Salvianolic Acid B Alleviates Liver Injury by Regulating Lactate-Mediated Histone Lactylation in Macrophages
Abstract
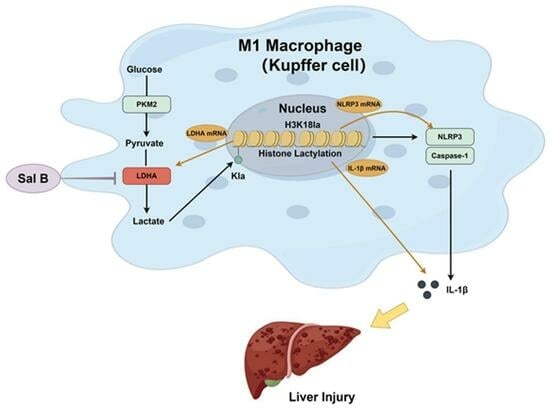
:1. Introduction
2. Results
2.1. Sal B Reduces the Inflammatory Responses and the Polarization of Macrophages Stimulated by LPS
2.2. Sal B Inhibits LPS Induced Glycolysis and NLRP3 in Macrophages
2.3. LDHA Overexpression Impairs the Inhibiting Effects of Sal B on Glycolysis, NLRP3, and M1 Polarization in LPS-Stimlated Macrophages
2.4. Sal B Regulated Histone Lactylation and Binding Ability of H3K18la to LDHA, NLRP3 and IL-1β Genes
2.5. Sal B Exerts Anti-Fibrosis Effects on CCl4-Induced Liver Injury
2.6. Sal B Reduces Lactylation in Liver Tissues in Mice Stimulating CCl4
2.7. Sal B Reduces Mechanisms Related to Histone Lactylation in Kupffer Cells
3. Discussion
4. Materials and Methods
4.1. Chemical and Reagents
4.2. Cell Culture
4.3. Animals
4.4. Western Blot Analysis
4.5. Flow Cytometry
4.6. Histological Analysis
4.7. Kupffer Cell Extraction
4.8. Immunohistochemistry Analysis
4.9. Cell Transfection
4.10. Chromatin Immunoprecipitation (ChIP) -qPCR
4.11. Quantitative Real-Time PCR Analysis (qRT-PCR)
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Chen, Y.H.; Li, L.Z.; Wang, F.; Song, W.; Alolga, R.N.; Zhou, W.; Yu, H.; Huang, F.-Q.; Yin, X. Omics and Transgenic Analyses Reveal That Salvianolic Acid B Exhibits Its Anti-Inflammatory Effects through Inhibiting the Mincle-Syk-Related Pathway in Macrophages. J. Proteome Res. 2021, 20, 3734–3748. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, S.; Chen, P.; Yue, X.; Wang, S.; Gu, Y.; Yuan, Y. Salvianolic Acid B Suppresses Hepatic Stellate Cell Activation and Liver Fibrosis by Inhibiting the Nf-Κb Signaling Pathway Via Mir-6499-3p/Lncrna-Ror. Phytomedicine 2022, 107, 154435. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lao, R.; Lei, J.; Chen, Y.; Zhou, Q.; Wang, T.; Tong, Y. Natural Products for Acetaminophen-Induced Acute Liver Injury: A Review. Molecules 2023, 28, 7901. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Wang, A.; Shi, M.; Chen, X.; Liu, R.; Li, T.; Zhang, C.; Zhang, Z.; Zhu, L.; Ju, Z.; et al. Analysis of Redox Landscapes and Dynamics In Living Cells and in Vivo Using Genetically Encoded Fluorescent Sensors. Nat. Protoc. 2018, 13, 2362–2386. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Fan, G.; Liu, Y.; Liu, L.; Zhang, T.; Liu, P.; Tu, Q.; Zhang, X.; Luo, S.; Yao, L.; et al. The Transcription Factor Klf14 Regulates Macrophage Glycolysis and Immune Function by Inhibiting Hk2 in Sepsis. Cell Mol. Immunol. 2022, 19, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Guo, M.; Huang, W.; Feng, L.; Zhu, J.; Luo, K.; Gao, J.; Zheng, B.; Kong, L.D.; Pang, T.; et al. Annexin A5 Regulates Hepatic Macrophage Polarization Via Directly Targeting Pkm2 and Ameliorates Nash. Redox Biol. 2020, 36, 101634. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.C.; Chen, Y.J.; Wei, Y.H.; Lin, H.A.; Chen, C.C.; Liu, T.F.; Hsieh, Y.L.; Huang, K.Y.; Lin, K.H.; Wang, H.H.; et al. Lactic Acid Fermentation Is Required for Nlrp3 Inflammasome Activation. Front. Immunol. 2021, 12, 630380. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Wang, B.; Chen, T.; Chen, Y.; Ma, W. Ldh-a Negatively Regulates Dmmr in Colorectal Cancer. Cancer Sci. 2021, 112, 3050–3063. [Google Scholar] [CrossRef]
- Lauterbach, M.A.; Hanke, J.E.; Serefidou, M.; Mangan, M.S.J.; Kolbe, C.C.; Hess, T.; Rothe, M.; Kaiser, R.; Hoss, F.; Gehlen, J.; et al. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation Via Atp-Citrate Lyase. Immunity 2019, 51, 997–1011.e7. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic Regulation of Gene Expression by Histone Lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Caslin, H.L.; Abebayehu, D.; Pinette, J.A.; Ryan, J.J. Lactate Is a Metabolic Mediator That Shapes Immune Cell Fate and Function. Front. Physiol. 2021, 12, 688485. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, S.; Lindenthal, L.; Zeitler, L.; Behnke, K.; Schlösser, D.; Strobl, B.; Scheller, J.; El Kasmi, K.C.; Murray, P.J. Lactate and Il6 Define Separable Paths of Inflammatory Metabolic Adaptation. Sci. Adv. 2021, 7, eabg3505. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, F.; Jian, Y.; Wang, X.; Yang, H.; Wang, J.; Su, J.; Lu, X.; Xi, M.; Wen, A.; et al. Salvianolic Acid B Regulates Macrophage Polarization in Ischemic/Reperfused Hearts by Inhibiting Mtorc1-Induced Glycolysis. Eur. J. Pharmacol. 2020, 871, 172916. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Huang, T.; Huang, J.; Quan, J.; Su, G.; Xiong, Z.; Lv, Y.; Li, S.; Lai, X.; Xiang, Y.; et al. The Hdac10 Instructs Macrophage M2 Program Via Deacetylation of Stat3 and Promotes Allergic Airway Inflammation. Theranostics 2023, 13, 3568–3581. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.M.; Tao, X.M.; Shi, Y.N.; Lu, Y.; Mei, J.Y. Salvianolic Acid B Exerts a Protective Effect in Acute Liver Injury by Regulating the Nrf2/Ho-1 Signaling Pathway. Can. J. Physiol. Pharmacol. 2020, 98, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.M.; Li, D.; Zhang, C.; Wen, G.H.; Wu, C.; Xu, Y.Y.; Kan, Y.; Lu, W.P.; Ding, H.Y.; Yang, Y. Salvianolic Acid B Protects against Acute and Chronic Liver Injury by Inhibiting Smad2c/L Phosphorylation. Exp. Ther. Med. 2021, 21, 341. [Google Scholar] [CrossRef]
- Cao, L.; Yin, G.; Du, J.; Jia, R.; Gao, J.; Shao, N.; Li, Q.; Zhu, H.; Zheng, Y.; Nie, Z.; et al. Salvianolic Acid B Regulates Oxidative Stress, Autophagy and Apoptosis against Cyclophosphamide-Induced Hepatic Injury in Nile Tilapia (Oreochromis niloticus). Animals 2023, 13, 341. [Google Scholar] [CrossRef]
- Liu, F.; Li, S.; Chen, P.; Gu, Y.; Wang, S.; Wang, L.; Chen, C.; Wang, R.; Yuan, Y. Salvianolic Acid B Inhibits Hepatic Stellate Cell Activation and Liver Fibrosis by Targeting Pdgfrβ. Int. Immunopharmacol. 2023, 122, 110550. [Google Scholar] [CrossRef]
- Pérez, S.; Rius-Pérez, S. Macrophage Polarization and Reprogramming in Acute Inflammation: A Redox Perspective. Antioxidants 2022, 11, 1394. [Google Scholar] [CrossRef]
- Marrocco, A.; Ortiz, L.A. Role of Metabolic Reprogramming in Pro-Inflammatory Cytokine Secretion from Lps or Silica-Activated Macrophages. Front. Immunol. 2022, 13, 936167. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Gao, S.; Yu, Z.; Zhang, F.; Yao, L.; Xu, M.; Li, J.; Wu, Z.; Huang, Y.; Wang, S. Salvianolic Acid B Inhibits Raw264.7 Cell Polarization towards the M1 Phenotype by Inhibiting Nf-Κb and Akt/Mtor Pathway Activation. Sci. Rep. 2022, 12, 13857. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Ewald, J.; Kaleta, C. Modeling the Energy Metabolism in Immune Cells. Curr. Opin. Biotechnol. 2021, 68, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Soto-Heredero, G.; de Las Heras, M.M.G.; Gabandé-Rodríguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis—A Key Player in the Inflammatory Response. Febs. J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef] [PubMed]
- Pajak, B.; Siwiak, E.; Sołtyka, M.; Priebe, A.; Zieliński, R.; Fokt, I.; Ziemniak, M.; Jaśkiewicz, A.; Borowski, R.; Domoradzki, T.; et al. 2-Deoxy-D-Glucose and Its Analogs: From Diagnostic to Therapeutic Agents. Int. J. Mol. Sci. 2019, 21, 234. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.; Wang, H.; Ni, M.; Wang, Z.; Wang, Z.; Wei, S.; Liu, M.; Wang, P.; Qiu, J.; Zhang, L.; et al. Fstl1 Promotes Liver Fibrosis by Reprogramming Macrophage Function through Modulating the Intracellular Function of Pkm2. Gut 2022, 71, 2539–2550. [Google Scholar] [CrossRef]
- Wei, J.; Wu, J.; Xu, W.; Nie, H.; Zhou, R.; Wang, R.; Liu, Y.; Tang, G.; Wu, J. Salvianolic Acid B Inhibits Glycolysis in Oral Squamous Cell Carcinoma Via Targeting Pi3k/Akt/Hif-1α Signaling Pathway. Cell Death Dis. 2018, 9, 599. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, Q. Uncoupled Pyroptosis and Il-1β Secretion Downstream of Inflammasome Signaling. Front. Immunol. 2023, 14, 1128358. [Google Scholar] [CrossRef]
- Comandatore, A.; Franczak, M.; Smolenski, R.T.; Morelli, L.; Peters, G.J.; Giovannetti, E. Lactate Dehydrogenase and Its Clinical Significance in Pancreatic and Thoracic Cancers. Semin. Cancer Biol. 2022, 86 Pt. 2, 93–100. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.C.; Liu, X.T.; Zhang, L.; Chen, Y.H.; Zhao, Q.; Gao, W.; Liu, B.; Yang, H.; Li, P. Blockage of Citrate Export Prevents Tca Cycle Fragmentation Via Irg1 Inactivation. Cell Rep. 2022, 38, 110391. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Guo, Y.; Shi, X.; Chen, X.; Feng, W.; Wu, L.L.; Zhang, J.; Yu, S.; Wang, Y.; et al. An Overview: The Diversified Role of Mitochondria in Cancer Metabolism. Int. J. Biol. Sci. 2023, 19, 897–915. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Wang, L.; Yang, J.; Chen, J.; Dou, X.; Chen, R.; Ge, Y.; Lin, Y. Ldha as a Regulator of T Cell Fate and Its Mechanisms in Disease. Biomed. Pharmacother. 2023, 158, 114164. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Jiang, Y.; Zhang, M. Crosstalk between Glucose Metabolism, Lactate Production and Immune Response Modulation. Cytokine Growth Factor. Rev. 2022, 68, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Izzo, L.T.; Wellen, K.E. Histone Lactylation Links Metabolism and Gene Regulation. Nature 2019, 574, 492–493. [Google Scholar] [CrossRef]
- Moon, J.S.; Hisata, S.; Park, M.A.; DeNicola, G.M.; Ryter, S.W.; Nakahira, K.; Choi, A.M.K. Mtorc1-Induced Hk1-Dependent Glycolysis Regulates Nlrp3 Inflammasome Activation. Cell Rep. 2015, 12, 102–115. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, W.; Zhou, R. Nlrp3 Inflammasome Activation and Cell Death. Cell Mol. Immunol. 2021, 18, 2114–2127. [Google Scholar] [CrossRef]
- Mridha, A.R.; Wree, A.; Robertson, A.A.B.; Yeh, M.M.; Johnson, C.D.; Van Rooyen, D.M.; Haczeyni, F.; Teoh, N.C.; Savard, C.; Ioannou, G.N.; et al. Nlrp3 Inflammasome Blockade Reduces Liver Inflammation and Fibrosis in Experimental Nash in Mice. J. Hepatol. 2017, 66, 1037–1046. [Google Scholar] [CrossRef]
- Sung, E.; Sim, H.; Cho, Y.C.; Lee, W.; Bae, J.S.; Tan, M.; Lee, S. Global Profiling of Lysine Acetylation and Lactylation in Kupffer Cells. J. Proteome Res. 2023, 22, 3683–3691. [Google Scholar] [CrossRef]
- Tu, H.; Ren, H.; Jiang, J.; Shao, C.; Shi, Y.; Li, P. Dying to Defend: Neutrophil Death Pathways and Their Implications in Immunity. Adv. Sci. 2023, e2306457. [Google Scholar] [CrossRef]
- Niu, Z.; Shi, Q.; Zhang, W.; Shu, Y.; Yang, N.; Chen, B.; Wang, Q.; Zhao, X.; Chen, J.; Cheng, N.; et al. Caspase-1 Cleaves Pparγ for Potentiating the Pro-Tumor Action of Tams. Nat. Commun. 2017, 8, 766. [Google Scholar] [CrossRef]
- Kumar, S.; Duan, Q.; Wu, R.; Harris, E.N.; Su, Q. Pathophysiological Communication between Hepatocytes and Non-Parenchymal Cells in Liver Injury from Nafld to Liver Fibrosis. Adv. Drug Deliv. Rev. 2021, 176, 113869. [Google Scholar] [CrossRef] [PubMed]
- Blériot, C.; Barreby, E.; Dunsmore, G.; Ballaire, R.; Chakarov, S.; Ficht, X.; De Simone, G.; Andreata, F.; Fumagalli, V.; Guo, W.; et al. A Subset of Kupffer Cells Regulates Metabolism through the Expression of Cd36. Immunity 2021, 54, 2101–2116.e6. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Zhou, Y.X.; Li, H.; Zhang, R.Z.; He, C.; Yang, X. The Mettl3/Malat1/Ptbp1/Usp8/Tak1 Axis Promotes Pyroptosis and M1 Polarization of Macrophages and Contributes to Liver Fibrosis. Cell Death Discov. 2021, 7, 368. [Google Scholar] [CrossRef]
- Li, G.; Luo, R.; Zhang, W.; He, S.; Wang, B.; Liang, H.; Song, Y.; Ke, W.; Shi, Y.; Feng, X.; et al. M6a Hypomethylation of Dnmt3b Regulated by Alkbh5 Promotes Intervertebral Disc Degeneration Via E4f1 Deficiency. Clin. Transl. Med. 2022, 12, e765. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, N.; Wu, Y.; Wang, X.; Sun, J.; Liu, L.; Zhang, F.; Gong, Y.; Zhang, Y.; Li, X.; et al. Hypoxia Regulates Fibrosis-Related Genes Via Histone Lactylation in the Placentas of Patients with Preeclampsia. J. Hypertens. 2022, 40, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, W.; Wang, X.; Mang, G.; Chen, J.; Yan, X.; Tong, Z.; Yang, Q.; Wang, M.; Chen, L.; et al. Histone Lactylation Boosts Reparative Gene Activation Post-Myocardial Infarction. Circ. Res. 2022, 131, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Lin, J.; Chen, X.; Zhou, X.; Zhang, Y.; Fan, M.; Xiang, J.; He, N.; Hu, Z.; Wang, F. Circxrn2 Suppresses Tumor Progression Driven by Histone Lactylation through Activating the Hippo Pathway in Human Bladder Cancer. Mol. Cancer 2023, 22, 151. [Google Scholar] [CrossRef]
- Wang, P.; Xie, D.; Xiao, T.; Cheng, C.; Wang, D.; Sun, J.; Wu, M.; Yang, Y.; Zhang, A.; Liu, Q. H3k18 Lactylation Promotes the Progression of Arsenite-Related Idiopathic Pulmonary Fibrosis Via Ythdf1/M6a/Nrep. J. Hazard. Mater. 2024, 461, 132582. [Google Scholar] [CrossRef]
- Fan, M.; Yang, K.; Wang, X.; Chen, L.; Gill, P.S.; Ha, T.; Liu, L.; Lewis, N.H.; Williams, D.L.; Li, C. Lactate Promotes Endothelial-to-Mesenchymal Transition Via Snail1 Lactylation after Myocardial Infarction. Sci. Adv. 2023, 9, eadc9465. [Google Scholar] [CrossRef]
- Gao, R.; Li, Y.; Xu, Z.; Zhang, F.; Xu, J.; Hu, Y.; Yin, J.; Yang, K.; Sun, L.; Wang, Q.; et al. Mitochondrial Pyruvate Carrier 1 Regulates Fatty Acid Synthase Lactylation and Mediates Treatment of Nonalcoholic Fatty Liver Disease. Hepatology 2023, 78, 1800–1815. [Google Scholar] [CrossRef]
- Cui, H.; Xie, N.; Banerjee, S.; Ge, J.; Jiang, D.; Dey, T.; Matthews, Q.L.; Liu, R.M.; Liu, G. Lung Myofibroblasts Promote Macrophage Profibrotic Activity through Lactate-Induced Histone Lactylation. Am. J. Respir. Cell Mol. Biol. 2021, 64, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Rho, H.; Terry, A.R.; Chronis, C.; Hay, N. Hexokinase 2-Mediated Gene Expression Via Histone Lactylation Is Required for Hepatic Stellate Cell Activation and Liver Fibrosis. Cell Metab. 2023, 35, 1406–1423.e8. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Li, Y.; Zhou, F.; Guo, J.; Wang, T.; Shi, Y.; Yang, Y.; Lu, J.; Pei, G. Administration of a Mixture of Triterpenoids from Yeyachun and Phenolic Acids from Danshen Ameliorates Carbon Tetrachloride-Induced Liver Fibrosis in Mice by the Regulation of Intestinal Flora. J. Pharmacol. Sci. 2020, 143, 165–175. [Google Scholar] [CrossRef] [PubMed]







| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| Arg1-PTM:92bp | Forward | AAGCTGTGGCCTCAGAACAT |
| Reverse | GGTAACCGCTGTGAAAGGAT | |
| Arg1-HRE-2kb:85bp | Forward | TGTCTCTCCCAGTTTCCCCA |
| Reverse | AGCAACTTGGCATCTGATGGA | |
| gene desert:79bp | Forward | CTGCCAGGGTTGTAGAGAGG |
| Reverse | GCCAGATCATATTGGCTTGG |
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| Mus GAPDH | Forward | ATGGGTGTGAACCACGAGA |
| Reverse | CAGGGATGATGTTCTGGGCA | |
| Mus LDHA | Forward | GTAACTGCGAACTCCAAGCT |
| Reverse | CAAGCCACGTAGGTCAAGAT | |
| Mus NLRP3 | Forward | CCATCAATGCTGCTTCGACA |
| Reverse | GAGCTCAGAACCAATGCGAG | |
| Mus IL-1β | Forward | TCAGGCAGGCAGTATCACTC |
| Reverse | AGCTCATATGGGTCCGACAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Yang, Z.; Li, L.; Yan, Q.; Hu, Y.; Zhou, F.; Tan, Y.; Pei, G. Salvianolic Acid B Alleviates Liver Injury by Regulating Lactate-Mediated Histone Lactylation in Macrophages. Molecules 2024, 29, 236. https://doi.org/10.3390/molecules29010236
Hu S, Yang Z, Li L, Yan Q, Hu Y, Zhou F, Tan Y, Pei G. Salvianolic Acid B Alleviates Liver Injury by Regulating Lactate-Mediated Histone Lactylation in Macrophages. Molecules. 2024; 29(1):236. https://doi.org/10.3390/molecules29010236
Chicago/Turabian StyleHu, Shian, Zehua Yang, Ling Li, Qinwen Yan, Yutong Hu, Feng Zhou, Yang Tan, and Gang Pei. 2024. "Salvianolic Acid B Alleviates Liver Injury by Regulating Lactate-Mediated Histone Lactylation in Macrophages" Molecules 29, no. 1: 236. https://doi.org/10.3390/molecules29010236
APA StyleHu, S., Yang, Z., Li, L., Yan, Q., Hu, Y., Zhou, F., Tan, Y., & Pei, G. (2024). Salvianolic Acid B Alleviates Liver Injury by Regulating Lactate-Mediated Histone Lactylation in Macrophages. Molecules, 29(1), 236. https://doi.org/10.3390/molecules29010236