Influence of the Maillard Reaction on the Allergenicity of Food Proteins and the Development of Allergic Inflammation. Curr. Allergy Asthma Rep. 2019, 19, 4. [Google Scholar] [CrossRef]
- Chung, S.Y.; Champagne, E.T. Association of end-product adducts with increased IgE binding of roasted peanuts. J. Agric. Food Chem. 2001, 49, 3911–3916. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.J.; Chung, S.-Y.; Champagne, E.T.; Raufman, J.-P. The effects of roasting on the allergenic properties of peanut proteins. J. Allergy Clin. Immunol. 2000, 106, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Toda, M.; Heilmann, M.; Ilchmann, A.; Vieths, S. The Maillard reaction and food allergies: Is there a link? Clin. Chem. Lab. Med. 2014, 52, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, B.; Maleki, S.J.; Rodriguez, J.; Burbano, C.; Muzquiz, M.; Jimenez, M.A.; Pedrosa, M.M.; Cuadrado, C.; Crespo, J.F. Heat and pressure treatments effects on peanut allergenicity. Food Chem. 2012, 132, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, B.; Cuadrado, C.; Rodriguez, J.; Hart, J.; Burbano, C.; Crespo, J.F.; Novak, N. Potential changes in the allergenicity of three forms of peanut after thermal processing. Food Chem. 2015, 183, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, C.; Sanchiz, A.; Arribas, C.; Pedrosa, M.M.; Gamboa, P.; Betancor, D.; Blanco, C.; Cabanillas, B.; Linacero, R. Mitigation of peanut allergenic reactivity by combined processing: Pressured heating and enzymatic hydrolysis. Innov. Food Sci. Emerg. Technol. 2023, 86, 103383. [Google Scholar] [CrossRef]
- Fatima, V.; Africa, S.; Rosa, R.-P.; Maria, P.; Santiago, Q.; Joseph, H.; Colette, B.; Rosario, L.; Karim, A.; Carmen, C. Influence of Instant Controlled Pressure Drop (DIC) on Allergenic Potential of Tree Nuts. Molecules 2020, 25, 1742. [Google Scholar]
- Sanchiz, A.; Cuadrado, C.; Dieguez, M.C.; Ballesteros, I.; Rodríguez, J.; Crespo, J.F.; de las Cuevas, N.; Rueda, J.; Linacero, R.; Cabanillas, B.; et al. Thermal processing effects on the IgE-reactivity of cashew and pistachio. Food Chem. 2018, 245, 595–602. [Google Scholar] [CrossRef]
- Cohen, C.G.; Toscano-Rivero, D.T.; Ahmed, E.A.; Zhao, W.; Dejgaard, K.; Berghuis, A.; Jean-Claude, B.J.; Mazer, B.D. Mechanism of reduced peanut-specific IgE binding following the high-pressure and temperature autoclaving of peanuts. 2023; manuscript in preparation. [Google Scholar]
- Augustijn, D.; de Groot, H.J.M.; Alia, A. HR-MAS NMR Applications in Plant Metabolomics. Molecules 2021, 26, 931. [Google Scholar] [CrossRef]
- Lucas-Torres, C.; Wong, A. Intact NMR spectroscopy: Slow high-resolution magic angle spinning chemical shift imaging. Analyst 2020, 145, 2520–2524. [Google Scholar] [CrossRef]
- Song, E.H.; Kim, H.J.; Jeong, J.; Chung, H.J.; Kim, H.Y.; Bang, E.; Hong, Y.S. A (1)H HR-MAS NMR-Based Metabolomic Study for Metabolic Characterization of Rice Grain from Various Oryza sativa L. Cultivars. J. Agric. Food Chem. 2016, 64, 3009–3016. [Google Scholar] [CrossRef] [PubMed]
- Ritota, M.; Marini, F.; Sequi, P.; Valentini, M. Metabolomic characterization of Italian sweet pepper (Capsicum annum L.) by means of HRMAS-NMR spectroscopy and multivariate analysis. J. Agric. Food Chem. 2010, 58, 9675–9684. [Google Scholar] [CrossRef] [PubMed]
- Munz, E.; Jakob, P.M.; Borisjuk, L. The potential of nuclear magnetic resonance to track lipids in planta. Biochimie 2016, 130, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Gouilleux, B.; Charrier, B.; Akoka, S.; Felpin, F.-X.; Rodriguez-Zubiri, M.; Giraudeau, P. Ultrafast 2D NMR on a benchtop spectrometer: Applications and perspectives. Trends Analyt. Chem. 2016, 83 Pt A, 65–75. [Google Scholar] [CrossRef]
- Silveira Junior, E.G.; Simionatto, E.; Perez, V.H.; Justo, O.R.; Zárate, N.A.H.; Vieira, M.D.C. Potential of Virginia-type peanut (Arachis hypogaea L.) as feedstock for biodiesel production. Ind. Crops Prod. 2016, 89, 448–454. [Google Scholar] [CrossRef]
- McDaniel, K.A.; White, B.L.; Dean, L.L.; Sanders, T.H.; Davis, J.P. Compositional and mechanical properties of peanuts roasted to equivalent colors using different time/temperature combinations. J. Food Sci. 2012, 77, C1293–C1299. [Google Scholar] [CrossRef]
- Senhaji Mouhri, Z.; Goodfellow, E.; Kelley, S.P.; Stein, R.S.; Rogers, R.D.; Jean-Claude, B.J. (15)N-, (13)C- and ¹H-NMR Spectroscopy Characterization and Growth Inhibitory Potency of a Combi-Molecule Synthesized by Acetylation of an Unstable Monoalkyltriazene. Molecules 2017, 22, 1183. [Google Scholar] [CrossRef]
- Settaluri, V.S.; Kandala, C.V.K.; Puppala, N.; Sundaram, J. Peanuts and Their Nutritional Aspects—A Review. Food Nutr. Sci. 2012, 3, 7. [Google Scholar] [CrossRef]
- Alessandri, S.; Sancho, A.; Vieths, S.; Mills, C.E.N.; Wal, J.-M.; Shewry, P.R.; Rigby, N.; Hoffmann-Sommergruber, K.; Uversky, V.N. High-Throughput NMR Assessment of the Tertiary Structure of Food Allergens. PLoS ONE 2012, 7, e39785. [Google Scholar] [CrossRef]
- Lehmann, K.; Schweimer, K.; Neudecker, P.; Rösch, P. Sequence-specific 1H, 13C and 15N resonance assignments of Ara h 6, an allergenic 2S albumin from peanut. J. Biomol. NMR 2004, 29, 93–94. [Google Scholar] [CrossRef]
- Schmitt, C.; Bastek, T.; Stelzer, A.; Schneider, T.; Fischer, M.; Hackl, T. Detection of Peanut Adulteration in Food Samples by Nuclear Magnetic Resonance Spectroscopy. J. Agric. Food Chem. 2020, 68, 14364–14373. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Cheng, K.; He, L.; Zhang, X.; Jiang, B.; Jiang, L.; Li, C.; Wang, G.; Yang, Y.; Liu, M. NMR-Based Methods for Protein Analysis. Anal. Chem. 2021, 93, 1866–1879. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.P.; McElroy, C.A.; Amero, C.D. Solution NMR of Large Molecules and Assemblies. Biochemistry 2007, 46, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Neumann, D. Protein bodies, storage organelles in plant seeds. Biochem. Physiol. Pflanz. 1980, 175, 279–306. [Google Scholar] [CrossRef]
- Cohen, C.; Jean-Claude, B.J.; Mazer, B. Peanut Hypoallergenic Formulations for Determining the Risk of. Anaphylaxis. Patent US17/510,772, 22 October 2020. [Google Scholar]

| Peanut Form | 1H NMR Technique | Molecules Detected | Signatures across Conditions (Raw, Roasted, Autoclaved) | ||
|---|---|---|---|---|---|
| Lipids (Triglycerides) | Sugars (Sucrose, Glucose) | Proteins (Peptides, Amino Acids) | |||
| Whole, intact peanut | HR-MAS | +++ | + | – | No major change |
| Defatted peanut flour | HR-MAS | + | +++ | +/– | No major change |
| Peanut-soaked solution | Solution | – | +++ | ++ | Distinct for autoclave (broad peaks) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohen, C.G.; Mazer, B.D.; Jean-Claude, B.J. Molecular Profiling of Peanut under Raw, Roasting, and Autoclaving Conditions Using High-Resolution Magic Angle Spinning and Solution 1H NMR Spectroscopy. Molecules 2024, 29, 162. https://doi.org/10.3390/molecules29010162
Cohen CG, Mazer BD, Jean-Claude BJ. Molecular Profiling of Peanut under Raw, Roasting, and Autoclaving Conditions Using High-Resolution Magic Angle Spinning and Solution 1H NMR Spectroscopy. Molecules. 2024; 29(1):162. https://doi.org/10.3390/molecules29010162
Chicago/Turabian StyleCohen, Casey G., Bruce D. Mazer, and Bertrand J. Jean-Claude. 2024. "Molecular Profiling of Peanut under Raw, Roasting, and Autoclaving Conditions Using High-Resolution Magic Angle Spinning and Solution 1H NMR Spectroscopy" Molecules 29, no. 1: 162. https://doi.org/10.3390/molecules29010162





