Performance of Classification Models of Toxins Based on Raman Spectroscopy Using Machine Learning Algorithms
Abstract
:1. Introduction
2. Results
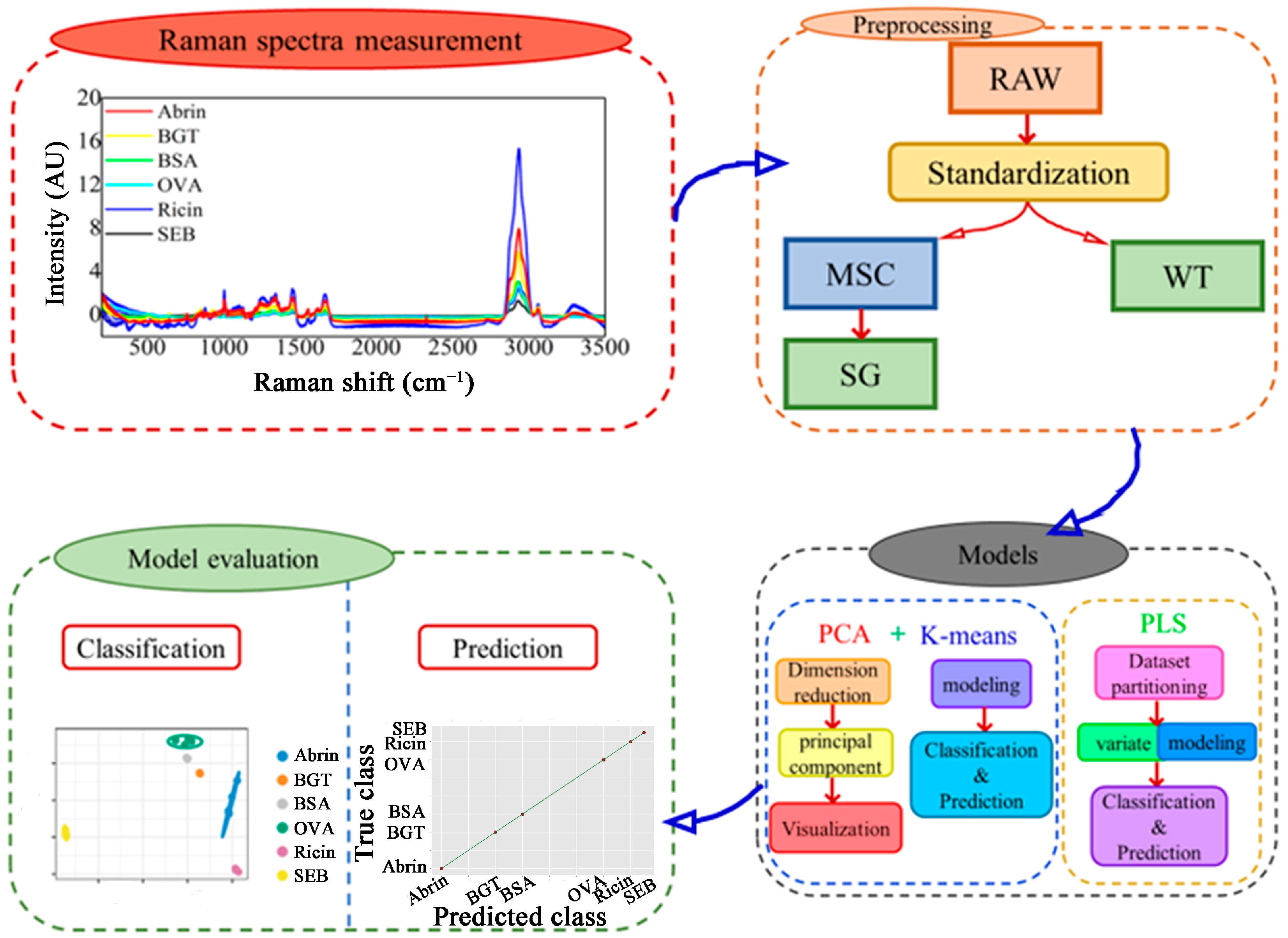
2.1. Raman Spectrum Preprocessing and Peak Assignments
2.1.1. Raman Spectrum Preprocessing
2.1.2. Peak Assignments
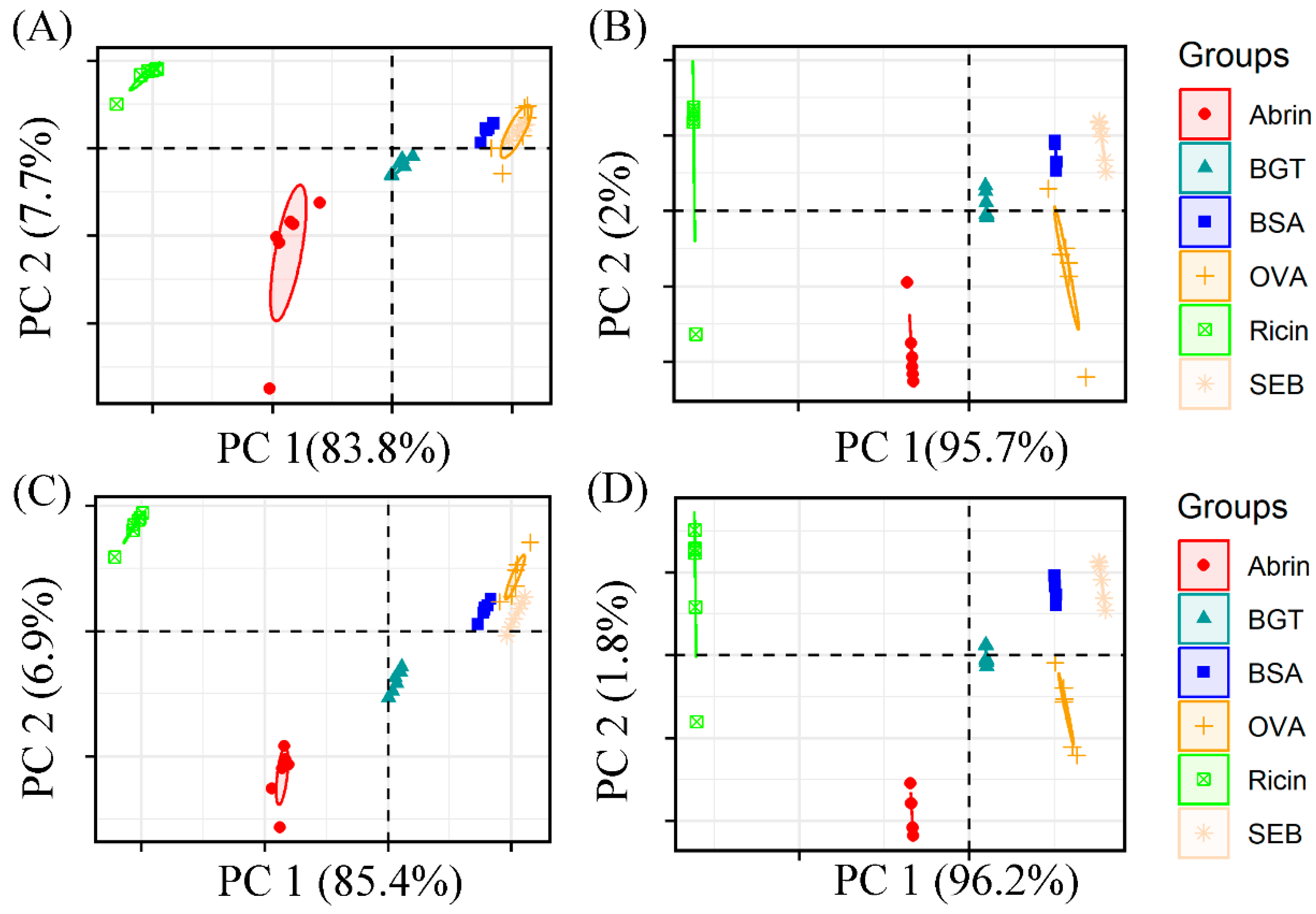
2.2. Evaluation of Preprocessing Methods Using K-Means and PCA
2.3. Evaluation of Two Raman Spectrum Regions Using PLS-DA
2.4. Evaluation of Two Raman Spectrum Regions Using PLSR
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Raman Spectra Acquisition
4.3. Spectra Preprocessing
4.4. Classification and Prediction Methods
4.5. Performance Evaluation of Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdalhamid, B.; McCutchen, E.L.; Bouska, A.C.; Weiwei, Z.; Loeck, B.; Hinrichs, S.H.; Iwen, P.C. Whole genome sequencing to characterize shiga toxin-producing Escherichia coli O26 in a public health setting. J. Infect. Public Health 2019, 12, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, X.; Zhao, M.; Huo, L.; Huang, M.; Li, D.; Zhang, S.; Cheng, X.; Gu, H.; Zhang, C.; et al. The gut-to-brain axis for toxin-induced defensive responses. Cell 2022, 185, 4298–4316. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, K.; Naggar, A.Y.; Heyden, Y.V.; Zhao, L.; Wu, L.; Xue, X. Natural plant toxins in honey: An ignored threat to human health. J. Hazard. Mater. 2022, 424, 127682. [Google Scholar] [CrossRef] [PubMed]
- Ler, S.G.; Lee, F.K.; Gopalakrishnakone, P. Trends in detection of warfare agents: Detection methods for ricin, staphylococcal enterotoxin B and T-2 toxin. J. Chromatogr. A 2006, 1133, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Zhang, Y.; Feng, X.S.; Gao, H.Y. Marine toxins in seafood: Recent updates on sample pretreatment and determination techniques. Food Chem. 2023, 438, 137995. [Google Scholar] [CrossRef]
- Alahi, M.E.E.; Mukhopadhyay, S.C. Detection methodologies for pathogen and toxins: A review. Sensors 2017, 17, 1885. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, B.; Murray, J.; Haverstick, J.; Chen, X.; Tripp, R.A.; Zhao, Y. Rapid and quantitative detection of respiratory viruses using surface-enhanced Raman spectroscopy and machine learning. Biosens. Bioelectron. 2022, 217, 114721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, K.; Wang, X.; Wang, H.; Yang, W.; Liu, J.; Li, D. Surface-enhanced Raman spectroscopy for environmental monitoring using gold clusters anchored on reduced graphene oxide. Sci. Total Environ. 2023, 856, 158879. [Google Scholar] [CrossRef]
- Golparvar, A.; Kim, J.; Boukhayma, A.; Briand, D.; Carrara, S. Highly accurate multimodal monitoring of lactate and urea in sweat by soft epidermal optofluidics with single-band Raman scattering. Sens. Actuators B Chem. 2023, 387, 133814. [Google Scholar] [CrossRef]
- Gogone, I.C.V.P.; Ferreira, G.H.; Gava, D.; Schaefer, R.; de Paula-Lopes, F.F.; Rocha, R.A.; de Barros, F.R.O. Applicability of Raman spectroscopy on porcine parvovirus and porcine circovirus type 2 detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 249, 119336. [Google Scholar] [CrossRef]
- Zheng, J.K.; Zhao, C.Y.; Tian, G.F.; He, L.L. Rapid screening for ricin toxin on letter papers using surface enhanced Raman spectroscopy. Talanta 2017, 162, 552–557. [Google Scholar] [CrossRef]
- Luna, A.S.; da Silva, A.P.; da Silva, C.S.; Lima, I.C.A.; de Gois, J.S. Chemometric methods for classification of clonal varieties of green coffee using Raman spectroscopy and direct sample analysis. J. Food Compos. Anal. 2019, 76, 44–50. [Google Scholar] [CrossRef]
- Hu, J.Q.; Zhang, D.; Zhao, H.T.; Sun, B.; Liang, P.; Ye, J.M.; Yu, Z.; Jin, S.Z. Intelligent spectral algorithm for pigments visualization, classification and identification based on Raman spectra. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 250, 119390. [Google Scholar] [CrossRef] [PubMed]
- You, R.Y.; Wang, H.N.; Wang, C.Y.; Huang, J.L.; Zhu, H.N.; Liu, Y.Z.; Zhang, J.H.; Liu, J.W.; Yu, X.W.; Lu, Y.D. Bacterial cellulose loaded with silver nanoparticles as a flexible, stable and sensitive SERS-active substrate for detection of the shellfish toxin DTX-1. Food Chem. 2023, 427, 136692. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Wasiewska, L.A.; Burgess, C.M.; Duffy, G.; Lovera, P.; O’Riordan, A. Detection of stx2 from Shiga toxin-producing Escherichia coli (STEC) by a Surface Enhanced Raman Spectroscopy (SERS) sensor using recycled silicon chips. Sens. Actuators B Chem. 2022, 373, 132618. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Zhang, J.N.; Gouda, M.; Zhang, C.H.; Lin, L.; Nie, P.C.; Ye, H.B.; Huang, W.; Ye, Y.X.; Zhou, C.Q.; et al. Structure analysis and non-invasive detection of cadmium-phytochelatin2 complexes in plant by deep learning Raman spectrum. J. Hazard. Mater. 2022, 427, 128152. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Wei, H.Y.; Gao, G.; Li, N.L.; Zou, W.; Liu, R.R.; Zhang, M.P.; Meng, X.; Chen, W.W.; Sun, Y.X.; et al. A portable kit for rapid detection of bromadiolone in human blood and urine via surface-enhanced raman scattering coupled with salt-induced liquid-liquid phase separation. Sen. Actuators B Chem. 2023, 374, 132809. [Google Scholar] [CrossRef]
- Zhang, Q.S.; Liu, T.T.; Hao, C.L.; Qu, Y.R.; Niu, J.H.; Wang, Q.; Chen, D. In situ Raman investigation on gas components and explosion risk of thermal runaway emission from lithium-ion battery. J. Energy Storage 2022, 56, 105905. [Google Scholar] [CrossRef]
- Sun, H.R.; Song, C.; Lin, X.M.; Gao, X. Identification of meat species by combined laser-induced breakdown and Raman spectroscopies. Spectrochim. Acta Part B At. Spectrosc. 2022, 194, 106456. [Google Scholar] [CrossRef]
- Olisah, C.C.; Smith, L.; Smith, M. Diabetes mellitus prediction and diagnosis from a data preprocessing and machine learning perspective. Comput. Methods Programs Biomed. 2022, 220, 106773. [Google Scholar] [CrossRef]
- Wei, Y.Y.; Chen, Z.Z.; Zhao, C.; Chen, X.; Yang, R.; He, J.H.; Zhang, C.Y.; Wu, S.T. Deterministic and probabilistic ship pitch prediction using a multi-predictor integration model based on hybrid data preprocessing, reinforcement learning and improved QRNN. Adv. Eng. Inform. 2022, 54, 101806. [Google Scholar] [CrossRef]
- Sun, J.Z.; Xu, X.F.; Feng, S.S.; Zhang, H.Y.; Xu, L.F.; Jiang, H.; Sun, B.B.; Meng, Y.Y.; Chen, W.Z. Rapid identification of salmonella serovars by using Raman spectroscopy and machine learning algorithm. Talanta 2023, 253, 123807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.S.; Hao, H.; Wang, Y.C.; Jiang, Y.; Shi, J.H.; Yu, J.; Cui, X.J.; Li, J.S.; Zhou, S.; Yu, B.L. Optimized adaptive Savitzky-Golay filtering algorithm based on deep learning network for absorption spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 263, 120187. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Peng, Y.K.; Li, Y.Y.; Wang, F. A new scattering correction method of different spectroscopic analysis for assessing complex mixtures. Anal. Chim. Acta 2019, 1087, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Nordon, A.; Roger, J.M. Improved prediction of tablet properties with near-infrared spectroscopy by a fusion of scatter correction techniques. J. Pharm. Biomed. Anal. 2021, 192, 113684. [Google Scholar] [CrossRef] [PubMed]
- Vrazhnov, D.; Mankova, A.; Stupak, E.; Kistenev, Y.; Shkurinov, A.; Cherkasova, O. Discovering Glioma Tissue through Its Biomarkers’ Detection in Blood by Raman Spectroscopy and Machine Learning. Pharmaceutics 2023, 15, 203. [Google Scholar] [CrossRef]
- Luo, R.; Popp, J.; Bocklitz, T. Deep learning for Raman spectroscopy: A review. Analytica 2022, 3, 287–301. [Google Scholar] [CrossRef]
- Flanagan, A.R.; Glavin, F.G. A Comparative Analysis of Data Synthesis Techniques to Improve Classification Accuracy of Raman Spectroscopy Data. J. Chem. Inf. Model. 2023. [Google Scholar] [CrossRef]
- Wang, P.X.; Zhan, K.; Wang, X.Q.; Peng, Y.C.; Liu, S.Y. Comparison of NIR and Raman spectrometries as quantitative methods to monitor polyethylene content in recycled polypropylene. Polym. Test. 2023, 119, 107938. [Google Scholar] [CrossRef]
- Hu, H.Z.; Liu, J.X.; Zhang, X.P.; Fang, M.G. An Effective and Adaptable K-means Algorithm for Big Data Cluster Analysis. Pattern Recognit. 2023, 139, 109404. [Google Scholar] [CrossRef]
- Deka, P.; Saha, U. Introduction of k-means clustering into random cascade model for disaggregation of rainfall from daily to 1-hour resolution with improved preservation of extreme rainfall. J. Hydrol. 2023, 620, 129478. [Google Scholar] [CrossRef]
- Huang, Y.C.; Bais, A. A novel PCA-based calibration algorithm for classification of challenging laser-induced breakdown spectroscopy soil sample data. Spectrochim. Acta B 2022, 193, 106451. [Google Scholar] [CrossRef]
- Xu, J.W.; Xu, J.J.; Tong, Z.Y.; Du, B.; Liu, B.; Mu, X.H.; Guo, T.X.; Yu, S.Q.; Liu, S.; Gao, C.; et al. Performance of feature extraction method for classification and identification of proteins based on three-dimensional fluorescence spectrometry. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 285, 121841. [Google Scholar] [CrossRef] [PubMed]
- Baddini, A.L.Q.; Santos, J.L.V.P.; Tavares, R.R.; Paula, L.S.; Filho, H.D.C.A.; Freitas, R.P. PLS-DA and data fusion of visible Reflectance, XRF and FTIR spectroscopy in the classification of mixed historical pigments. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 265, 120384. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Tahirov, T.H.; Lu, T.H.; Liaw, Y.C.; Chen, Y.L.; Lin, J.Y. Crystal structure of abrin-a at 2.14 Å. J. Mol. Biol. 1995, 250, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Goto, M.; Sakamoto, N.; Higashi, S.; Kawata, R.; Nagatsu, K.; Saito, R. Crystal structure of ricin toxin A chain complexed with a highly potent pterin-based small-molecular inhibitor. J. Enzyme. Inhib. Med. Chem. 2023, 38, 2219038. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.D.; Papageorgiou, A.C.; Titball, R.W.; Miller, J.; White, S.; Lingard, B.; Lee, J.J.; Cavanagh, D.; Kehoe, M.A.; Robinson, J.H.; et al. Structural and functional role of threonine 112 in a superantigen Staphylococcus aureus enterotoxin B. J. Biol. Chem. 2002, 277, 2756–2762. [Google Scholar] [CrossRef]
- Kwong, P.D.; McDonald, N.Q.; Sigler, P.B.; Hendrickson, W.A. Structure of beta 2-bungarotoxin: Potassium channel binding by Kunitz modules and targeted phospholipase action. Structure 1995, 3, 1109–1119. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Shepherd, N.; Crow, P.; Barr, H. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J. Raman Spectrosc. 2002, 33, 564–573. [Google Scholar] [CrossRef]
- Huang, Z.W.; McWilliams, A.; Lui, H.; McLean, D.I.; Lam, S.; Zeng, H.S. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer 2003, 107, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Laska, J.; Widlarz, J. Spectroscopic and structural characterization of low molecular weight fractions of polyaniline. Polymer 2005, 46, 1485–1495. [Google Scholar] [CrossRef]
- Notingher, I.; Green, C.; Dyer, C.; Perkins, E.; Hopkins, N.; Lindsay, C.; Hench, L.L. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. R. Soc. Interface 2004, 1, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Shetty, G.; Kendall, C.; Shepherd, N.; Stone, N.; Barr, H. Raman spectroscopy: Elucidation of biochemical changes in carcinogenesis of oesophagus. Br. J. Cancer 2006, 94, 1460–1464. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, R.J.; Kartha, V.B.; Krishna, C.M.; Solomon, J.G.R.; Ullas, G.; Devi, P.U. Tissue Raman spectroscopy for the study of radiation damage: Brain irradiation of mice. Radiat. Res. 2002, 157, 175–182. [Google Scholar] [CrossRef]
- Yang, H.; Deng, M.; Ga, S.; Chen, S.H.; Kang, L.; Wang, J.H.; Xin, W.W.; Zhang, T.; You, Z.L.; An, Y.; et al. Capillary-driven surface-enhanced Raman scattering (SERS)-based microfluidic chip for abrin detection. Nanoscale Res. Lett. 2014, 9, 138. [Google Scholar] [CrossRef]
- Faolain, E.O.; Hunter, M.B.; Byrne, J.M.; Kelehan, P.; McNamara, M.; Byrne, H.J.; Lyng, F.M. A study examining the effects of tissue processing on human tissue sections using vibrational spectroscopy. Vib. Spectrosc. 2005, 38, 121–127. [Google Scholar] [CrossRef]
- Jia, X.F.; Wang, K.L.; Li, X.Y.; Liu, Z.Z.; Liu, Y.; Xiao, R.; Wang, S.Q. Highly sensitive detection of three protein toxins via SERS-lateral flow immunoassay based on SiO2@ Au nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2022, 41, 102522. [Google Scholar] [CrossRef]
- Chan, J.W.; Taylor, D.S.; Zwerdling, T.; Lane, S.M.; Ihara, K.; Huser, T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 2006, 90, 648–656. [Google Scholar] [CrossRef]
- Hu, Y.G.; Jiang, T.; Shen, A.G.; Li, W.; Wang, X.P.; Hu, J.M. A background elimination method based on wavelet transform for Raman spectra. Chemometr. Intell. Lab. 2007, 85, 94–101. [Google Scholar] [CrossRef]
- Li, X.N.; Cheng, X.; Wu, W.J.; Wang, Q.H.; Tong, Z.Y.; Zhang, X.Q.; Deng, D.H.; Li, Y.H.; Gao, X.B. An improved wavelet de-noising-based back propagation neural network model to forecast the bioaerosol concentration. Aerosol Sci. Technol. 2020, 55, 352–360. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.Q.; Chen, Q.; Zhang, J.; Dunham, M.E.; McWhorter, A.J.; Feng, J.M.; Li, Y.P.; Yao, S.M.; Xu, J. Machine-learning-assisted spontaneous Raman spectroscopy classification and feature extraction for the diagnosis of human laryngeal cancer. Comput. Biol. Med. 2022, 146, 105617. [Google Scholar] [CrossRef] [PubMed]
- Bellou, E.; Gyftokostas, N.; Stefas, D.; Gazeli, O.; Couris, S. Laser-induced breakdown spectroscopy assisted by machine learning for olive oils classification: The effect of the experimental parameters. Spectrochim. Acta Part B At. Spectrosc. 2020, 163, 105746. [Google Scholar] [CrossRef]
- Malakouti, S.M. Improving the prediction of wind speed and power production of SCADA system with ensemble method and 10-fold cross-validation. Case Stud. Chem. Environ. Eng. 2023, 8, 100351. [Google Scholar] [CrossRef]
- Leng, T.; Li, F.; Xiong, L.A.; Xiong, Q.; Zhu, M.T.; Chen, Y. Quantitative detection of binary and ternary adulteration of minced beef meat with pork and duck meat by NIR combined with chemometrics. Food Control 2020, 113, 107203. [Google Scholar] [CrossRef]
- Liu, S.Y.; Wang, S.T.; Hu, C.H.; Kong, D.M.; Yuan, Y.Y. Series fusion of scatter correction techniques coupled with deep convolution neural network as a promising approach for NIR modeling. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 291, 122371. [Google Scholar] [CrossRef]



| Assignment | Wavenumber (cm−1) | This Work (cm−1) | References |
|---|---|---|---|
| Tryptophan/cytosine (C), guanine (G) | 573 | 578 | [40] |
| Ring breathing mode of tyrosine | 853 | 853 | [40,41] |
| Phenylalanine, symmetric ring breathing | 1002–1005 | 1004 | [42,43] |
| C-H in-plane phenylalanine (proteins) | 1033 | 1033 | [44] |
| Tryptophan and phenylalanine ν(C-C6H5) | 1209 | 1209 | [44] |
| Amide III | 1230–1300 | 1231 | [45,46] |
| CH2 bending (proteins) | 1450 | 1450 | [46,47] |
| Amide II | 1480–1575 | 1484–1545 | [48,49] |
| Amide I (proteins), C=O stretching (lipids) | 1655–1680 | 1655–1680 | [50] |
| Methods | Actual Class | Spectral Data | PC Data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| * Abrin | * BGT | * BSA | * OVA | * Ricin | * SEB | * Abrin | * BGT | * BSA | * OVA | * Ricin | * SEB | ||
| MSC | Abrin | 100 | 100 | ||||||||||
| BGT | 100 | 100 | |||||||||||
| BSA | 100 | 100 | |||||||||||
| OVA | 83.3 | 66.7 | |||||||||||
| Ricin | 100 | 83.3 | |||||||||||
| SEB | 100 | 0 | |||||||||||
| MSC-SG | Abrin | 100 | 83.3 | ||||||||||
| BGT | 100 | 100 | |||||||||||
| BSA | 100 | 100 | |||||||||||
| OVA | 100 | 83.3 | |||||||||||
| Ricin | 100 | 83.3 | |||||||||||
| SEB | 100 | 50 | |||||||||||
| Model | RMSEC | RMSECV | Rc | Rcv |
|---|---|---|---|---|
| FS—Raw | 1.567 | 4.271 | 0.959 | 0.636 |
| FS—MSC | 0.938 | 4.256 | 0.985 | 0.639 |
| FS—MSC—SG | 0.192 | 3.488 | 0.999 | 0.776 |
| OS—Raw | 0.897 | 4.602 | 0.987 | 0.555 |
| OS—MSC | 2.087 | 5.087 | 0.926 | 0.392 |
| OS—MSC—SG | 0.212 | 4.087 | 0.999 | 0.674 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Liu, B.; Mu, X.; Xu, J.; Du, B.; Wang, J.; Liu, Z.; Tong, Z. Performance of Classification Models of Toxins Based on Raman Spectroscopy Using Machine Learning Algorithms. Molecules 2024, 29, 197. https://doi.org/10.3390/molecules29010197
Zhang P, Liu B, Mu X, Xu J, Du B, Wang J, Liu Z, Tong Z. Performance of Classification Models of Toxins Based on Raman Spectroscopy Using Machine Learning Algorithms. Molecules. 2024; 29(1):197. https://doi.org/10.3390/molecules29010197
Chicago/Turabian StyleZhang, Pengjie, Bing Liu, Xihui Mu, Jiwei Xu, Bin Du, Jiang Wang, Zhiwei Liu, and Zhaoyang Tong. 2024. "Performance of Classification Models of Toxins Based on Raman Spectroscopy Using Machine Learning Algorithms" Molecules 29, no. 1: 197. https://doi.org/10.3390/molecules29010197
APA StyleZhang, P., Liu, B., Mu, X., Xu, J., Du, B., Wang, J., Liu, Z., & Tong, Z. (2024). Performance of Classification Models of Toxins Based on Raman Spectroscopy Using Machine Learning Algorithms. Molecules, 29(1), 197. https://doi.org/10.3390/molecules29010197






