Computational Predictive and Electrochemical Detection of Metabolites (CP-EDM) of Piperine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Electrochemical Behavior of Piperine: Cyclic Voltammetry Studies
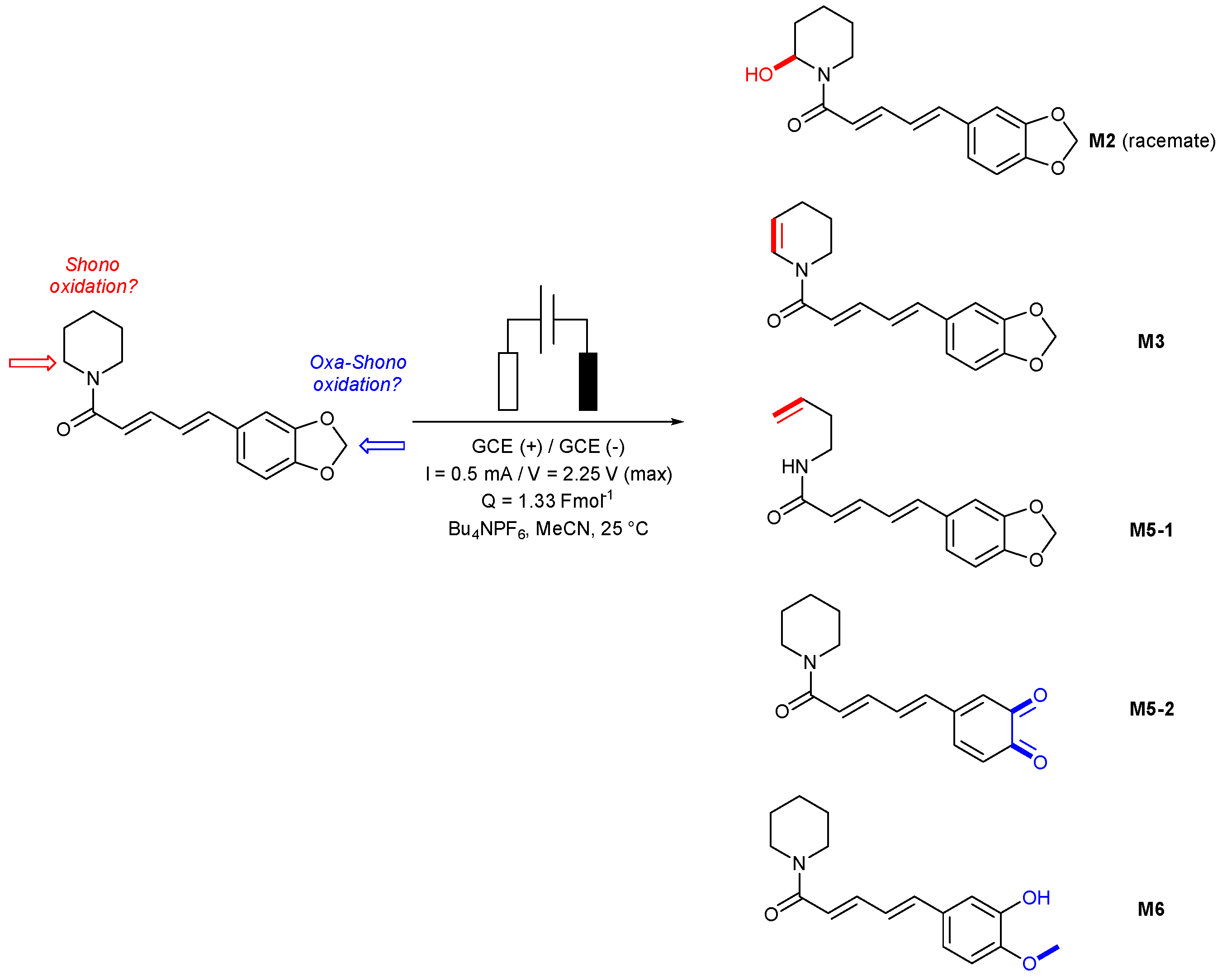
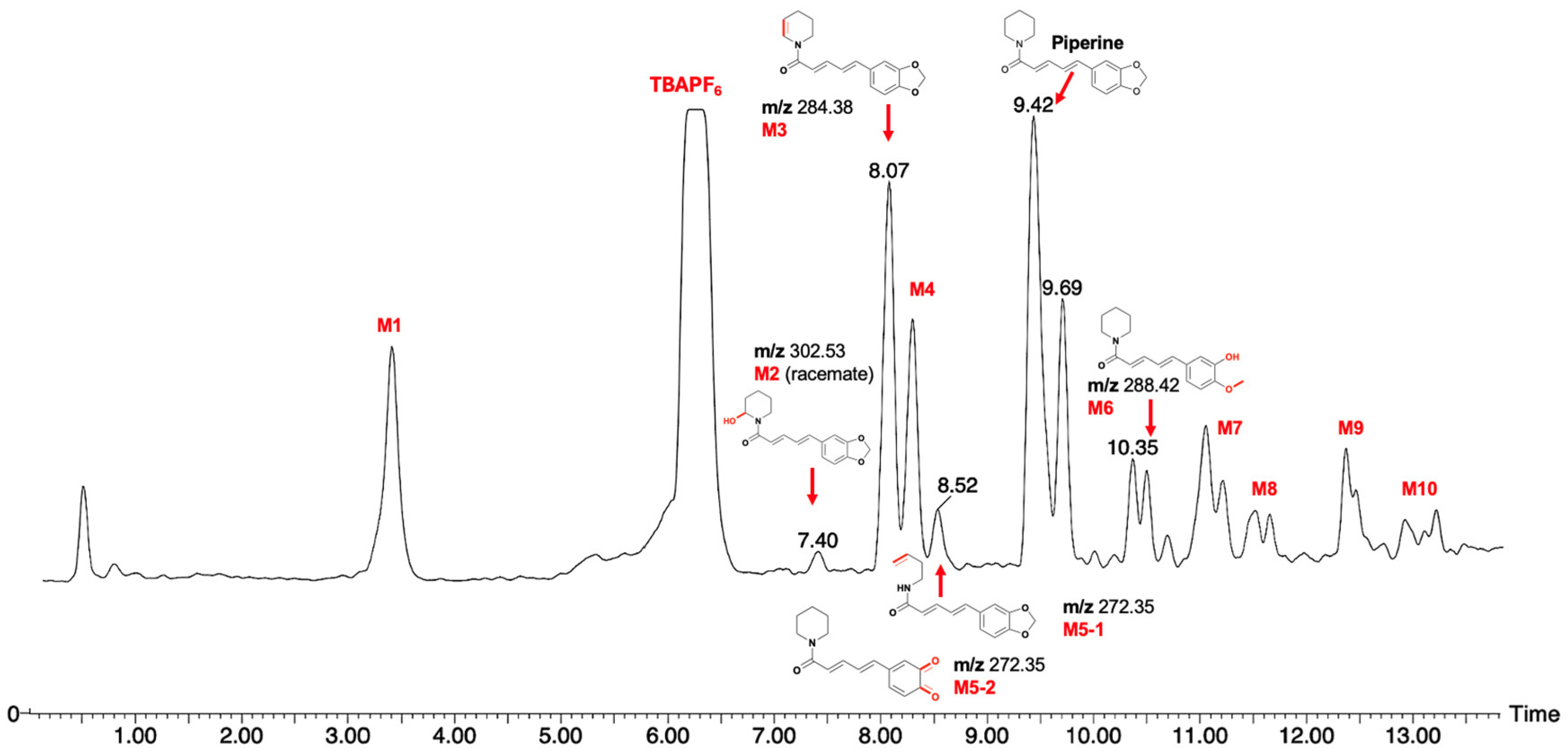
2.2. A Comparison of Electrosynthesis and Hepatocyte Incubation Metabolites
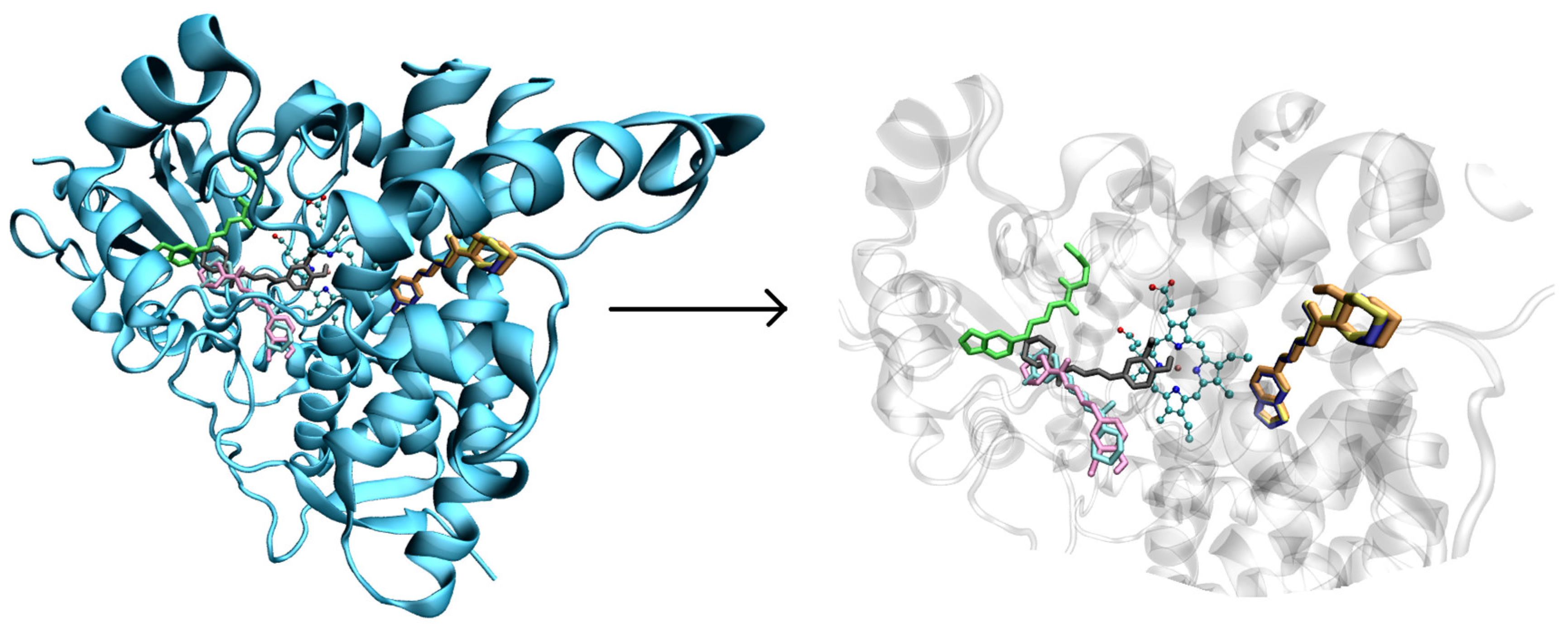
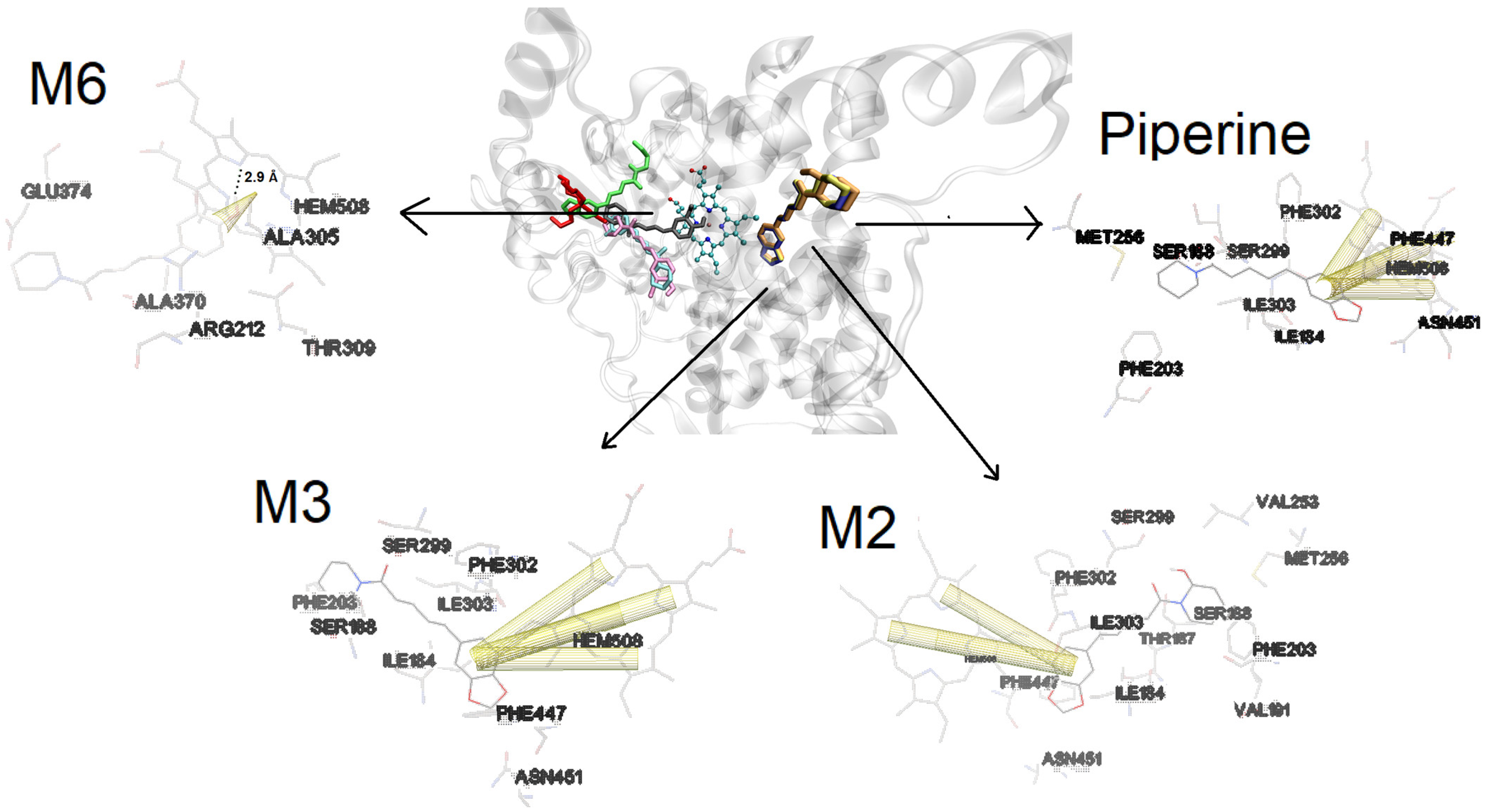
2.3. Molecular Docking Approaches in Understanding Piperine Metabolism
3. Materials and Methods
3.1. Sample Preparation
3.2. Electrosynthesis of Piperine
3.3. Purification and Analysis of Piperine Metabolites
3.4. Cyclic Voltammetry Procedure
3.5. Recrystallization of TBAPF6
3.6. LCMS Analysis of Piperine Metabolites
3.7. Docking Procedures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schadt, S.; Bister, B.; Chowdhury, S.K.; Funk, C.; Hop, C.E.C.A.; Humphreys, W.G.; Igarashi, F.; Alexander, D.J.; Kagan, M.; Khojasteh, S.C.; et al. A Decade in the MIST: Learnings from Investigations of Drug Metabolites in Drug Development under the “Metabolites in Safety Testing” Regulatory Guidance. Drug Metab. Dispos. 2018, 46, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Nikzad, N.; Rafiee, M. Electrochemical Study of Drug Metabolism. Curr. Opin. Electrochem. 2024, 44, 101446. [Google Scholar] [CrossRef]
- Madsen, K.G.; Grönberg, G.; Skonberg, C.; Jurva, U.; Hansen, S.H.; Olsen, J. Electrochemical Oxidation of Troglitazone: Identification and Characterization of the Major Reactive Metabolite in Liver Microsomes. Chem. Res. Toxicol. 2008, 21, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Asra, R.; Jones, A.M. Green Electrosynthesis of Drug Metabolites. Toxicol. Res. 2023, 12, 150–177. [Google Scholar] [CrossRef] [PubMed]
- Kuzikov, A.V.; Masamrekh, R.A.; Filippova, T.A.; Shumyantseva, V.V. Electrochemical Analysis of Metabolites as a Method for Cytochromes P450 Activity Determination. Biomed. Chem. Res. Methods 2022, 5, e00176. [Google Scholar] [CrossRef]
- Jurva, U.; Weidolf, L. Electrochemical Generation of Drug Metabolites with Applications in Drug Discovery and Development. TrAC Trends Anal. Chem. 2015, 70, 92–99. [Google Scholar] [CrossRef]
- Madsen, K.G.; Olsen, J.; Skonberg, C.; Hansen, S.H.; Jurva, U. Development and Evaluation of an Electrochemical Method for Studying Reactive Phase-I Metabolites: Correlation to In Vitro Drug Metabolism. Chem. Res. Toxicol. 2007, 20, 821–831. [Google Scholar] [CrossRef]
- Khera, S.; Hu, N. Generation of Statin Drug Metabolites through Electrochemical and Enzymatic Oxidations. Anal. Bioanal. Chem. 2013, 405, 6009–6018. [Google Scholar] [CrossRef]
- Yao, H.; Sherer, E.C.; Lu, M.; Small, J.; Martin, G.E.; Lam, Y.H.; Chen, Q.; Helmy, R.; Liu, Y.; Chen, H. One-Step Regio- and Stereoselective Electrochemical Synthesis of Orexin Receptor Antagonist Oxidative Metabolites. J. Org. Chem. 2022, 87, 15011. [Google Scholar] [CrossRef]
- Walgama, C.; Nerimetla, R.; Materer, N.F.; Schildkraut, D.; Elman, J.F.; Krishnan, S. A Simple Construction of Electrochemical Liver Microsomal Bioreactor for Rapid Drug Metabolism and Inhibition Assays. Anal. Chem. 2015, 87, 4712–4718. [Google Scholar] [CrossRef]
- Potęga, A.; Garwolińska, D.; Nowicka, A.M.; Fau, M.; Kot-Wasik, A.; Mazerska, Z. Phase I and Phase II Metabolism Simulation of Antitumor-Active 2-Hydroxyacridinone with Electrochemistry Coupled on-Line with Mass Spectrometry. Xenobiotica 2019, 49, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, A.; Julian Wesenberg, L.; Peez, N.; Waldvogel, S.R.; Hoffmann, T.; Wesenberg, L.J.; Peez, N.; Waldvogel, S.R.; Hoffmann, T. Charged Tags for the Identification of Oxidative Drug Metabolites Based on Electrochemistry and Mass Spectrometry. ChemistryOpen 2020, 9, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Stalder, R.; Roth, G.P. Preparative Microfluidic Electrosynthesis of Drug Metabolites. ACS Med. Chem. Lett. 2013, 4, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tang, W. Drug Metabolism in Drug Discovery and Development. Acta Pharm. Sin. B 2018, 8, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gong, C.; Zhu, J.; Liu, G.; Tang, Y.; Li, W. Prediction of Sites of Metabolism of CYP3A4 Substrates Utilizing Docking-Derived Geometric Features. J. Chem. Inf. Model. 2023, 63, 4158–4169. [Google Scholar] [CrossRef] [PubMed]
- Lokwani, D.K.; Sarkate, A.P.; Karnik, K.S.; Nikalje, A.P.G.; Seijas, J.A. Structure-Based Site of Metabolism (SOM) Prediction of Ligand for CYP3A4 Enzyme: Comparison of Glide XP and Induced Fit Docking (IFD). Molecules 2020, 25, 1622. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.C.; Mckinnon, R.A.; Miners, J.O. Computational Prediction of the Site(s) of Metabolism and Binding Modes of Protein Kinase Inhibitors Metabolized by CYP3A4 s. Drug Metab. Dispos. 2019, 47, 616–631. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; Le Duff, C.S.; Tang, B.; Cornelio, M.L.; Jones, A.M. Synthesis and Spectroscopic Analysis of Piperine- and Piperlongumine-Inspired Natural Product Scaffolds and Their Molecular Docking with IL-1β and NF-ΚB Proteins. Molecules 2020, 25, 2841. [Google Scholar] [CrossRef]
- Povinelli, A.P.R.; Zazeri, G.; Jones, A.M.; Cornélio, M.L. A Computational-Experimental Investigation of the Molecular Mechanism of Interleukin-6-Piperine Interaction. Int. J. Mol. Sci. 2022, 23, 7994. [Google Scholar] [CrossRef]
- Rito, D.S.; Vieira, E.F.T.; de Menezes, I.C.; Lameira, O.A.; Poltronieri, M.C.; de Lemos, O.F.; Rodrigues, S.D.M. Morphological Characterization of Native Piperacea Conserved in Vegetation House. Res. Soc. Dev. 2021, 10, e33410615686. [Google Scholar] [CrossRef]
- Koul, S.; Koul, J.L.; Taneja, S.C.; Dhar, K.L.; Jamwal, D.S.; Singh, K.; Reen, R.K.; Singh, J. Structure–Activity Relationship of Piperine and Its Synthetic Analogues for Their Inhibitory Potentials of Rat Hepatic Microsomal Constitutive and Inducible Cytochrome P450 Activities. Bioorganic Med. Chem. 2000, 8, 251–268. [Google Scholar] [CrossRef]
- Suresh, D.; Srinivasan, K. Tissue Distribution & Elimination of Capsaicin, Piperine & Curcumin Following Oral Intake in Rats. Indian J. Med. Res. 2010, 131, 682–691. [Google Scholar] [PubMed]
- Pradeepa, B.R.; Vijayakumar, T.M.; Manikandan, K.; Kammala, A.K. Cytochrome P450-Mediated Alterations in Clinical Pharmacokinetic Parameters of Conventional Drugs Coadministered with Piperine: A Systematic Review and Meta-Analysis. J. Herb. Med. 2023, 41, 100713. [Google Scholar] [CrossRef]
- Bhardwaj, R.K.; Glaeser, H.; Becquemont, L.; Klotz, U.; Gupta, S.K.; Fromm, M.F. Piperine, a Major Constituent of Black Pepper, Inhibits Human P-Glycoprotein and CYP3A4. J. Pharmacol. Exp. Ther. 2002, 302, 645–650. [Google Scholar] [CrossRef] [PubMed]
- New Provided No Predicted Metabolic Site on Piperine. Available online: https://Biotransformer.ca/ (accessed on 9 March 2024).
- Azam, S.; Park, J.Y.; Kim, I.S.; Choi, D.K. Piperine and Its Metabolite’s Pharmacology in Neurodegenerative and Neurological Diseases. Biomedicines 2022, 10, 154. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Wang, Z.; Wen, M.; Tang, J. Identification of the Metabolites of Piperine via Hepatocyte Incubation and Liquid Chromatography Combined with Diode-Array Detection and High-Resolution Mass Spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8947. [Google Scholar] [CrossRef]
- Praneetha, P.; Balhara, A.; Ladumor, M.K.; Singh, D.K.; Patil, A.; Preethi, J.; Pokharkar, S.; Deshpande, A.Y.; Giri, S.; Singh, S. Characterization of Stable and Reactive Metabolites of Piperine Formed on Incubation with Human Liver Microsomes. J. Mass Spectrom. 2019, 54, 738–749. [Google Scholar] [CrossRef]
- Shang, Z.; Cai, W.; Cao, Y.; Wang, F.; Wang, Z.; Lu, J.; Zhang, J. An Integrated Strategy for Rapid Discovery and Identification of the Sequential Piperine Metabolites in Rats Using Ultra High-Performance Liquid Chromatography/High Resolution Mass Spectrometery. J. Pharm. Biomed. Anal. 2017, 146, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Asra, R.; Malmakova, A.E.; Jones, A.M. Electrochemical Synthesis of the in Human S-Oxide Metabolites of Phenothiazine Containing Anti-Psychotic Medications. ChemRxiv 2024. [Google Scholar] [CrossRef]
- Carp, O.E.; Moraru, A.; Pinteala, M.; Arvinte, A. Electrochemical Behaviour of Piperine. Comparison with Control Antioxidants. Food Chem. 2021, 339, 128110. [Google Scholar] [CrossRef]
- Fuchigami, H.; Bal, M.K.; Brownson, D.A.C.; Banks, C.E.; Jones, A.M. Voltammetric Behaviour of Drug Molecules as a Predictor of Metabolic Liabilities. Sci. Pharm. 2020, 88, 46. [Google Scholar] [CrossRef]
- Aljabali, A.A.A.; Barclay, J.E.; Butt, J.N.; Lomonossoff, G.P.; Evans, D.J. Redox-Active Ferrocene-Modified Cowpea Mosaic Virus Nanoparticles. Dalton Trans. 2010, 39, 7569. [Google Scholar] [CrossRef] [PubMed]
- Teli, A.M.; Bhat, T.S.; Beknalkar, S.A.; Mane, S.M.; Chaudhary, L.S.; Patil, D.S.; Pawar, S.A.; Efstathiadis, H.; Cheol Shin, J. Bismuth Manganese Oxide Based Electrodes for Asymmetric Coin Cell Supercapacitor. Chem. Eng. J. 2022, 430, 133138. [Google Scholar] [CrossRef]
- Bal, M.K.; Banks, C.E.; Jones, A.M. Metabolism Mimicry: An Electrosynthetic Method for the Selective Deethylation of Tertiary Benzamides. ChemElectroChem 2019, 6, 4284–4291. [Google Scholar] [CrossRef]
- Jones, A.M.; Banks, C.E. The Shono-Type Electroorganic Oxidation of Unfunctionalised Amides. Carbon-Carbon Bond Formation via Electrogenerated N-Acyliminium Ions. Beilstein J. Org. Chem. 2014, 10, 3056–3072. [Google Scholar] [CrossRef]
- Barone, M.R.; Jones, A.M. Selective C-H Bond Electro-Oxidation of Benzylic Acetates and Alcohols to Benzaldehydes. Org. Biomol. Chem. 2017, 15, 10010–10015. [Google Scholar] [CrossRef] [PubMed]
- Zazeri, G.; Povinelli, A.P.R.; Pavan, N.M.; de Carvalho, D.R.; Cardoso, C.L.; Ximenes, V.F. Experimental Studies and Computational Modeling on Cytochrome c Reduction by Quercetin: The Role of Oxidability and Binding Affinity. J. Mol. Struct. 2021, 1244, 130995. [Google Scholar] [CrossRef]
- Damghani, T.; Sedghamiz, T.; Sharifi, S.; Pirhadi, S. Critical C-Met-Inhibitor Interactions Resolved from Molecular Dynamics Simulations of Different c-Met Complexes. J. Mol. Struct. 2020, 1203, 127456. [Google Scholar] [CrossRef]
- Sharma, N.; Prosser, O.; Kumar, P.; Tuplin, A.; Giri, R. Small Molecule Inhibitors Possibly Targeting the Rearrangement of Zika Virus Envelope Protein. Antivir. Res. 2020, 182, 104876. [Google Scholar] [CrossRef]
- Iwaloye, O.; Elekofehinti, O.O.; Momoh, A.I.; Babatomiwa, K.; Ariyo, E.O. In Silico Molecular Studies of Natural Compounds as Possible Anti-Alzheimer’s Agents: Ligand-Based Design. Netw. Model. Anal. Health Inform. Bioinform. 2020, 9, 54. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; Lima, M.d.F.; Cornélio, M.L. Experimental Approaches and Computational Modeling of Rat Serum Albumin and Its Interaction with Piperine. Int. J. Mol. Sci. 2019, 20, 2856. [Google Scholar] [CrossRef] [PubMed]

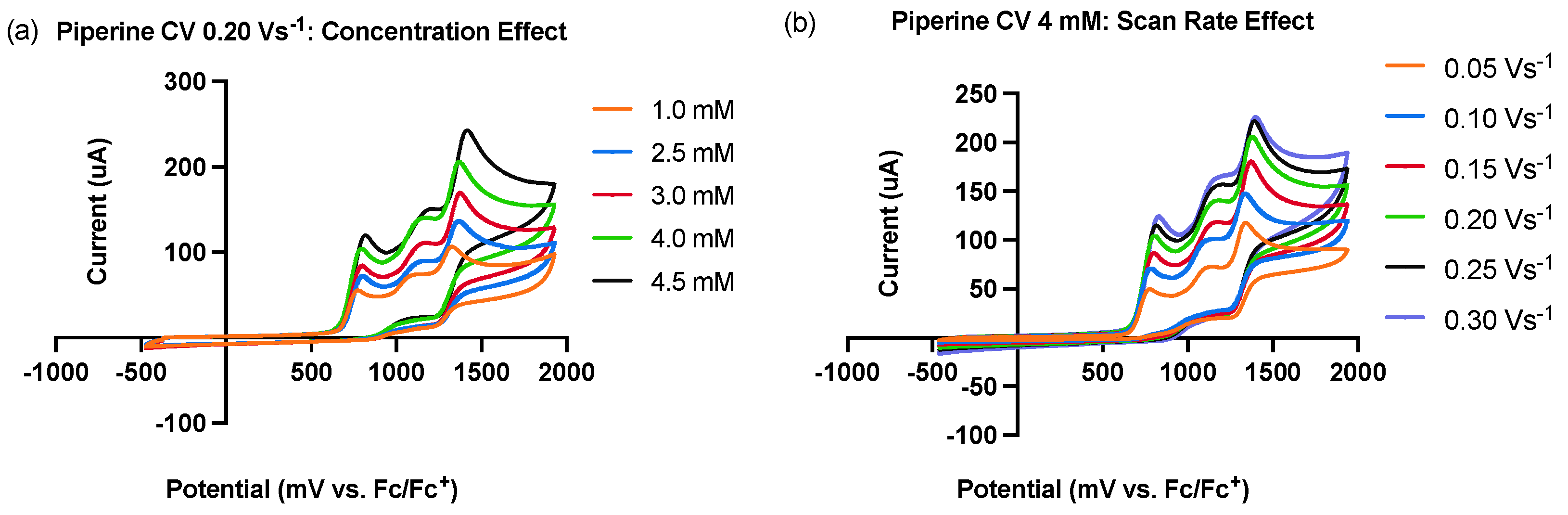

| Scan Rate (Vs−1) | Anodic Peak Potential Ep1 (mV) | Anodic Peak Current Ip1 (µA) | Square Root of Scan Rate v1/2 (V/s)1/2 | Diffusion Coefficient (10−6 cm2 s−1) |
|---|---|---|---|---|
| 0.05 | 783 | 49 | 0.224 | 1.0312 |
| 0.10 | 791 | 71 | 0.316 | 1.0825 |
| 0.15 | 811 | 86 | 0.387 | 1.0588 |
| 0.20 | 812 | 104 | 0.447 | 1.1613 |
| 0.25 | 825 | 114 | 0.500 | 1.1163 |
| 0.30 | 832 | 124 | 0.548 | 1.1006 |
| No | Data Plot | Linear Regression |
|---|---|---|
| 1 | Scan rate (mVs−1) vs. peak current (Ip1) | Y = 0.2983 x + 39.13 R2 = 0.9792 |
| 2 | Square root of scan rate v1/2 (V/s)1/2 vs. peak current (Ip1) (µA) | Y = 234.3 x − 3.248 R2 = 0.9975 |
| 3 | Log of scan rate (mVs−1) vs. Log of peak potential (Ep1) mV | Y = 0.03439 x + 2.833 R2 = 0.9401 |
| 4 | Log of scan rate (mVs−1) vs. Log of peak current (Ip1) (µA) | Y = 0.5230 x + 0.8030 R2 = 0.9983 |
| 5 | Scan rate (mVs−1) vs. peak potential (Ep1) mV | Y = 0.1989 x + 774.2 R2 = 0.9983 |
| 6 | Concentration vs. peak current (Ip1) (µA) | Y = 18.13 x + 32.80 R2 = 0.9585 |
| 7 | Concentration vs. peak potential (Ep1) mV | Y = 11.07 x + 767.8 R2 = 0.7492 |
| Predicted Oxidative Piperine Metabolites | Electrochemical Azam et al. [26] | Hepatocytes Li et al. [27] | This Work |
|---|---|---|---|
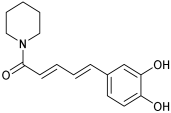 | C12 | M12 | n.d. |
 | C4 | n.d. | n.d. |
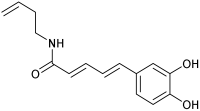 | C8 | M8 | n.d. |
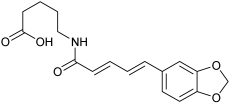 | n.d. | M14 | n.d. |
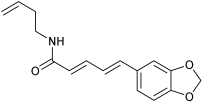 | n.d. | M19 | M5-1 |
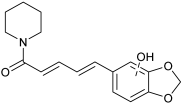 | n.d. | M18 | n.d. |
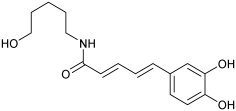 | n.d. | M3 | n.d. |
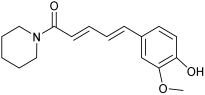 | C17 | n.d. | M6 |
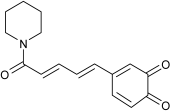 | GSH-adduct Detected via ortho-quinone | n.d. | M5-2 |
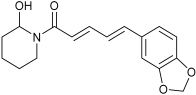 | n.d. | n.d. | M2 |
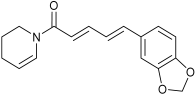 | n.d. | n.d. | M3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asra, R.; Povinelli, A.P.R.; Zazeri, G.; Jones, A.M. Computational Predictive and Electrochemical Detection of Metabolites (CP-EDM) of Piperine. Molecules 2024, 29, 2406. https://doi.org/10.3390/molecules29102406
Asra R, Povinelli APR, Zazeri G, Jones AM. Computational Predictive and Electrochemical Detection of Metabolites (CP-EDM) of Piperine. Molecules. 2024; 29(10):2406. https://doi.org/10.3390/molecules29102406
Chicago/Turabian StyleAsra, Ridho, Ana P. R. Povinelli, Gabriel Zazeri, and Alan M. Jones. 2024. "Computational Predictive and Electrochemical Detection of Metabolites (CP-EDM) of Piperine" Molecules 29, no. 10: 2406. https://doi.org/10.3390/molecules29102406
APA StyleAsra, R., Povinelli, A. P. R., Zazeri, G., & Jones, A. M. (2024). Computational Predictive and Electrochemical Detection of Metabolites (CP-EDM) of Piperine. Molecules, 29(10), 2406. https://doi.org/10.3390/molecules29102406








