Advances in Nucleic Acid Assays for Infectious Disease: The Role of Microfluidic Technology
Abstract
:1. Introduction
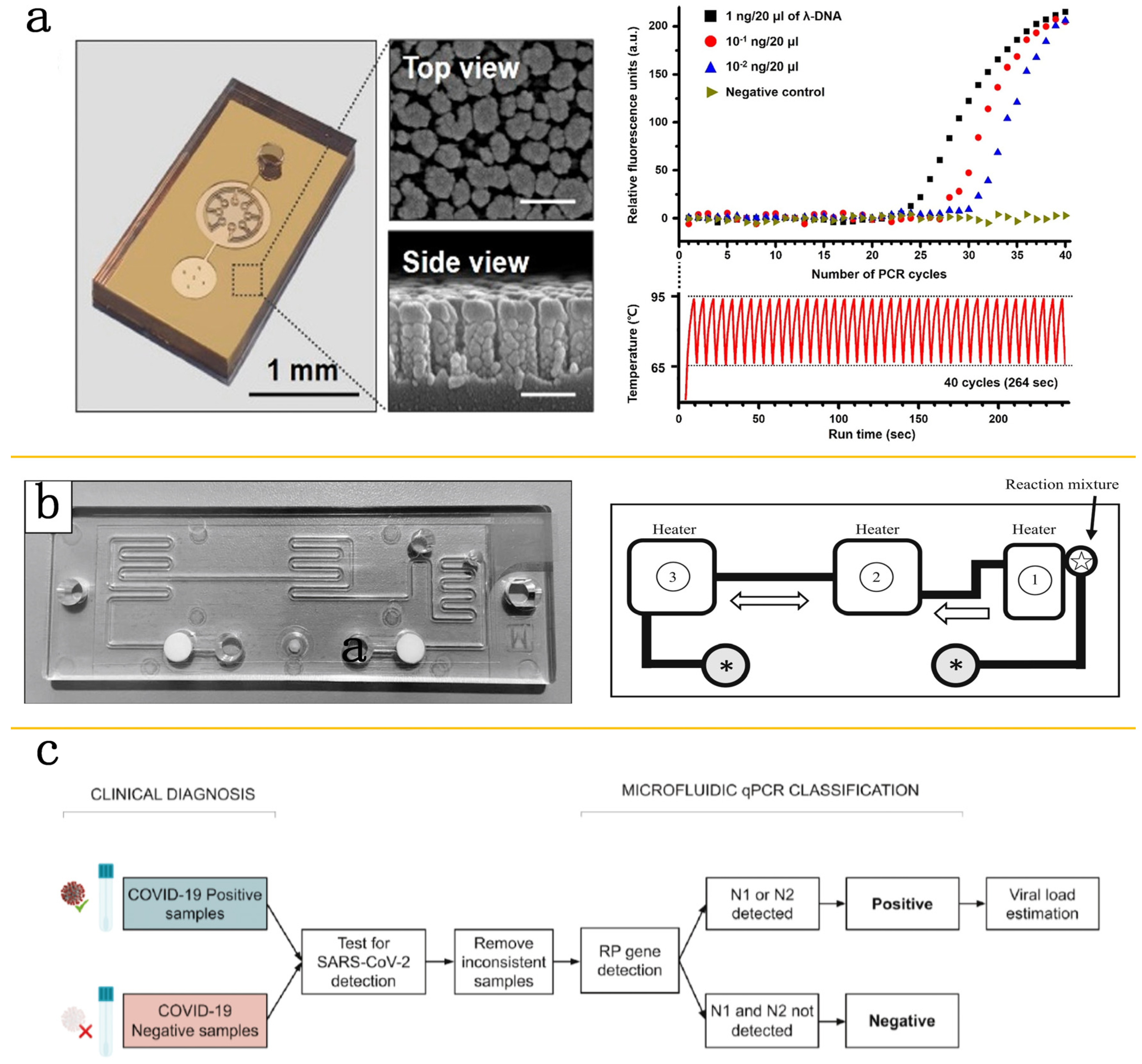
2. The Application of PCR Technology in Microfluidic Platforms
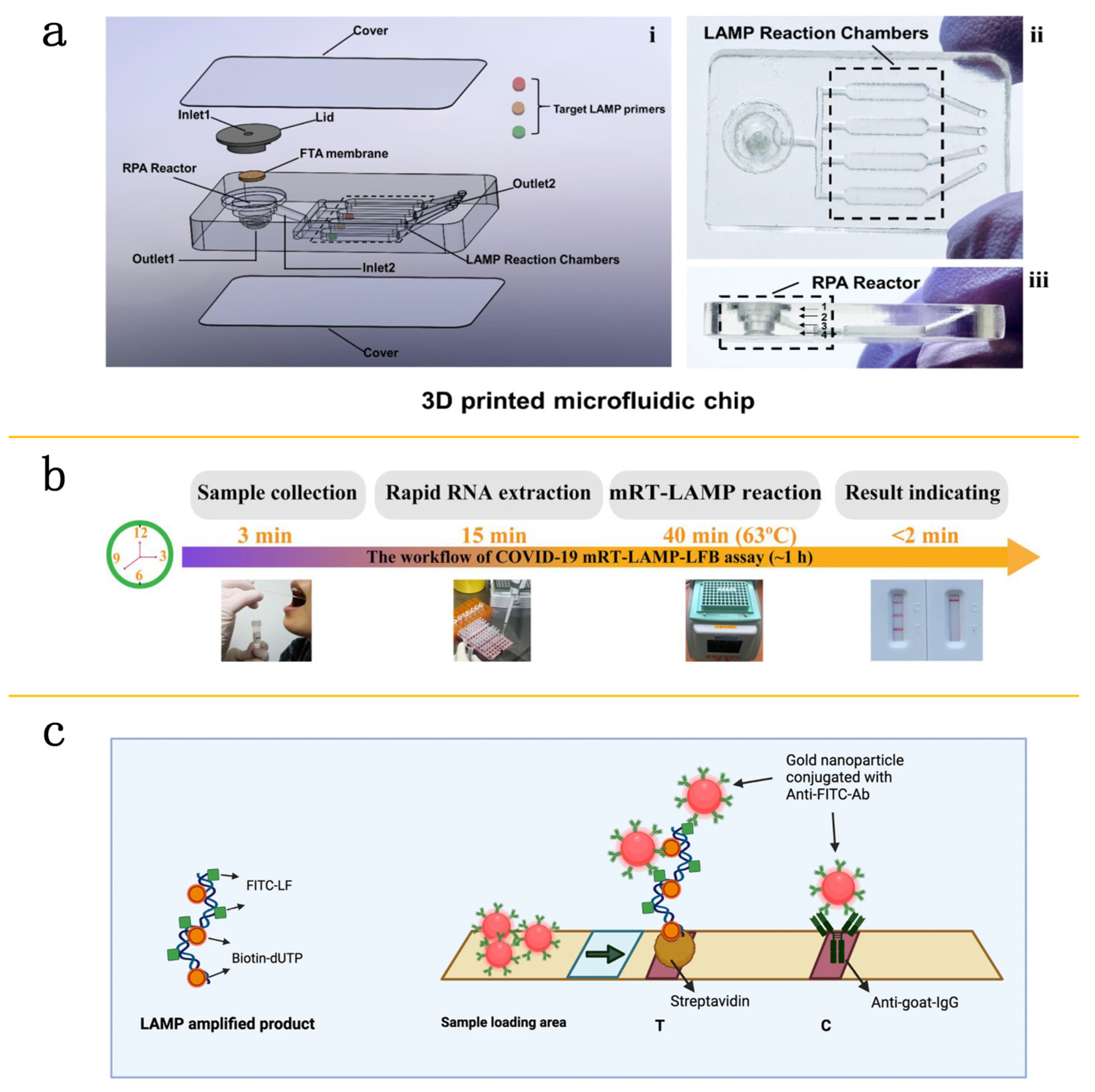
3. Isothermal Amplification Techniques on Microfluidic Platforms
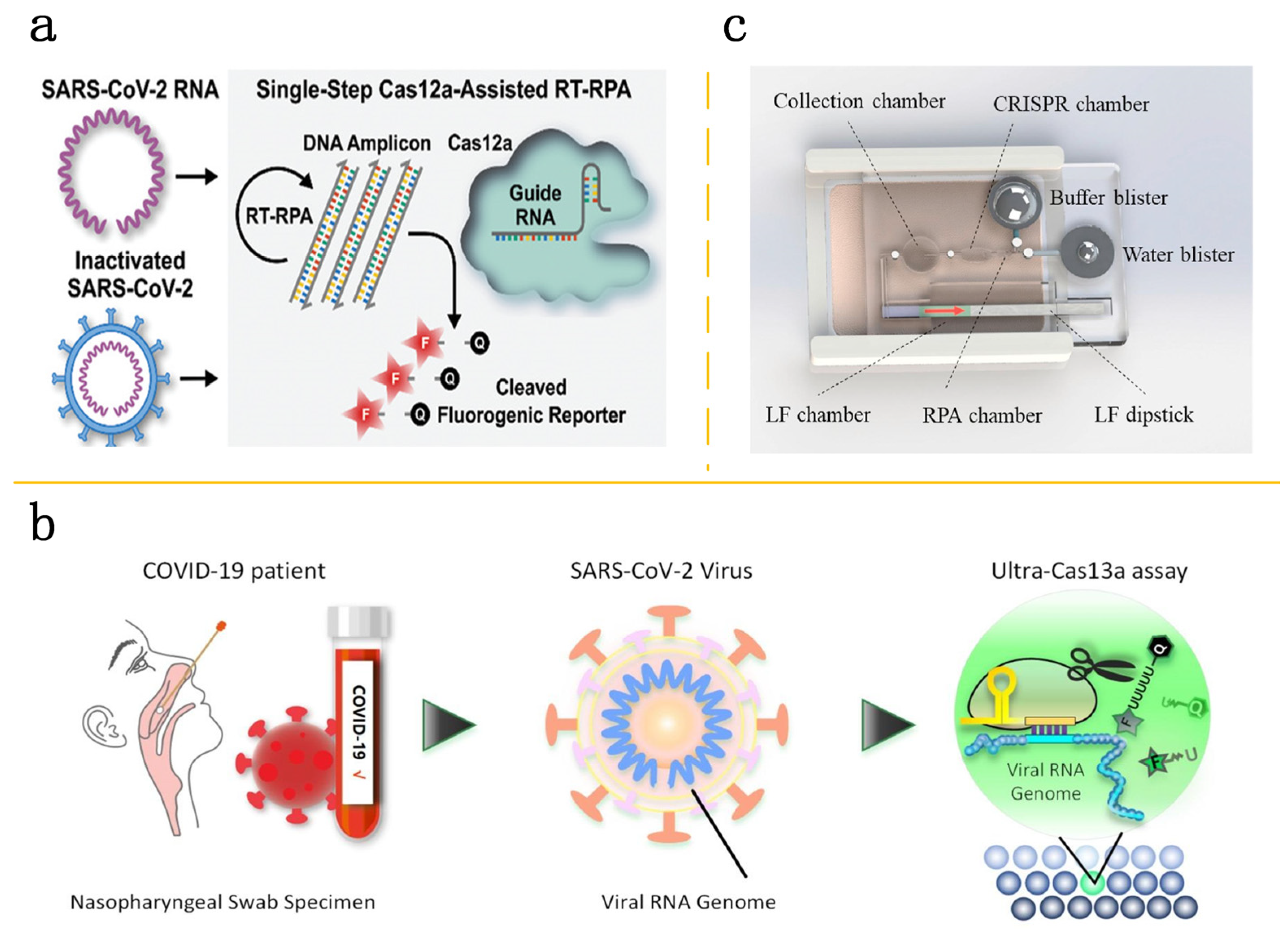
4. Application of CRISPR-Cas Technology in Microfluidic Platforms
5. Smartphone Applications in Microfluidic Platforms
6. AI-Assisted Microfluidic Technologies
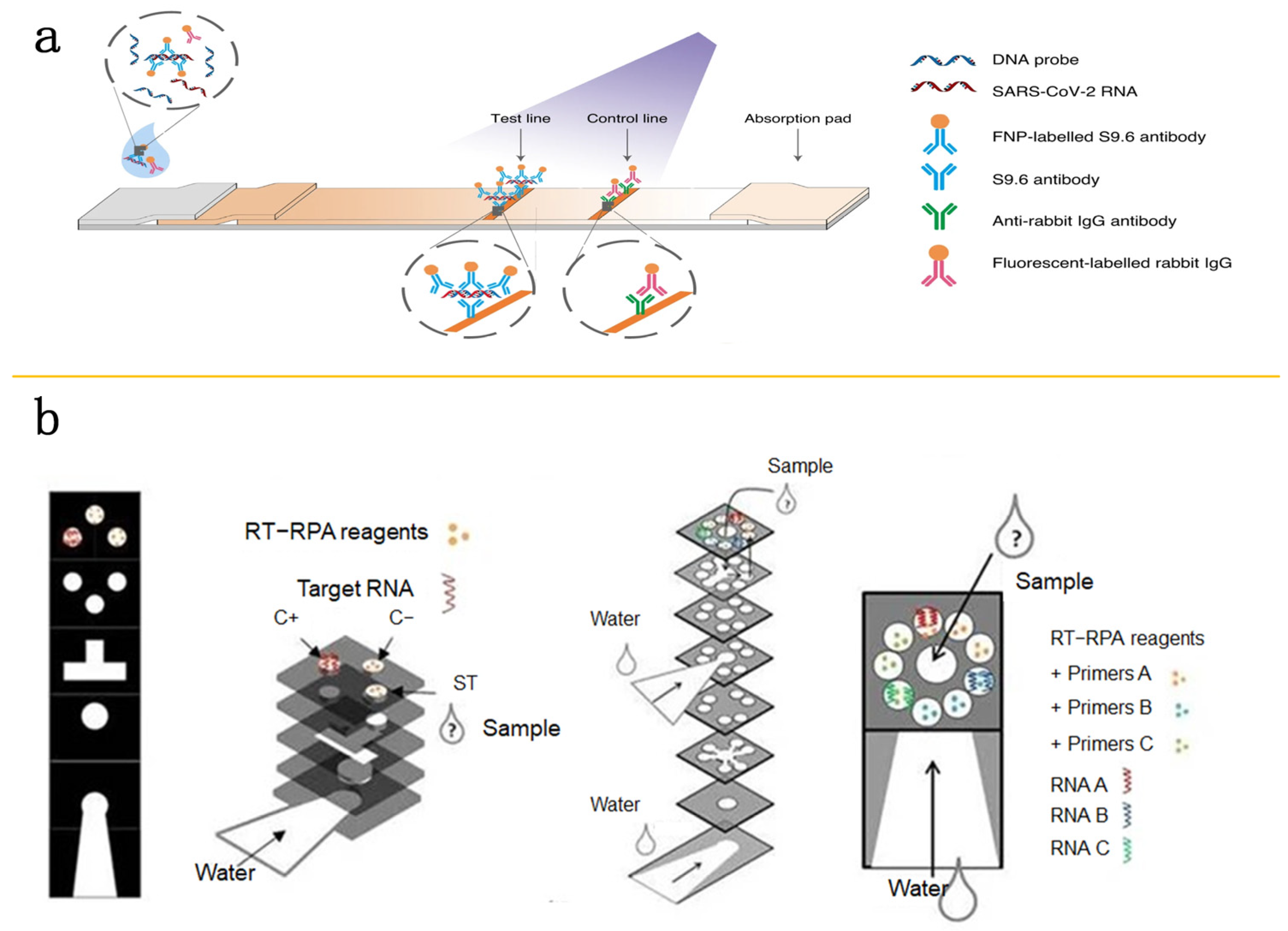
7. Paper-Based POC Testing Applications in Microfluidic Platforms
8. Outlook
- (1)
- Developing modular microfluidic system components and procedures is essential for creating clinically meaningful integrated products.
- (2)
- Formulating unified market supervision standards is necessary. Varying regulatory requirements across regions not only limit investor incentives but may also prevent technology from passing regulations and evolving into a clinical diagnostic system.
- (3)
- Ensuring data connection to electronic medical record systems is crucial. With the increasing use of digital devices such as mobile phones, establishing remote data sharing platforms is very important.
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Janikddfghx, E.; Ceremuga, M.; Niemcewicz, M.; Bijak, M. Dangerous pathogens as a potential problem for public health. Medicina 2020, 56, 591. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.F.; et al. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2022, 20, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Onder, G.; Rezza, G.; Brusaferro, S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. JAMA 2022, 323, 1775–1776. [Google Scholar] [CrossRef] [PubMed]
- Plourde, A.R.; Bloch, E.M. A literature review of zika virus. Emerg. Infect. Dis. 2016, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Santiago, G.A.; Vázquez, J.; Courtney, S.; Matías, K.Y.; Andersen, L.E.; Colón, C.; Butler, A.E.; Roulo, R.; Bowzard, J.; Villanueva, J.M.; et al. Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat. Commun. 2018, 9, 1391. [Google Scholar] [CrossRef]
- Kinsman, J.; de Bruijne, K.; Jalloh, A.M.; Harris, M.; Abdullah, H.; Boye-Thompson, T.; Sankoh, O.; Jalloh, A.K.; Jalloh-Vos, H. Development of a set of community-informed Ebola messages for Sierra Leone. PLoS Negl. Trop. Dis. 2017, 11, e0005742. [Google Scholar] [CrossRef] [PubMed]
- Guha-Sapir, D.; Schimmer, B. Dengue fever: New paradigms for a changing epidemiology. Emerg. Themes Epidemiol. 2005, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Mallah, S.I.; Ghorab, O.K.; Al-Salmi, S.; Abdellatif, O.S.; Tharmaratnam, T.; Iskandar, M.A.; Sefen, J.A.N.; Sidhu, P.; Atallah, B.; El-Lababidi, R.; et al. COVID-19: Breaking down a global health crisis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 35. [Google Scholar] [CrossRef]
- Shin, D.J.; Andini, N.; Hsieh, K.; Yang, S.; Wang, T.H. Emerging Analytical Techniques for Rapid Pathogen Identification and Susceptibility Testing. Annu. Rev. Anal. Chem. 2019, 12, 41–67. [Google Scholar] [CrossRef]
- Yach, D.; Hawkes, C.; Gould, C.L.; Hofman, K.J. The global burden of chronic diseases: Overcoming impediments to prevention and control. JAMA 2004, 291, 2616–2622. [Google Scholar] [CrossRef]
- Bloom, D.E.; Cadarette, D. Infectious disease threats in the twenty-first century: Strengthening the global response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ji, D.; Cai, W.; Hu, Y.; Bai, Y.; Wu, J.; Xu, J. Clinical characteristics, cause analysis and infectivity of COVID-19 nucleic acid repositive patients: A literature review. J. Med. Virol. 2021, 93, 1288–1295. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef] [PubMed]
- Razzini, K.; Castrica, M.; Menchetti, L.; Maggi, L.; Negroni, L.; Orfeo, N.V.; Pizzoccheri, A.; Stocco, M.; Muttini, S.; Balzaretti, C.M. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. Sci. Total Environ. 2020, 742, 140540. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Gao, Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta 2015, 853, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.N.D.; La, H.C.; Lee, N.Y. Fully Integrated and Foldable Microdevice Encapsulated with Agarose for Long-Term Storage Potential for Point-of-Care Testing of Multiplex Foodborne Pathogens. ACS Sens. 2019, 4, 2754–2762. [Google Scholar] [CrossRef] [PubMed]
- Mitsakakis, K.; D’Acremont, V.; Hin, S.; von Stetten, F.; Zengerle, R. Diagnostic tools for tackling febrile illness and enhancing patient management. Microelectron. Eng. 2018, 201, 26–59. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Xia, Y.; Loo, J.F.C.; Li, L.; Ho, H.P.; He, J.; Gu, D. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform. RSC Adv. 2020, 10, 34088–34098. [Google Scholar] [CrossRef]
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar] [CrossRef]
- Schumacher, S.; Nestler, J.; Otto, T.; Wegener, M.; Ehrentreich-Förster, E.; Michel, D.; Wunderlich, K.; Palzer, S.; Sohn, K.; Weber, A.; et al. Highly-integrated lab-on-chip system for point-of-care multiparameter analysis. Lab Chip 2012, 12, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Jin, X.; Xu, B.; Wang, R.; Fu, R.; Su, Y.; Jiang, K.; Yang, H.; Lu, Y.; Guo, Y.; et al. Fast and parallel detection of Four Ebola Virus species on a microfluidic-chip-based portable reverse transcription loop-mediated isothermal amplification system. Micromachines 2019, 10, 777. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, Z.; Rashid, Z.; Ali, A.; Arif, A.; Ameen, F.; AlTami, M.S.; Yousaf, M.Z. Prospects of Microfluidic Technology in Nucleic Acid Detection Approaches. Biosensors 2023, 13, 584. [Google Scholar] [CrossRef] [PubMed]
- Pittman, T.W.; Decsi, D.B.; Punyadeera, C.; Henry, C.S. Saliva-based microfluidic point-of-care diagnostic. Theranostics 2023, 13, 1091–1108. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.S.; Agrawal, S.; Fanton, A.; Jangid, A.R.; Charrez, B.; Escajeda, A.M.; Son, S.; Mcintosh, R.; Tran, H.; Bhuiya, A.; et al. Rapid detection of SARS-CoV-2 RNA in saliva via Cas13. Nat. Biomed. Eng. 2022, 6, 944–956. [Google Scholar] [CrossRef]
- Song, W.; Zhang, T.; Lin, H.; Yang, Y.; Zhao, G.; Huang, X. Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2. Micromachines 2022, 13, 636. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Pineda, O.G.; Rodriguez-Moncayo, R.; Cedillo-Alcantar, D.F.; Guevara-Pantoja, P.E.; Amador-Hernandez, J.U.; Garcia-Cordero, J.L. Microfluidic systems for the analysis of blood-derived molecular biomarkers. Electrophoresis 2022, 43, 1667–1700. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lee, L.P. Toward Rapid and Accurate Molecular Diagnostics at Home. Adv. Mater. 2023, 35, e2206525. [Google Scholar] [CrossRef] [PubMed]
- Mimmi, S.; Zimbo, A.M.; Rotundo, S.; Cione, E.; Nisticò, N.; Aloisio, A.; Maisano, D.; Tolomeo, A.M.; Dattilo, V.; Lionello, R.; et al. SARS-CoV-2 spike protein-guided exosome isolation facilitates detection of potential miRNA biomarkers in COVID-19 infections. Clin. Chem. Lab. Med. 2023, 61, 1518–1524. [Google Scholar] [CrossRef]
- Kang, B.H.; Lee, Y.; Yu, E.S.; Na, H.; Kang, M.; Huh, H.J.; Jeong, K.H. Ultrafast and Real-Time Nanoplasmonic On-Chip Polymerase Chain Reaction for Rapid and Quantitative Molecular Diagnostics. ACS Nano 2021, 15, 10194–10202. [Google Scholar] [CrossRef]
- Sakai, J.; Tarumoto, N.; Orihara, Y.; Kawamura, R.; Kodana, M.; Matsuzaki, N.; Matsumura, R.; Ogane, K.; Kawamura, T.; Takeuchi, S.; et al. Evaluation of a high-speed but low-throughput RT-qPCR system for detection of SARS-CoV-2. J. Hosp. Infect. 2020, 105, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Fevraleva, I.; Glinshchikova, O.; Makarik, T.; Sudarikov, A. How to Avoid False-Negative and False-Positive COVID-19 PCR Testing. Int. J. Transl. Med. 2022, 2, 204–209. [Google Scholar] [CrossRef]
- Benevides Lima, L.; Mesquita, F.P.; Brasil de Oliveira, L.L.; Andréa da Silva Oliveira, F.; Elisabete Amaral de Moraes, M.; Souza, P.F.N.; Montenegro, R.C. True or false: What are the factors that influence COVID-19 diagnosis by RT-qPCR? Expert Rev. Mol. Diagn. 2022, 22, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Garzarelli, V.; Chiriacò, M.S.; Cereda, M.; Autuori, I.; Ferrara, F. Miniaturized Real-Time PCR systems for SARS-CoV-2 detection at the Point-of-Care. Clin. Chim. Acta 2022, 536, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Gjorgjieva, T.; Attieh, Z.; Dieng, M.M.; Arnoux, M.; Khair, M.; Moussa, Y.; Jallaf, F.A.; Rahiman, N.; Jackson, C.A.; et al. Microfluidic nano-scale QPCR enables ultra-sensitive and quantitative detection of SARS-CoV-2. Processes 2020, 8, 1425. [Google Scholar] [CrossRef]
- Fassy, J.; Lacoux, C.; Leroy, S.; Noussair, L.; Hubac, S.; Degoutte, A.; Vassaux, G.; Leclercq, V.; Rouquié, D.; Marquette, C.H.; et al. Versatile and flexible microfluidic qPCR test for high-throughput SARS-CoV-2 and cellular response detection in nasopharyngeal swab samples. PLoS ONE 2021, 16, e0243333. [Google Scholar] [CrossRef]
- Tian, F.; Liu, C.; Deng, J.; Han, Z.; Zhang, L.; Chen, Q.; Sun, J. A fully automated centrifugal microfluidic system for sample-to-answer viral nucleic acid testing. Sci. China Chem. 2020, 63, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Ye, X.; Li, Y.; Wang, L.; Zhang, J.; Fang, X.; Kong, J. Rapid Differential Diagnosis of Seven Human Respiratory Coronaviruses Based on Centrifugal Microfluidic Nucleic Acid Assay. Anal. Chem. 2020, 92, 14297–14302. [Google Scholar] [CrossRef] [PubMed]
- Zayed, B.A.; Ali, A.N.; Elgebaly, A.A.; Talaia, N.M.; Hamed, M.; Mansour, F.R. Smartphone-based point-of-care testing of the SARS-CoV-2: A systematic review. Sci. Afr. 2023, 21, e01757. [Google Scholar] [CrossRef]
- Kabay, G.; DeCastro, J.; Altay, A.; Smith, K.; Lu, H.W.; Capossela, A.M.D.; Moarefian, M.; Aran, K.; Dincer, C. Emerging Biosensing Technologies for the Diagnostics of Viral Infectious Diseases. Adv. Mater. 2022, 34, e2201085. [Google Scholar] [CrossRef]
- Yin, B.; Wan, X.; Sohan, A.S.M.M.F.; Lin, X. Microfluidics-Based POCT for SARS-CoV-2 Diagnostics. Micromachines 2022, 13, 1238. [Google Scholar] [CrossRef] [PubMed]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W. Principles and Applications of Loop-Mediated Isothermal Amplification to Point-of-Care Tests. Biosensors 2022, 12, 857. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Xu, Z.; Li, Z.; Wang, X.; Zhao, H.; Otis, C.; Li, B.; Liu, C. Multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in wastewater on a 3D printed integrated microfluidic chip. Sens. Actuators B Chem. 2021, 344, 130242. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Warmt, C.; Henkel, J.; Schrick, L.; Nitsche, A.; Bier, F.F. Lateral flow–based nucleic acid detection of SARS-CoV-2 using enzymatic incorporation of biotin-labeled dUTP for POCT use. Anal. Bioanal. Chem. 2022, 414, 3177–3186. [Google Scholar] [CrossRef]
- Xing, W.; Liu, Y.; Wang, H.; Li, S.; Lin, Y.; Chen, L.; Zhao, Y.; Chao, S.; Huang, X.; Ge, S.; et al. A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2. Engineering 2020, 6, 1130–1140. [Google Scholar] [CrossRef]
- Liu, D.; Shen, H.; Zhang, Y.; Shen, D.; Zhu, M.; Song, Y.; Zhu, Z.; Yang, C. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab. Chip 2021, 21, 2019–2026. [Google Scholar] [CrossRef]
- Yeh, E.; Fu, C.; Hu, L.; Thakur, R.; Feng, J.; Lee, L.P. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017, 3, e1501645. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Silva, S.J.R.d.; Paiva, M.H.S.; Guedes, D.R.D.; Krokovsky, L.; Melo, F.L.d.; Silva, M.A.L.d.; Silva, A.d.; Ayres, C.F.J.; Pena, L.J. Development and Validation of Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) for Rapid Detection of ZIKV in Mosquito Samples from Brazil. Sci. Rep. 2019, 9, 4494. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, A.; Ornob, A.; Yu, H.; Damhorst, G.L.; Chen, W.; Sun, F.; Bhuiya, A.; Cunningham, B.T.; Bashir, R. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed. Microdevices 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Kaarj, K.; Akarapipad, P.; Yoon, J.Y. Simpler, Faster, and Sensitive Zika Virus Assay Using Smartphone Detection of Loop-mediated Isothermal Amplification on Paper Microfluidic Chips. Sci. Rep. 2018, 8, 12438. [Google Scholar] [CrossRef] [PubMed]
- Batule, B.S.; Seok, Y.; Kim, M.G. Paper-based nucleic acid testing system for simple and early diagnosis of mosquito-borne RNA viruses from human serum. Biosens. Bioelectron. 2020, 151, 111998. [Google Scholar] [CrossRef] [PubMed]
- Oliver, G.F.; Carr, J.M.; Smith, J.R. Emerging infectious uveitis: Chikungunya, dengue, Zika and Ebola: A review. Clin. Exp. Ophthalmol. 2019, 47, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Harapan, H.; Michie, A.; Sasmono, R.T.; Imrie, A. Dengue: A Minireview. Viruses 2020, 1, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Mendes, G.M.; Oliveira, K.G.; Borba, J.C.; Oliveira, T.S.; Fiaccadori, F.S.; Nogueira, M.L.; Bailão, A.M.; Soares, C.M.A.; Carrilho, E.; Duarte, G.R.M. Molecular diagnostics of dengue by reverse transcription-loop mediated isothermal amplification (RT-LAMP) in disposable polyester-toner microdevices. J. Braz. Chem. Soc. 2019, 30, 1841–1849. [Google Scholar] [CrossRef]
- Nguyen, T.; Chidambara, V.A.; Andreasen, S.Z.; Golabi, M.; Huynh, V.N.; Linh, Q.T.; Bang, D.D.; Wolff, A. Point-of-care devices for pathogen detections: The three most important factors to realise towards commercialization. TrAC Trends Anal. Chem. 2020, 131, 116004. [Google Scholar] [CrossRef]
- Shao, Y.; Zhu, S.; Jin, C.; Chen, F. Development of multiplex loop-mediated isothermal amplification-RFLP (mLAMP-RFLP) to detect Salmonella spp. and Shigella spp. in milk. Int. J. Food Microbiol. 2011, 148, 75–79. [Google Scholar] [CrossRef]
- Watts, M.R.; James, G.; Sultana, Y.; Ginn, A.N.; Outhred, A.C.; Kong, F.; Verweij, J.J.; Iredell, J.R.; Chen, S.C.; Lee, R. A loop-mediated isothermal amplification (LAMP) assay for Strongyloides stercoralis in stool that uses a visual detection method with SYTO-82 fluorescent dye. Am. J. Trop. Med. Hyg. 2014, 90, 306–311. [Google Scholar] [CrossRef]
- Park, J.S.; Hsieh, K.; Chen, L.; Kaushik, A.; Trick, A.Y.; Wang, T.H. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv. Sci. 2021, 8, 2003564. [Google Scholar] [CrossRef] [PubMed]
- Welch, N.L.; Zhu, M.; Hua, C.; Weller, J.; Mirhashemi, M.E.; Nguyen, T.G.; Mantena, S.; Bauer, M.R.; Shaw, B.M.; Ackerman, C.M.; et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat. Med. 2022, 28, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Shu, B.; Jiang, Y.; Ye, M.; Liu, L.; Guo, Z.; Han, Z.; Wang, Z.; Zhou, X. An Ultralocalized Cas13a Assay Enables Universal and Nucleic Acid Amplification-Free Single-Molecule RNA Diagnostics. ACS Nano 2021, 15, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ding, X.; Yin, K.; Avery, L.; Ballesteros, E.; Liu, C. Instrument-free, CRISPR-based diagnostics of SARS-CoV-2 using self-contained microfluidic system. Biosens. Bioelectron. 2022, 199, 113865. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Park, M.; Alfson, K.J.; Tamhankar, M.; Carrion, R.; Patterson, J.L.; Griffiths, A.; He, Q.; Yildiz, A.; Mathies, R.; et al. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Yu, D.; Bao, M.; Korensky, G.; Chen, J.; Shin, M.; Kim, J.; Park, M.; Qin, P.; Du, K. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens. Bioelectron. 2020, 154, 112068. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yadav, V.; Zhang, C.; Huo, X.; Wang, C.; Senapati, S.; Chang, H.C. Elliptical Pipette Generated Large Microdroplets for POC Visual ddPCR Quantification of Low Viral Load. Anal. Chem. 2021, 93, 6456–6462. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Bui, H.K.; Phan, V.M.; Seo, T.S. An internet of things-based point-of-care device for direct reverse-transcription-loop mediated isothermal amplification to identify SARS-CoV-2. Biosens. Bioelectron. 2022, 195, 113655. [Google Scholar] [CrossRef]
- Silva, F.S.R.; Erdogmus, E.; Shokr, A.; Kandula, H.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Hardie, J.M.; Pacheco, L.G.C.; Li, J.Z.; Kuritzkes, D.R.; et al. SARS-CoV-2 RNA Detection by a Cellphone-Based Amplification-Free System with CRISPR/CAS-Dependent Enzymatic (CASCADE) Assay. Adv. Mater. Technol. 2021, 6, 2100602. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, J.; Hu, A.; Pan, J.; Deng, G.; Hua, C.; Zhu, C.; Liu, Y.; Yang, K.; Zhu, L. A novel microfluidic device integrated with chitosan-modified capillaries for rapid ZIKV detection. Micromachines 2020, 11, 186. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xiong, L.; Huang, Y.; Chen, X.; Yu, Y.; Ye, S.; Dong, H.; Jia, Y.; Zhang, W. AI-aided on-chip nucleic acid assay for smart diagnosis of infectious disease. Fundam. Res. 2022, 2, 476–486. [Google Scholar] [CrossRef]
- Wang, B.; Li, Y.; Zhou, M.; Han, Y.; Zhang, M.; Gao, Z.; Liu, Z.; Chen, P.; Du, W.; Zhang, X.; et al. Smartphone-based platforms implementing microfluidic detection with image-based artificial intelligence. Nat. Commun. 2023, 14, 1341. [Google Scholar] [CrossRef] [PubMed]
- Solmaz, M.E.; Mutlu, A.Y.; Alankus, G.; Kılıç, V.; Bayram, A.; Horzum, N. Quantifying colorimetric tests using a smartphone app based on machine learning classifiers. Sens. Actuators B Chem. 2018, 255, 1967–1973. [Google Scholar] [CrossRef]
- Guo, X.; Khalid, M.A.; Domingos, I.; Michala, A.L.; Adriko, M.; Rowel, C.; Ajambo, D.; Garrett, A.; Kar, S.; Yan, X.X.; et al. Smartphone-based DNA diagnostics for malaria detection using deep learning for local decision support and blockchain technology for security. Nat. Electron. 2021, 4, 615–624. [Google Scholar] [CrossRef]
- Wang, D.; He, S.; Wang, X.; Yan, Y.; Liu, J.; Wu, S.; Liu, S.; Lei, Y.; Chen, M.; Li, L.; et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, S.; Sayama, Y.; Nagata, N.; Ikegami, T.; Miranda, M.E.; Watanabe, S.; Iizuka, I.; Fukushi, S.; Mizutani, T.; Ishii, Y.; et al. Analysis of the humoral immune responses among cynomolgus macaque naturally infected with Reston virus during the 1996 outbreak in the Philippines. BMC Vet. Res. 2012, 8, 4–7. [Google Scholar] [CrossRef]
- Magro, L.; Jacquelin, B.; Escadafal, C.; Garneret, P.; Kwasiborski, A.; Manuguerra, J.C.; Monti, F.; Sakuntabhai, A.; Vanhomwegen, J.; Lafaye, P.; et al. Paper-based RNA detection and multiplexed analysis for Ebola virus diagnostics. Sci. Rep. 2017, 7, 1347. [Google Scholar] [CrossRef]
- Das, S.; Rundell, M.S.; Mirza, A.H.; Pingle, M.R.; Shigyo, K.; Garrison, A.R.; Paragas, J.; Smith, S.K.; Olson, V.A.; Larone, D.H.; et al. A Multiplex PCR/LDR Assay for the simultaneous identification of category a infectious pathogens: Agents of viral hemorrhagic fever and Variola virus. PLoS ONE 2015, 10, e0138484. [Google Scholar] [CrossRef]
- Du, K.; Cai, H.; Park, M.; Wall, T.A.; Stott, M.A.; Alfson, K.J.; Griffiths, A.; Carrion, R.; Patterson, J.L.; Hawkins, A.R.; et al. Multiplexed efficient on-chip sample preparation and sensitive amplification-free detection of Ebola virus. Biosens. Bioelectron. 2017, 91, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Parks, J.W.; Wall, T.A.; Stott, M.A.; Stambaugh, A.; Alfson, K.; Griffiths, A.; Mathies, R.A.; Carrion, R.; Patterson, J.L.; et al. Optofluidic analysis system for amplification-free, direct detection of Ebola infection. Sci. Rep. 2015, 5, 14494. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Chen, J.; Yang, Z.; Wang, X.; Zhang, Y.; Chen, M.; Ming, Z.; Zhang, K.; Zhang, D.; Zheng, L. Advances in Nucleic Acid Assays for Infectious Disease: The Role of Microfluidic Technology. Molecules 2024, 29, 2417. https://doi.org/10.3390/molecules29112417
Wang Y, Chen J, Yang Z, Wang X, Zhang Y, Chen M, Ming Z, Zhang K, Zhang D, Zheng L. Advances in Nucleic Acid Assays for Infectious Disease: The Role of Microfluidic Technology. Molecules. 2024; 29(11):2417. https://doi.org/10.3390/molecules29112417
Chicago/Turabian StyleWang, Yiran, Jingwei Chen, Zhijin Yang, Xuanyu Wang, Yule Zhang, Mengya Chen, Zizhen Ming, Kaihuan Zhang, Dawei Zhang, and Lulu Zheng. 2024. "Advances in Nucleic Acid Assays for Infectious Disease: The Role of Microfluidic Technology" Molecules 29, no. 11: 2417. https://doi.org/10.3390/molecules29112417
APA StyleWang, Y., Chen, J., Yang, Z., Wang, X., Zhang, Y., Chen, M., Ming, Z., Zhang, K., Zhang, D., & Zheng, L. (2024). Advances in Nucleic Acid Assays for Infectious Disease: The Role of Microfluidic Technology. Molecules, 29(11), 2417. https://doi.org/10.3390/molecules29112417





