Changes of Active Substances in Ganoderma lucidum during Different Growth Periods and Analysis of Their Molecular Mechanism
Abstract
:1. Introduction
2. Results
2.1. Changes in Bioactive Nutrient of G. lucidum at Various Growth Periods
2.2. Metabolic Characteristics of G. lucidum at Various Growth Periods
2.3. Changes in Triterpenoid Content of G. lucidum during Different Growth Periods
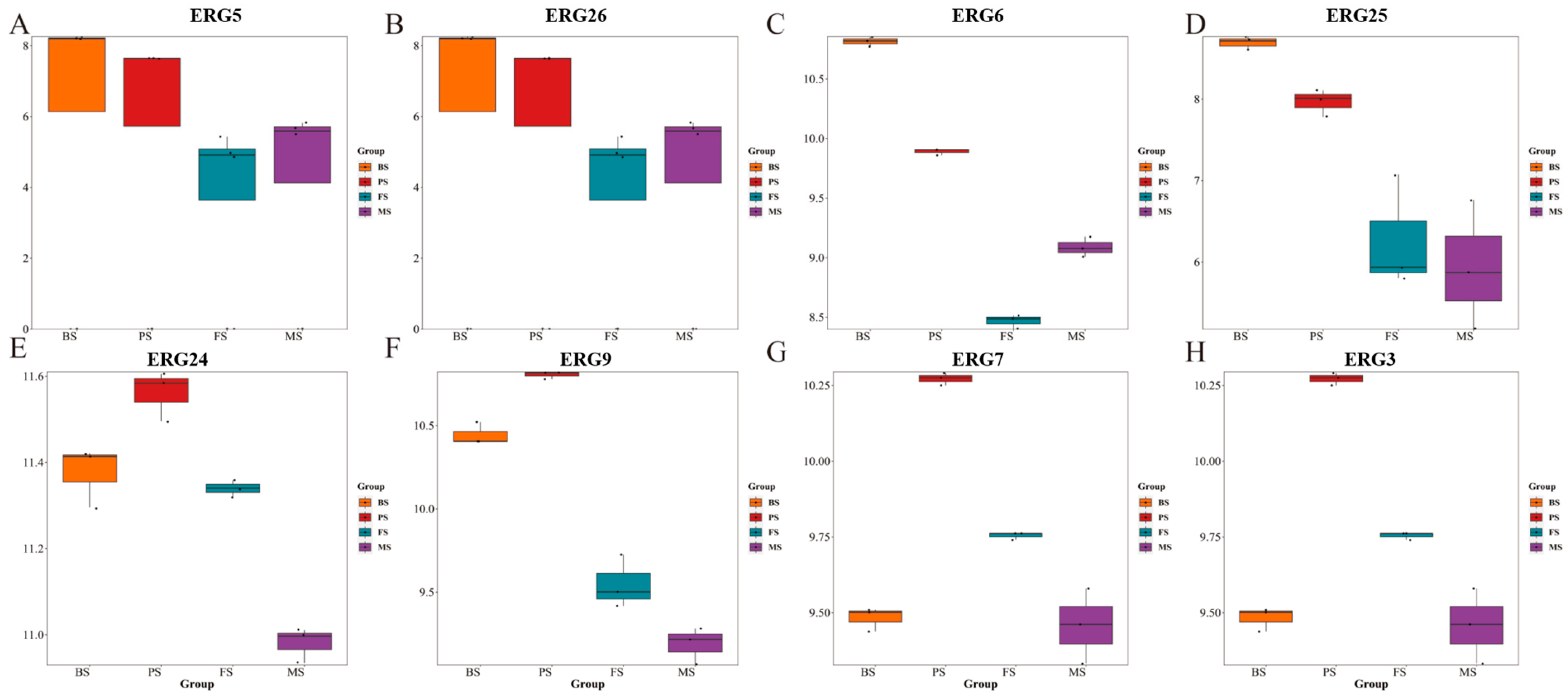
2.4. Changes in Steroid Content of G. lucidum during Different Growth Periods
2.5. Proteomic Results of G. lucidum at Different Growth Periods
2.6. Changes in Proteins Related to Triterpene and Steroid Synthesis Content of G. lucidum during Different Growth Periods
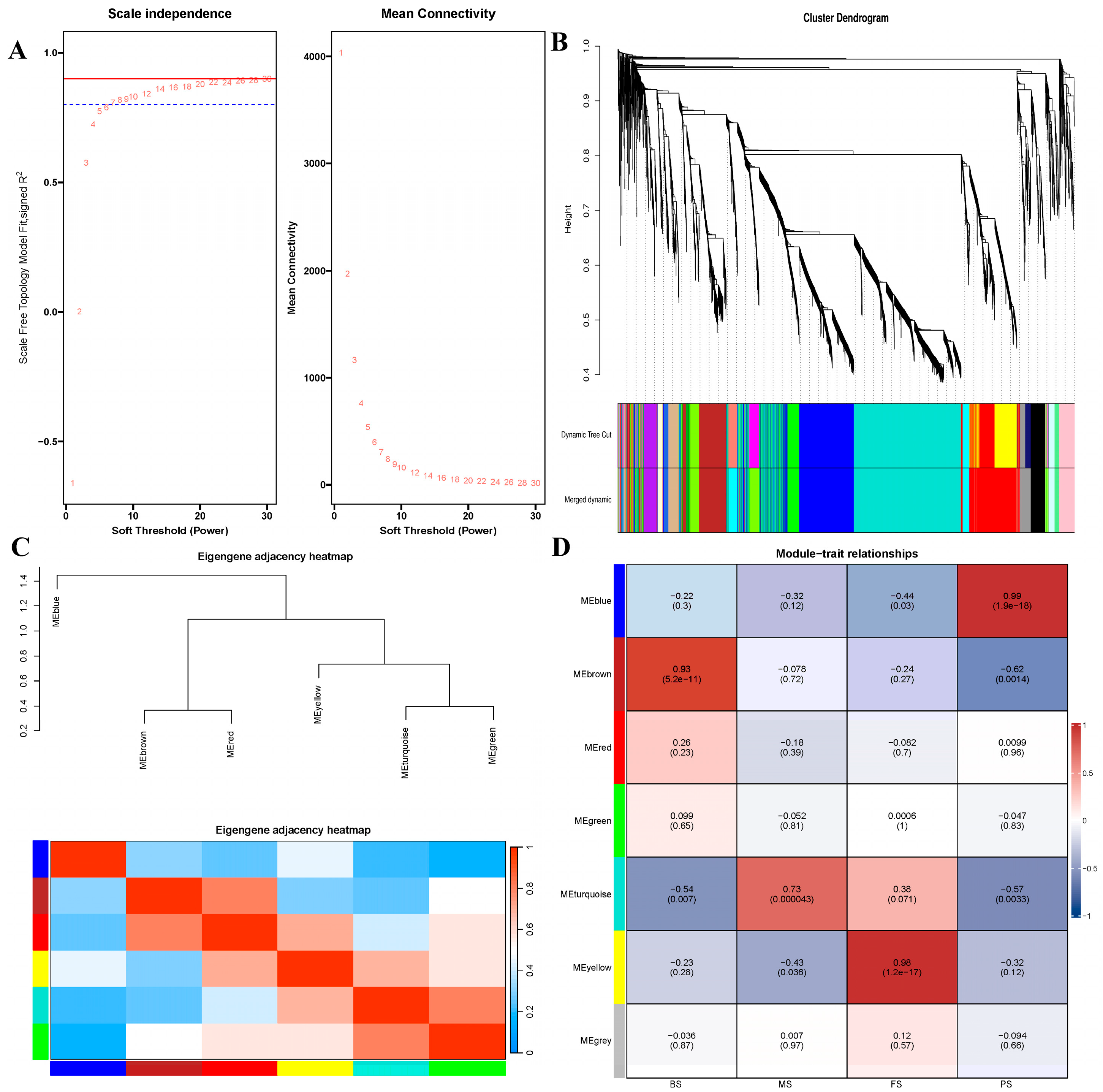
2.7. WGCNA and Color Module Identification of Metabolomic at Different Growth Periods
2.8. WGCNA and Color Module Identification of Proteins at Various Growth Periods
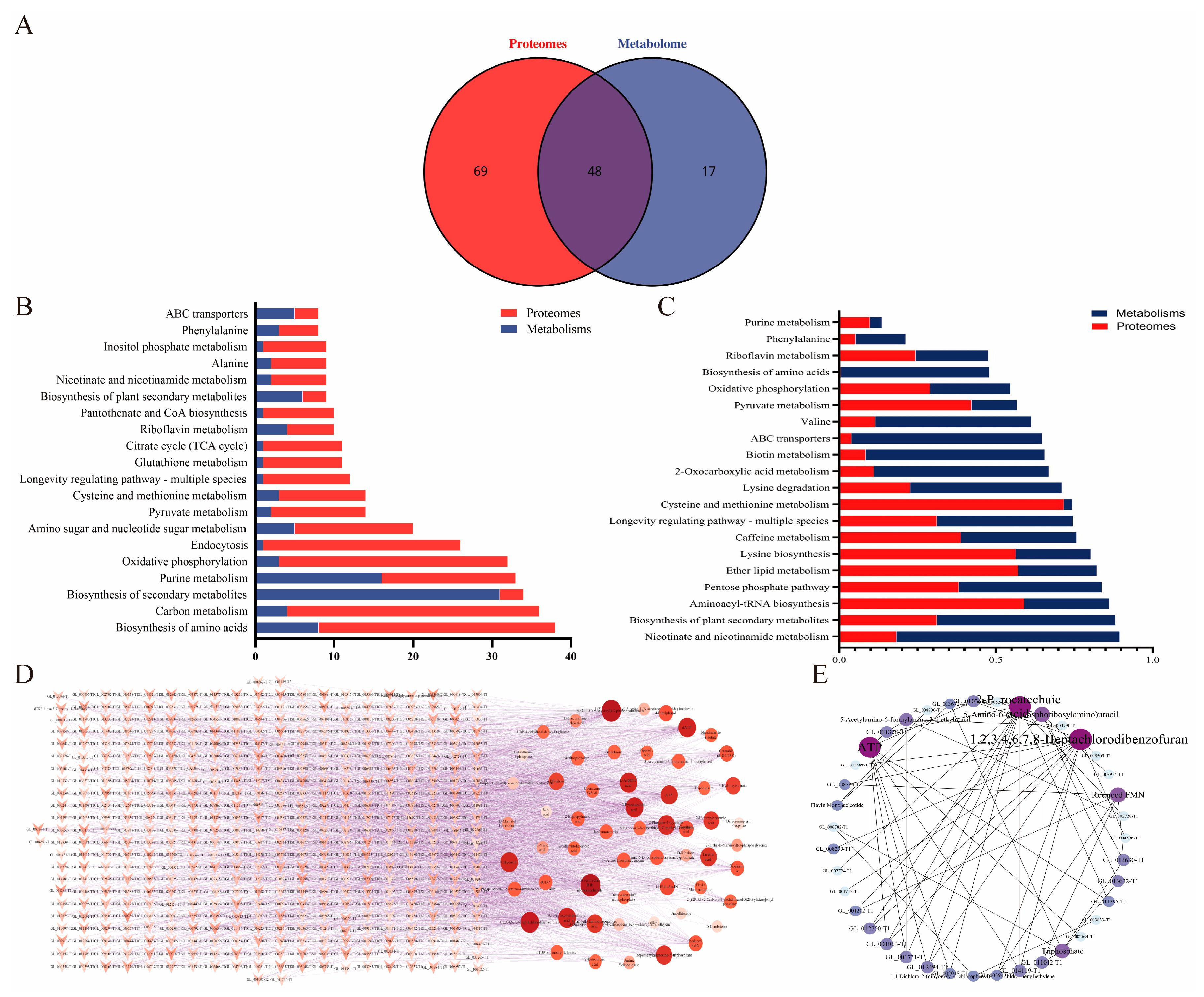
2.9. Integrated Analysis of the Metabolome and Proteome
3. Discussion
3.1. Impact of G. lucidum at Different Growth Periods on Phenylalanine, Tyrosine, and Tryptophan Biosynthesis
3.2. Impact of G. lucidum at Different Growth Periods on Energy Production
3.3. Impact of G. lucidum at Different Growth Periods on ABC Transporters
3.4. Variations in Triterpenes and Their Synthesis during the Development of G. lucidum
4. Materials and Methods
4.1. Materials and Chemicals
4.2. Sample Extraction
4.2.1. Metabolite Extraction
4.2.2. Protein Extraction
4.3. Determination of Nutrient Composition and Micro- and Macro-Elements
4.4. Metabolomic Analysis
4.5. HPLC-MS/MS Gel-Free Proteomic Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El Sheikha, A.F. Nutritional Profile and Health Benefits of Ganoderma lucidum “Lingzhi, Reishi, or Mannentake” as Functional Foods: Current Scenario and Future Perspectives. Foods 2022, 11, 1030. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.F. Ganoderma lucidum: A rational pharmacological approach to surmount cancer. J. Ethnopharmacol. 2020, 260, 113047. [Google Scholar] [CrossRef] [PubMed]
- Campos Ziegenbein, F.; Hanssen, H.P.; König, W.A. Secondary metabolites from Ganoderma lucidum and Spongiporus leucomallellus. Phytochemistry 2006, 67, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Wittstein, K.; Kirk, P.M.; Stadler, M. An assessment of the taxonomy and chemotaxonomy of Ganoderma. Fungal Divers. 2015, 71, 1–15. [Google Scholar] [CrossRef]
- Li, Y.; Bao, H. Mycomedicines in China, 1st ed.; Central Plains Farmers Publishing House: Zhengzhou, China, 2020. [Google Scholar]
- Bao, H.; Li, Z. “Zhi” Culture and Medicinal Historical Evolution of Lingzhi and Its Research Ideas. Ginseng Res. 2022, 34, 40–44. [Google Scholar]
- Hirotani, M.; Furuya, T.; Shiro, M. Cryptoporic acids H and I, drimane sesquiterpenes from Ganoderma neo-japonicum and Cryptoporus volvatus. Phytochemistry 1991, 30, 1555–1559. [Google Scholar] [CrossRef]
- Wang, X.M.; Yang, M.; Guan, S.H.; Liu, R.X.; Xia, J.M.; Bi, K.S.; Guo, D.A. Quantitative determination of six major triterpenoids in Ganoderma lucidum and related species by high performance liquid chromatography. J. Pharm. Biomed. Anal. 2006, 41, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kurashiki, K.; Fukuta, A.; Kaneko, S.; Suimi, Y.; Shimizu, K.; Kondo, R. Quantitative determination of the representative triterpenoids in the extracts of Ganoderma lucidum with different growth stages using high-performance liquid chromatography for evaluation of their 5α-reductase inhibitory properties. Food Chem. 2012, 133, 1034–1038. [Google Scholar] [CrossRef]
- Nakagawa, T.; Zhu, Q.; Tamrakar, S.; Amen, Y.; Mori, Y.; Suhara, H.; Kaneko, S.; Kawashima, H.; Okuzono, K.; Inoue, Y.; et al. Changes in content of triterpenoids and polysaccharides in Ganoderma lingzhi at different growth stages. J. Nat. Med. 2018, 72, 734–744. [Google Scholar] [CrossRef]
- Ren, X.; Wang, J.; Huang, L.; Cheng, K.; Zhang, M.; Yang, H. Comparative Studies on Bioactive Compounds, Ganoderic Acid Biosynthesis, and Antioxidant Activity of Pileus and Stipes of Lingzhi or Reishi Medicinal Mushroom, Ganoderma lucidum (Agaricomycetes) Fruiting Body at Different Growth Stages. Int. J. Med. Mushrooms 2020, 22, 133–144. [Google Scholar] [CrossRef]
- Li, Z.; Bao, H. Comparative Analysis of Metabolic Compositions and Trace Elements of Inonotus hispidus Mushroom Grown on Five Different Tree Species. ACS Omega 2022, 7, 9343–9358. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; He, X.; Yang, W.; Song, H.; Yang, J.; Zhang, G.; Yang, Z.; Chen, H.; Liang, Z.; Kollie, L.; et al. Unveiling the distribution of chemical constituents at different body parts and maturity stages of Ganoderma lingzhi by combining metabolomics with desorption electrospray ionization mass spectrometry imaging (DESI). Food Chem. 2024, 436, 137737. [Google Scholar] [CrossRef] [PubMed]
- Barros, B.H.R.; da Silva, S.H.; Marques, E.d.R.; Rosa, J.C.; Yatsuda, A.P.; Roberts, D.W.; Braga, G.U.L. A proteomic approach to identifying proteins differentially expressed in conidia and mycelium of the entomopathogenic fungus Metarhizium acridum. Fungal Biol. 2010, 114, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, R.; Vitorino, R.; Domingues, P.; Radhouani, H.; Carvalho, C.; Poeta, P.; Torres, C.; Igrejas, G. Proteome of a methicillin-resistant Staphylococcus aureus clinical strain of sequence type ST398. J. Proteom. 2012, 75, 2892–2915. [Google Scholar] [CrossRef] [PubMed]
- Scherl, A.; François, P.; Bento, M.; Deshusses, J.M.; Charbonnier, Y.; Converset, V.; Huyghe, A.; Walter, N.; Hoogland, C.; Appel, R.D.; et al. Correlation of proteomic and transcriptomic profiles of Staphylococcus aureus during the post-exponential phase of growth. J. Microbiol. Methods 2005, 60, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wen, X.; Zhang, Y.; Zou, P.; Cheng, L.; Gan, R.; Li, X.; Liu, D.; Geng, F. Quantitative proteomic and metabolomic analysis of Dictyophora indusiata fruiting bodies during post-harvest morphological development. Food Chem. 2021, 339, 127884. [Google Scholar] [CrossRef] [PubMed]
- Buts, K.; Michielssens, S.; Hertog, M.L.A.T.M.; Hayakawa, E.; Cordewener, J.; America, A.H.P.; Nicolai, B.M.; Carpentier, S.C. Improving the identification rate of data independent label-free quantitative proteomics experiments on non-model crops: A case study on apple fruit. J. Proteom. 2014, 105, 31–45. [Google Scholar] [CrossRef]
- Li, R.; Zhang, Z.; Su, X.; Yu, J.; Lu, L.; Liu, T. Nontargeted metabolomics study and pharmacodynamic evaluation of bidirectional fermentation for Ganoderma lucidum with Marsdenia tenacissima. Front. Pharmacol. 2022, 13, 1012063. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Deckers, P.; Swennen, R.; Panis, B. Preparation of protein extracts from recalcitrant plant tissues: An evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 2005, 5, 2497–2507. [Google Scholar] [CrossRef]
- Tian, Y.Z.; Wang, Z.F.; Liu, Y.D.; Zhang, G.Z.; Li, G. The whole-genome sequencing and analysis of a Ganoderma lucidum strain provide insights into the genetic basis of its high triterpene content. Genomics 2021, 113, 840–849. [Google Scholar] [CrossRef]
- Balic, I.; Vizoso, P.; Nilo-Poyanco, R.; Sanhueza, D.; Olmedo, P.; Sepúlveda, P.; Arriagada, C.; Defilippi, B.G.; Meneses, C.; Campos-Vargas, R. Transcriptome analysis during ripening of table grape berry cv. Thompson Seedless. PLoS ONE 2018, 13, e0190087. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea (Cicer arietinum) metabolome following long-term drought stress. Plant Cell Environ. 2019, 42, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Dong, F.; Lao, X.; Zheng, H. Strategies to Increase the Production of Biosynthetic Riboflavin. Mol. Biotechnol. 2021, 63, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Pompe, N.; Chen, J.; Illarionov, B.; Panter, S.; Fischer, M.; Bacher, A.; Weber, S. Methyl groups matter: Photo-CIDNP characterizations of the semiquinone radicals of FMN and demethylated FMN analogs. J. Chem. Phys. 2019, 151, 235103. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Cör, D.; Knez, Ž.; Knez Hrnčič, M. Antitumour, Antimicrobial, Antioxidant and Antiacetylcholinesterase Effect of Ganoderma Lucidum Terpenoids and Polysaccharides: A Review. Molecules 2018, 23, 649. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.S.; Guo, J.J.; Bao, J.L.; Li, X.W.; Chen, X.P.; Lu, J.J.; Wang, Y.T. Anti-cancer properties of triterpenoids isolated from Ganoderma lucidum—A review. Expert Opin. Investig. Drugs 2013, 22, 981–992. [Google Scholar] [CrossRef]
- Ma, R.; Yang, P.; Jing, C.; Fu, B.; Teng, X.; Zhao, D.; Sun, L. Comparison of the metabolomic and proteomic profiles associated with triterpene and phytosterol accumulation between wild and cultivated ginseng. Plant Physiol. Biochem. 2023, 195, 288–299. [Google Scholar] [CrossRef]
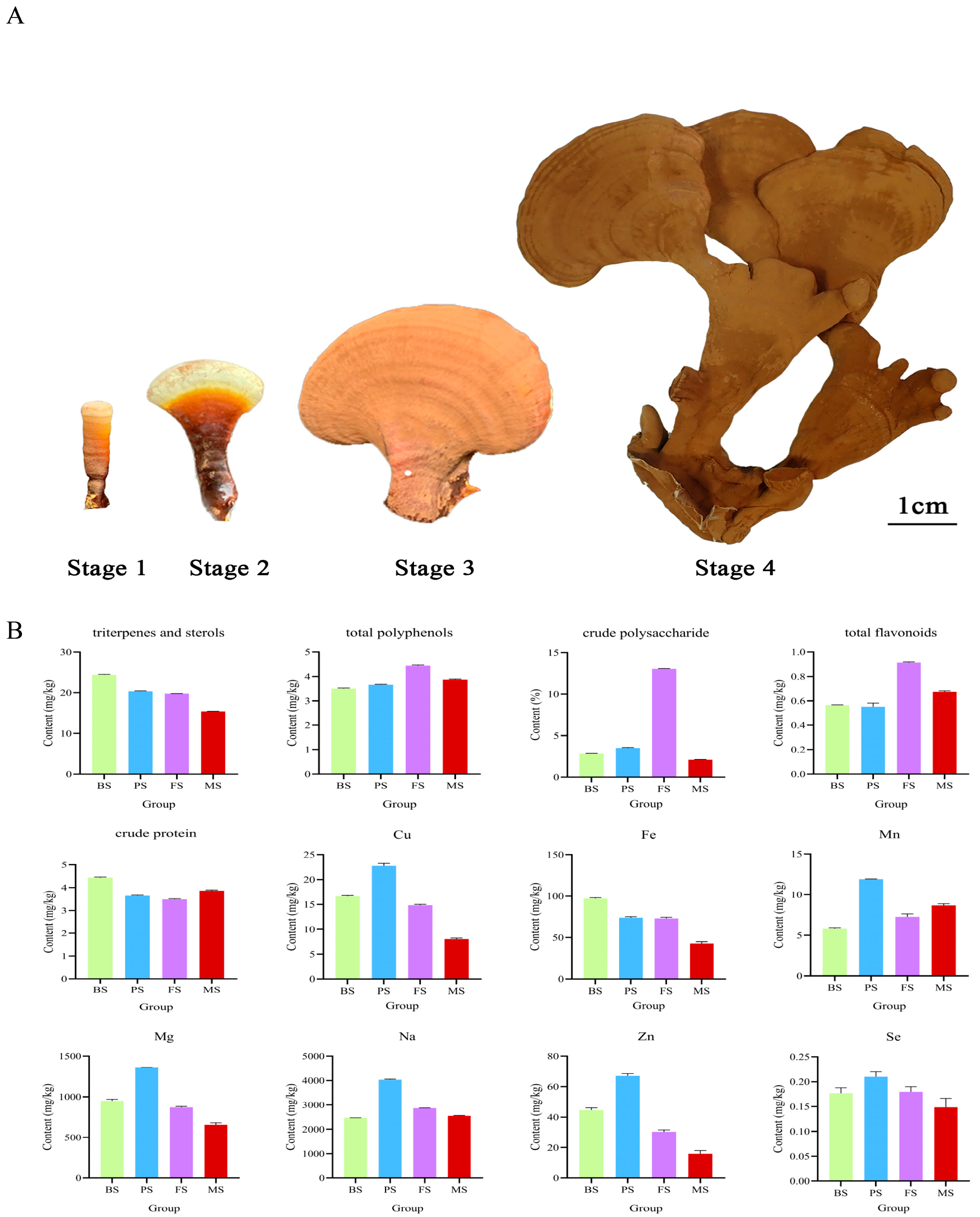
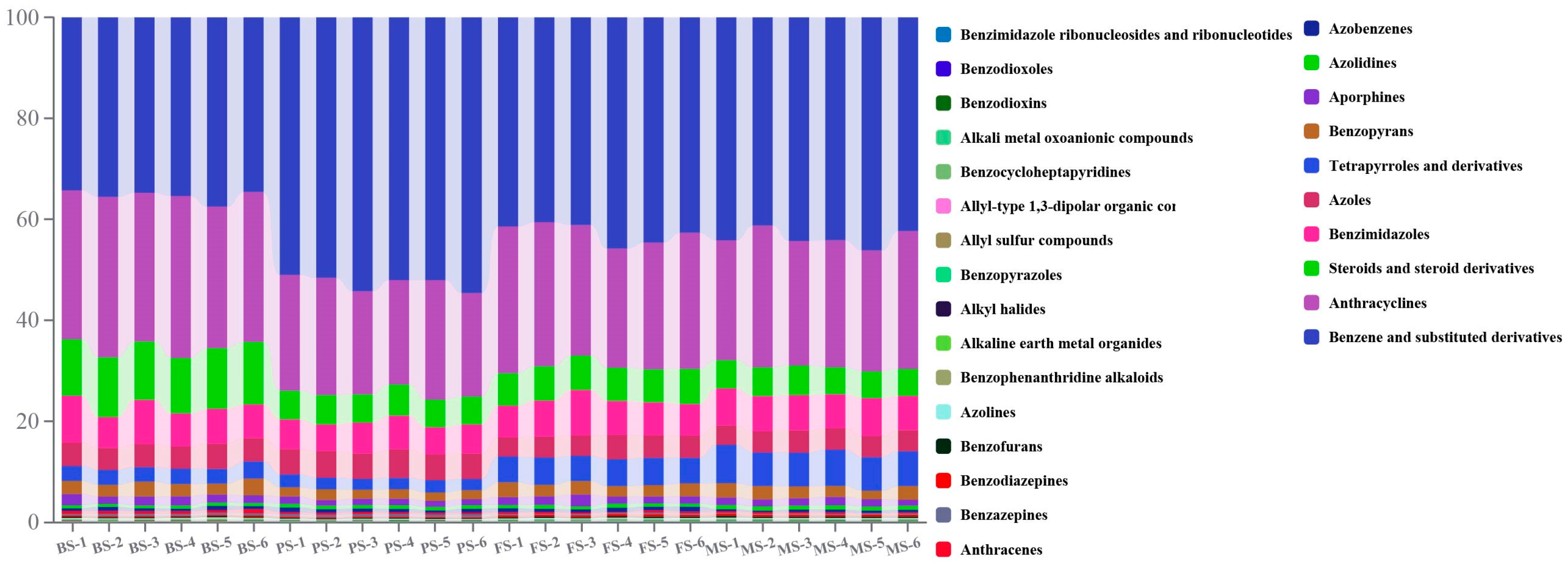
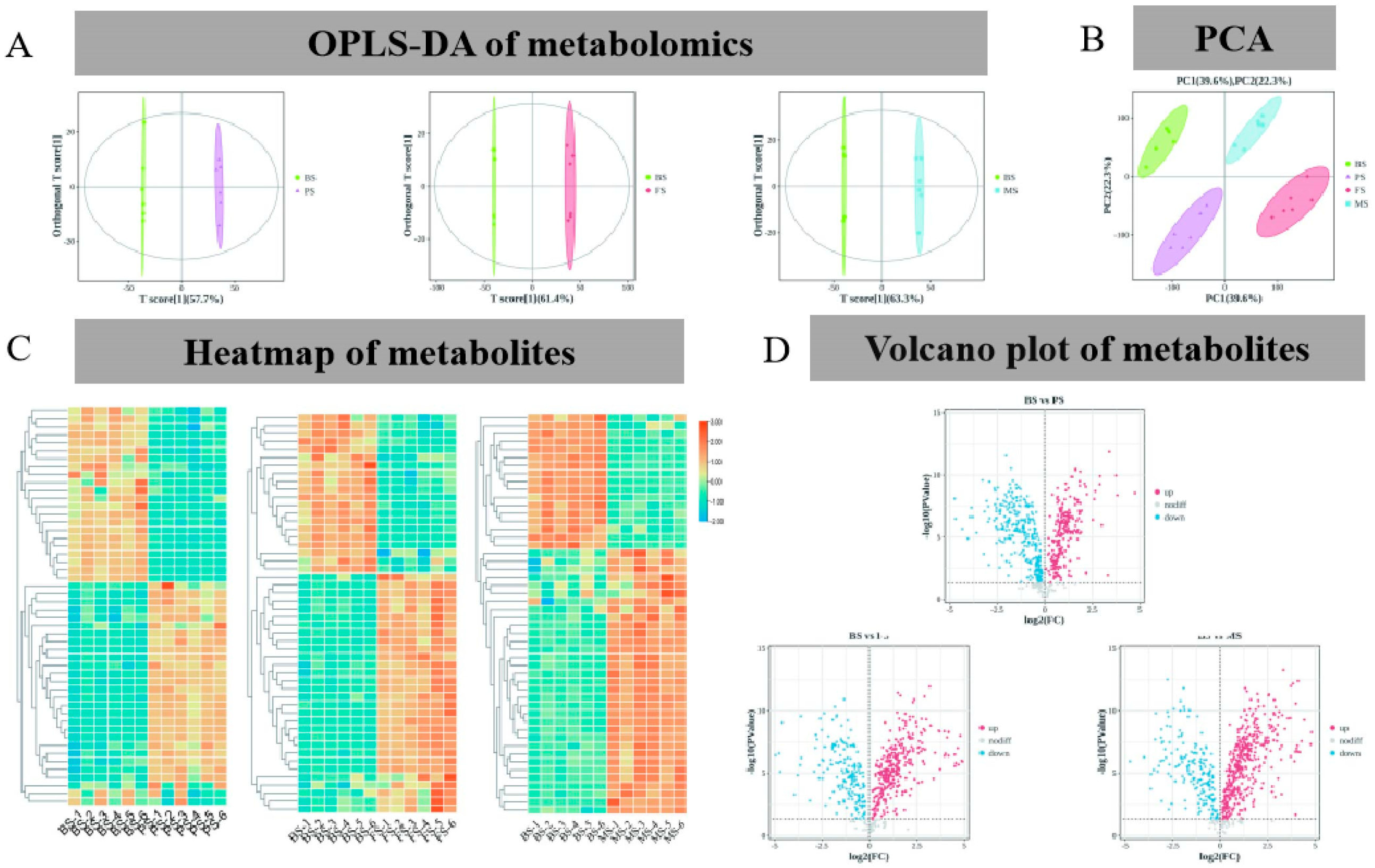

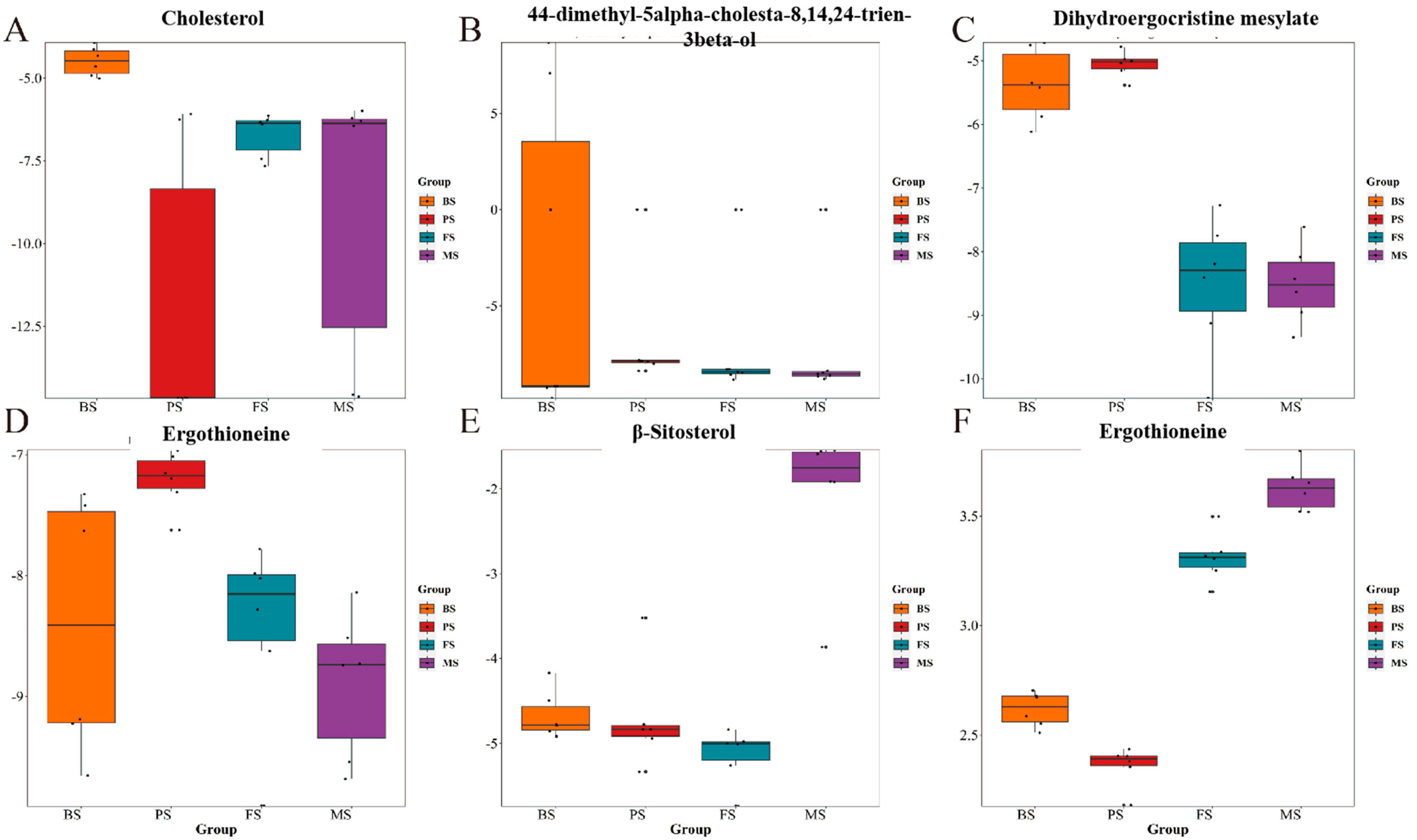



| Group | BS (mg/g) | PS (mg/g) | FS (mg/g) | MS (mg/g) | |
|---|---|---|---|---|---|
| Amino Acid | |||||
| Asp | 1.307 + 0.0036 a | 1.1796 + 0.0009 b | 1.0762 + 0.0006 c | 0.7313 + 0.0023 d | |
| Thr | 0.8141 + 0.0015 a | 0.7759 + 0.0014 b | 0.7324 + 0.001 c | 0.5369 + 0.0017 d | |
| Ser | 0.8563 + 0.0023 a | 0.7623 + 0.001 b | 0.6824 + 0.0004 c | 0.4841 + 0.0011 d | |
| Glu | 2.1855 + 0.0031 a | 2.0083 + 0.0029 b | 1.5459 + 0.0015 c | 0.9228 + 0.0014 d | |
| Gly | 0.7297 + 0.0017 a | 0.6283 + 0.0004 b | 0.5733 + 0.0001 c | 0.3955 + 0.0005 d | |
| Ala | 0.8326 + 0.0016 a | 0.7454 + 0.0022 b | 0.6687 + 0.0009 c | 0.4575 + 0.0005 d | |
| Cys | 0.1502 + 0.003 a | 0.1375 + 0.0006 b | 0.1339 + 0.0008 c | 0.1186 + 0.0011 d | |
| Val | 0.7861 + 0.0026 a | 0.6959 + 0.0026 b | 0.6491 + 0.001 c | 0.4561 + 0.0015 d | |
| Met | 3.6717 + 0.004 a | 3.457 + 0.005 b | 3.9251 + 0.0035 c | 4.285 + 0.0046 d | |
| Ile | 0.5833 + 0.0027 a | 0.5098 + 0.0009 b | 0.501 + 0.0016 c | 0.3549 + 0.0017 d | |
| Leu | 1.2311 + 0.0044 a | 1.4158 + 0.0013 b | 1.3719 + 0.0033 c | 1.1637 + 0.0017 d | |
| Tyr | 0.3854 + 0.0021 a | 0.3278 + 0.0014 b | 0.2957 + 0.0026 c | 0.1908 + 0.0008 d | |
| Phe | 0.6179 + 0.0025 a | 0.6574 + 0.0006 b | 0.5976 + 0.0018 c | 0.4198 + 0.0016 d | |
| Lys | 0.7186 + 0.0035 a | 0.6813 + 0.0019 b | 0.6279 + 0.0011 c | 0.3673 + 0.0219 d | |
| His | 0.273 + 0.0017 a | 0.2654 + 0.0012 b | 0.2286 + 0.001 c | 0.1435 + 0.0006 d | |
| Arg | 0.7021 + 0.003 a | 0.643 + 0.0014 b | 0.5532 + 0.0032 c | 0.3115 + 0.0027 d | |
| pro | 0.6063 + 0.0046 a | 0.5379 + 0.003 b | 0.4886 + 0.0038 c | 0.3205 + 0.0029 d | |
| Metabolite Category | BS–PS | BS–FS | BS–MS | |||
|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |
| Fatty Acyls | 27 | 4 | 19 | 3 | 22 | 5 |
| Benzene and substituted derivatives | 18 | 14 | 24 | 11 | 30 | 12 |
| Carboxylic acids and derivatives | 15 | 17 | 34 | 10 | 36 | 7 |
| Organooxygen compounds | 9 | 12 | 5 | 6 | 38 | 8 |
| Glycerolipids | 8 | 6 | 2 | 15 | 8 | 14 |
| Prenol lipids | 7 | 13 | 10 | 12 | 16 | 10 |
| Organonitrogen compounds | 6 | 12 | 5 | 6 | 4 | 8 |
| Flavonoids | 5 | 4 | 3 | 4 | 5 | 4 |
| Purine nucleosides | 4 | 8 | 9 | 4 | 10 | 4 |
| Others | 135 | 214 | 214 | 118 | 231 | 122 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Huo, H.; Bao, H.; Wang, J.; Gao, D. Changes of Active Substances in Ganoderma lucidum during Different Growth Periods and Analysis of Their Molecular Mechanism. Molecules 2024, 29, 2591. https://doi.org/10.3390/molecules29112591
Gao X, Huo H, Bao H, Wang J, Gao D. Changes of Active Substances in Ganoderma lucidum during Different Growth Periods and Analysis of Their Molecular Mechanism. Molecules. 2024; 29(11):2591. https://doi.org/10.3390/molecules29112591
Chicago/Turabian StyleGao, Xusheng, Huimin Huo, Haiying Bao, Jialu Wang, and Dan Gao. 2024. "Changes of Active Substances in Ganoderma lucidum during Different Growth Periods and Analysis of Their Molecular Mechanism" Molecules 29, no. 11: 2591. https://doi.org/10.3390/molecules29112591







