Surface-Enhanced Raman Spectroscopy (SERS)-Based Sensors for Deoxyribonucleic Acid (DNA) Detection
Abstract
:1. Introduction
1.1. Raman Spectroscopy and SERS
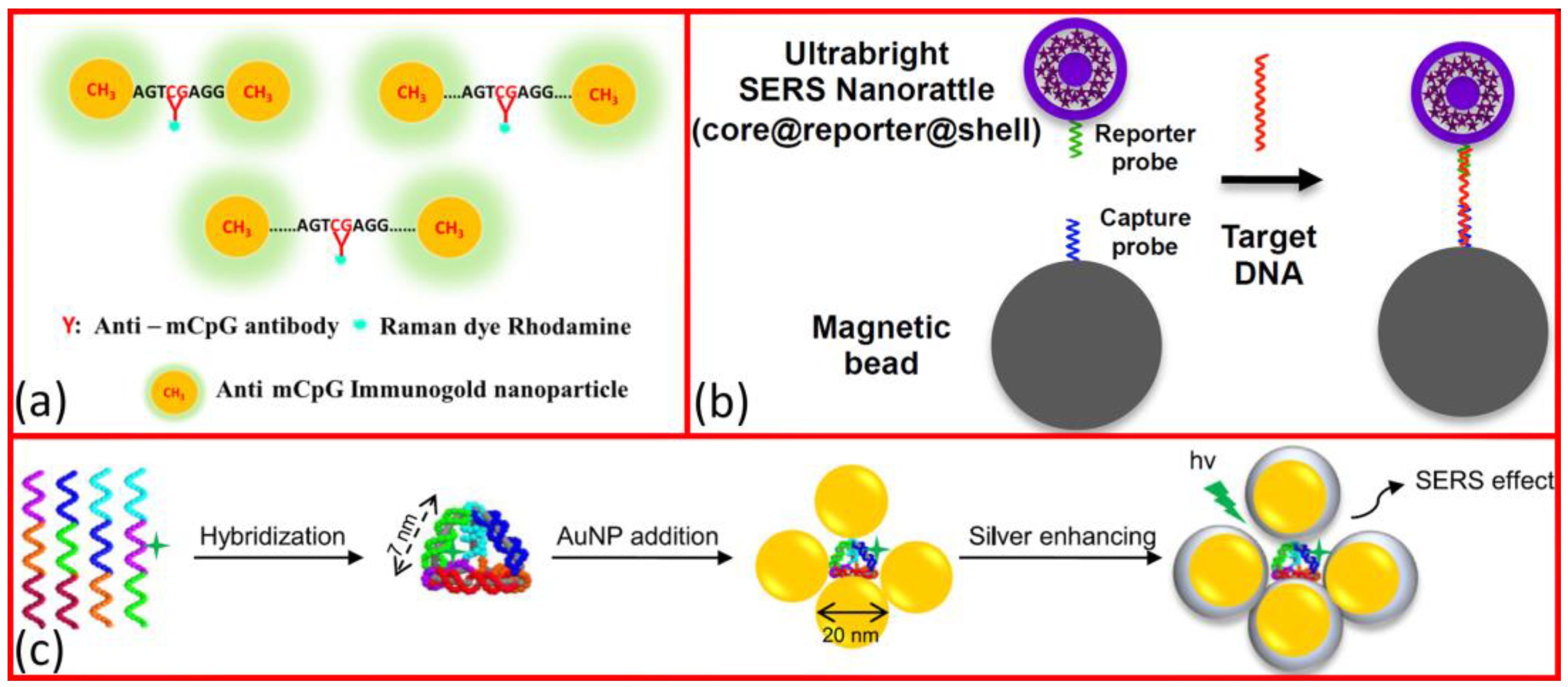
1.2. SERS-Based DNA Sensing
1.3. Factors Affecting DNA Sensing Performance with SERS
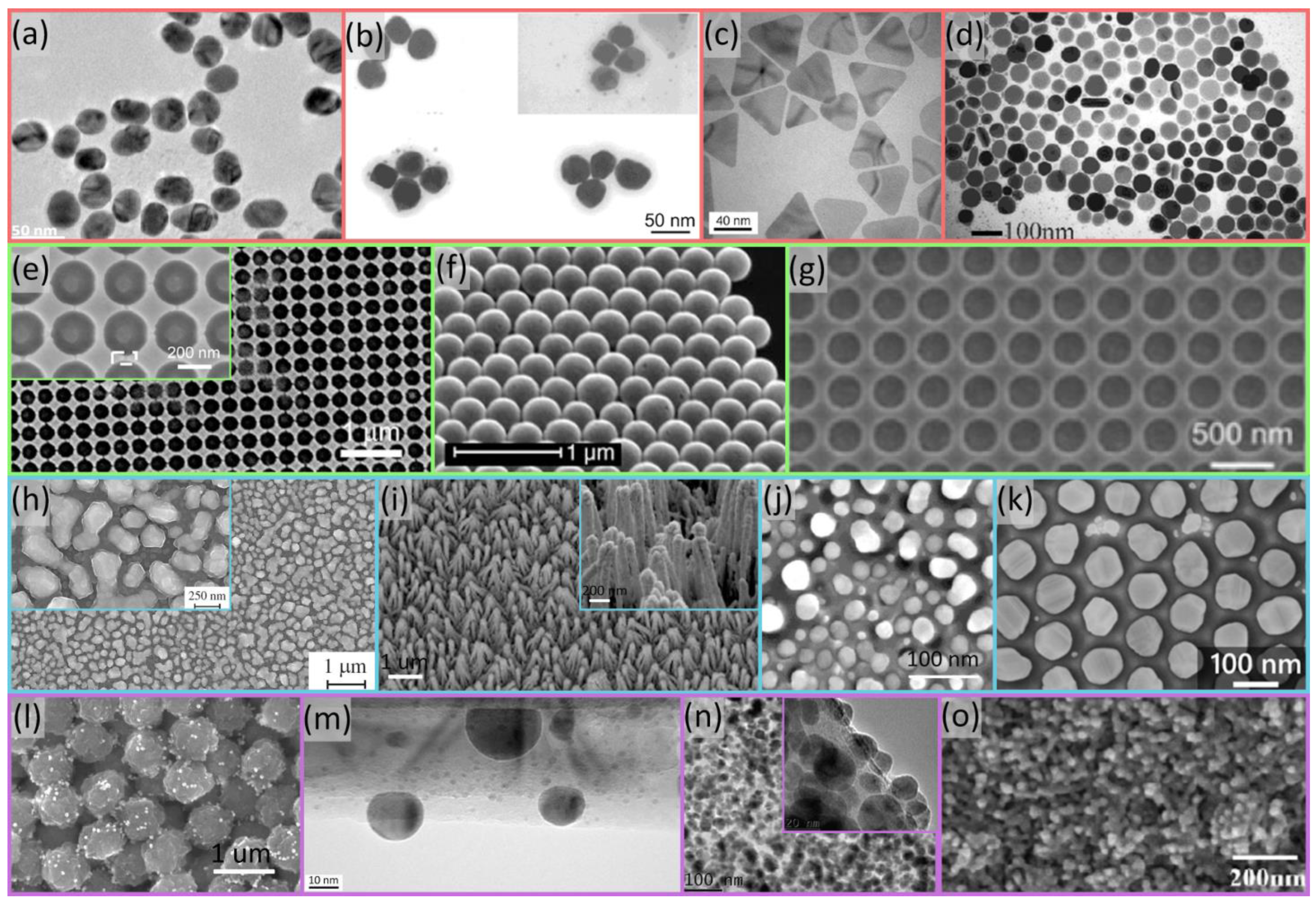
2. SERS Substrate Fabrication Technologies
2.1. Chemical Methods
2.2. Self-Assembly
2.3. Physical Methods
2.4. Hybrid Nanostructures
3. Conditions Affecting SERS Measurements
4. Designing SERS Experiments for DNA Detection
| Ref | Material Type | Shape/ Structure | Size/ Geometry | Fabrication Technology | DNA Concentration Range/Value | Limit of Detection (LOD) | Target Type, Length, Raman Label | Enhancement Factor (EF) | SERS Parameters: Wavelength (λ), Power (p), and Acquisition Time (t) |
|---|---|---|---|---|---|---|---|---|---|
| [47] | Au | nanoplate | edge length = 134 ± 6 nm, density = 916 ± 40 GNPs/spot | Wet chemical synthesis (seed-mediated growth) of nanoplates + self-assembly on glass | 10−2–10−6 mg/mL | 10−6 mg/mL | nucleobases (A,T,G,C) label-free | 5.4 × 107 | λ = 785 nm, p = 9.5 mW |
| [50] | Au + Ag | nanoprism/ nanosphere | Ag nanoprism edge length = 110 ± 10 nm Au nanosphere dimeter = 25 ± 5 nm | Wet chemical synthesis (seed-mediated growth) of Ag nanoprism + hybridization with AuNP-modified DNA | 10−8–10−11 M | 10−11 M | ss-DNA 30 bases Raman labels: DTNB and 4-MBA | - | λ = 632.8 nm, p = 2.3 mW |
| [53] | Au + Ag coated GaN | nanopillars | thickness = 90 nm | GaN epitaxial growth (MOCVD) + photo-etching + physical deposition (Au + Ag) + dealloying | 10−5 M | - | ss-DNA 22 bases Raman label: MGITC | 1 × 107 | λ = 632.8 nm, p = 5 mW, t = 10–30 s |
| [56] | Au | nanosphere dimers | gap = 1 nm, dimeter = 100 nm | Nanotrench-guided self-assembly on patterned Si | - | 10−11 M | ss-DNA 8 bases label-free | 1011 | λ = 632.8 nm, p = 8.2 mW, t = 0.5 s |
| [49] | Ag | nanospheres (self-similar chains) | d1 = 148 ± 30 nm, d2 = 64 ± 6 nm, d3 = 27 ± 5 nm, gap23 = 10 nm, gap12 = 31 nm | E-beam lithography on Si + electroless Ag deposition | 10−8 M | - | ss-DNA 6–9 bases label-free | 1012 | λ = 514 nm, p = 0.012 mW |
| [57] | Au | nanospheres | dimeter = 60 nm, gap ≅ 5 nm | Wet chemical synthesis + DNA-controlled aggregation | - | 10−7 M | ss-DNA (12 bases) ds-DNA (24 bases) label-free | 2.2 × 105 | λ = 632.8 nm, p = 10 mW |
| [58] | Au + GO + Ag | AuNPs@ GO mesh@ AgNPs | AuNP diameter = 40 nm; AgNP diameter = 50 nm | Chemical reduction of hAuCl4 on GO/MoS2 + chemical synthesis and deposition of Ag NPs | - | 10−13 M | ss-DNA (9 bases) ds-DNA (12 bases) label-free | 4.2 × 108 | λ = 532 nm, p = 0.5 mW |
| [59] | Au + Ag on Si | Nanogaps | gaps = 15 ± 10 nm, Ag thickness = 30 nm, Au thickness = 15 nm | Photolithography + reactive ion etching for the Si substrate + physical deposition (Au, Ag) | 10−8–10−12 M | 10−12 M (using RB label) | PNA-DNA complex (22 bases) Raman label: RB | - | λ = 785 nm, p = 300 mW |
| [43] | Ag | Ag NPs on AAO substrate | Ag NP diameter = 20–50 nm, AAO pore size = 20–40 nm, gap = 10–20 nm | Anodic oxidation and wet chemical etching (AAO) + Ag electrochemical plating | 104–10−4 ppm | 10−3 ppm | nucleobases (T,G,C) label-free | 1.9 × 108 | λ = 632.8 nm, p = - t = 60 s |
| [60] | Au + Ag | Au@Ag core–shell structures | Au nanosphere diameter = 20 nm Au@Ag core–shell diameter = 35 nm | Wet chemical synthesis + DNA self-assembly into nanopyramids | 10−9 M | - | ds-DNA (DNA-pyramid) Raman label: Cy3 | - | λ = 514.5 nm, p = 20 mW, t = 10 s |
| [61] | Au on Si | coated nanopillar | Si nanopillar height and diameter: 500 nm × 100 nm Au layer thickness: 200 nm (2D arrays 4 mm × 4 mm) | Reactive ion etching of Si (randomly distributed nanopillars) + physical deposition (Au) | 5 × 10−6 M | - | ss-DNA, 75 bases, label-free | - | λ = 780 nm, p = 0.1 mW, t = 1 s |
| [48] | Ag | iodide-modified Ag nanospheres | diameter = 50 nm | Wet chemical synthesis + iodide-modification | 3.5 × 10−6 M | - | ss-DNA and ds-DNA (10–51 bases) label-free | - | λ = 532 nm, p = 5 mW, t = 10 s |
| [86] | Au | nanospheres | diameter = 15 nm | Wet chemical synthesis (agarose-stabilized nanoparticles) | 10−4 M | - | nucleosides (dA, dT, dCMP, and dGMP) label-free | - | λ = 785 nm, p = 5 mW |
| [39] | Au | Au nanoshell on glass sphere | core diameter = 120 nm | Stöber method for silica nanoparticles, wet chemical synthesis (AuNPs) + surface chemistry to create nanoshells | 4 × 10−5 M | - | ss-DNA and ds-DNA (20–70 bases) label-free | - | λ = 785 nm, p = 0.57 mW, t = 20 s |
| [79] | Au + Ag | nanorattles (Au–Ag porous cages) | diameter ≅ 60 nm | Wet chemical synthesis (seed-mediated growth, galvanic replacement) | 10−9–10−13 M | 3 × 10−12 M | ss-DNA and ds-DNA (25–80 bases) Raman label: HITC | - | λ = 785 nm, p = 300 mW, t = 1 s |
| [81] | Ag | Ag NP + GO nano- composite | Ag NP diameter = 57.5 nm | Wet chemical synthesis (Ag NP), modified Hummers’ method (GO nanosheets) | 10−6–10−12 M | 10−12 M | ds-DNA, 20 bases, Raman label: 4-MBA, TP | - | λ = 532 nm, p = 1.5 mW, t = 3 s |
| [80] | Ag + Si | Si NW + Ag NP | Si NW length = 2.4 μm, diameter = 20–60 nm, Ag NP diameter = 40 nm | Metal-assisted chemical etching (Si NWs), Ag thermal evaporation, and pulsed laser ablation for Ag NPs | 10−10–10−15 M | 10−15 M | ss-DNA 25 bases label-free | ~106 | λ = 488 nm, p = 100 μW, t = 10 s |
| [73] | Au + Ag | Ag NW + AuNP | Ag NW length = 24 μm, diameter = 121 nm, Au NP diameter = ~35 nm | Wet chemical synthesis for both Ag NWs and AuNPs | 10−9 mg/mL – 1.8 × 10−11 mg/mL | 1.8 × 10−11 mg/mL | ds-DNA (long oligomer) Raman label: R6G | - | λ = 785 nm, p = 8.5 mW t = 0.3 s |
| [119] | Ag | Ag DIANPs | Ag NP diameter ≅ 100 nm | Chemical synthesis: DCM-modified Ag IANPs (DIANPs) | 1 × 10−6 M | - | ss-DNA and ds-DNA (10–50 bases) label-free | - | λ = 633 nm, p = - t = 10 s |
| [32] | Ag | AgZNPs | Ag NP diameter ≅ 36 nm | Chemical synthesis: Zr ion-modified Ag NPs (AgZNPs) | 1 × 10−5 M | - | ss-DNA (12–35 bases) label-free | - | λ = 633 nm, p = 20 mW t = 30 s |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-Enhanced Raman Spectroscopy: Benefits, Trade-Offs and Future Developments. Chem. Sci. 2020, 11, 4563–4577. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, J.; Li, D. Applications of Raman Spectroscopy in Detection of Water Quality. Appl. Spectrosc. Rev. 2016, 51, 333–357. [Google Scholar] [CrossRef]
- Popp, J.; Mayerhöfer, T. (Eds.) Micro-Raman Spectroscopy; De Gruyter: Berlin, Germany, 2020; ISBN 9783110515312. [Google Scholar]
- Mao, P.; Liu, C.; Favraud, G.; Chen, Q.; Han, M.; Fratalocchi, A.; Zhang, S. Broadband Single Molecule SERS Detection Designed by Warped Optical Spaces. Nat. Commun. 2018, 9, 5428. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-F.; Huang, Y.-F.; Duan, S.; Pang, R.; Wu, D.-Y.; Ren, B.; Xu, X.; Tian, Z.-Q. SERS and DFT Study of Water on Metal Cathodes of Silver, Gold and Platinum Nanoparticles. Phys. Chem. Chem. Phys. 2010, 12, 2493. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Emory, S.R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 1997, 275, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Polubotko, A.M.; Chelibanov, V.P. The Theory of SERS on Semiconductor and Dielectric Substrates. Opt. Spectrosc. 2017, 122, 937–943. [Google Scholar] [CrossRef]
- Plieth, W.; Dietz, H.; Anders, A.; Sandmann, G.; Meixner, A.; Weber, M.; Kneppe, H. Electrochemical Preparation of Silver and Gold Nanoparticles: Characterization by Confocal and Surface Enhanced Raman Microscopy. Surf. Sci. 2005, 597, 119–126. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface Raman Spectroelectrochemistry. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef]
- Albrecht, M.G.; Creighton, J.A. Anomalously Intense Raman Spectra of Pyridine at a Silver Electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. [Google Scholar] [CrossRef]
- Etchegoin, P.G.; Galloway, C.; Le Ru, E.C. Polarization-Dependent Effects in Surface-Enhanced Raman Scattering (SERS). Phys. Chem. Chem. Phys. 2006, 8, 2624. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Lin, X.; Jiang, C.; Li, C.; Lin, H.; Huang, J.; Wang, S.; Liu, G.; Yan, X.; Zhong, Q.; et al. Reliable Quantitative SERS Analysis Facilitated by Core–Shell Nanoparticles with Embedded Internal Standards. Angew. Chem. 2015, 127, 7416–7420. [Google Scholar] [CrossRef]
- Shi, R.; Liu, X.; Ying, Y. Facing Challenges in Real-Life Application of Surface-Enhanced Raman Scattering: Design and Nanofabrication of Surface-Enhanced Raman Scattering Substrates for Rapid Field Test of Food Contaminants. J. Agric. Food Chem. 2018, 66, 6525–6543. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and Applications of SERS-Based Bioanalytical Sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef]
- Rahman, M.M.; Ali, M.E.; Abd Hamid, S.B. Gold Nanoparticles—An Enhanced DNA Sensing Tools Using Surface Enhance Raman Scattering. Adv. Mat. Res. 2015, 1109, 439–443. [Google Scholar] [CrossRef]
- Zhang, Y.; Chu, W.; Foroushani, A.; Wang, H.; Li, D.; Liu, J.; Barrow, C.; Wang, X.; Yang, W. New Gold Nanostructures for Sensor Applications: A Review. Materials 2014, 7, 5169–5201. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, V.; Sahbafar, H.; Zeinalizad, L.; Hadi, A. A Review of Applications of Surface-Enhanced Raman Spectroscopy Laser for Detection of Biomaterials and a Quick Glance into Its Advances for COVID-19 Investigations. ISSS J. Micro Smart Syst. 2022, 11, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, M.; Huang, Q. A Review of Aptamer-Based SERS Biosensors: Design Strategies and Applications. Talanta 2021, 227, 122188. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhu, C.; Meng, G.; Wu, N. Review—Surface-Enhanced Raman Scattering Sensors for Food Safety and Environmental Monitoring. J. Electrochem. Soc. 2018, 165, B3098–B3118. [Google Scholar] [CrossRef]
- Celis, F.; Garcia, M.; Diaz-Fleming, G.; Campos-Vallette, M. A Review of Raman, Surface-Enhanced Raman Scattering (Sers) and Related Spectroscopic Techniques Applied to Biomolecules in Biomaterials. J. Chil. Chem. Soc. 2017, 62, 3627–3632. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Aldeanueva-Potel, P.; Mateo-Mateo, C.; Rodríguez-Lorenzo, L.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M. Surface-Enhanced Raman Scattering Biomedical Applications of Plasmonic Colloidal Particles. J. R. Soc. Interface 2010, 7 (Suppl. 4), S435–S450. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Andrade, G.F.S.; Brolo, A.G. A Review on the Fabrication of Substrates for Surface Enhanced Raman Spectroscopy and Their Applications in Analytical Chemistry. Anal. Chim. Acta 2011, 693, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Willets, K.A.; Van Duyne, R.P. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mi, X.; Tan, X.; Xiang, R. Recent Progress on Liquid Biopsy Analysis Using Surface-Enhanced Raman Spectroscopy. Theranostics 2019, 9, 491–525. [Google Scholar] [CrossRef] [PubMed]
- Harper, M.M.; Dougan, J.A.; Shand, N.C.; Graham, D.; Faulds, K. Detection of SERS Active Labelled DNA Based on Surface Affinity to Silver Nanoparticles. Analyst 2012, 137, 2063–2068. [Google Scholar] [CrossRef] [PubMed]
- Braun, G.; Seung, J.L.; Dante, M.; Nguyen, T.Q.; Moskovits, M.; Reich, N. Surface-Enhanced Raman Spectroscopy for DNA Detection by Nanoparticle Assembly onto Smooth Metal Films. J. Am. Chem. Soc. 2007, 129, 6378–6379. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Ren, X.; Obata, K.; Ito, Y.; Sugioka, K. Label-Free Trace Detection of Bio-Molecules by Liquid-Interface Assisted Surface-Enhanced Raman Scattering Using a Microfluidic Chip. Opto-Electron. Adv. 2022, 5, 210121-1–210121-10. [Google Scholar] [CrossRef]
- Freeman, L.M.; Pang, L.; Fainman, Y. Self-Reference and Random Sampling Approach for Label-Free Identification of DNA Composition Using Plasmonic Nanomaterials. Sci. Rep. 2018, 8, 7398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Quan, X.; Cao, N.; Zhang, Z.; Li, Y. Label-Free Detection of DNA via Surface-Enhanced Raman Spectroscopy Using Au@Ag Nanoparticles. Nanomaterials 2022, 12, 3119. [Google Scholar] [CrossRef]
- Mahmood, M.H.; Jaafar, A.; Himics, L.; Péter, L.; Rigó, I.; Zangana, S.; Bonyár, A.; Veres, M. Nanogold-Capped Poly(DEGDMA) Microparticles as Surface-Enhanced Raman Scattering Substrates for DNA Detection. J. Phys. D Appl. Phys. 2022, 55, 405401. [Google Scholar] [CrossRef]
- .Zhang, Y.; Zhan, D.; Xu, X.; Zhang, Z.; Hafez, M.; He, Y.; Li, Y.; Li, D. Label-free detection of DNA methylation by surface-enhanced Raman spectroscopy using zirconium-modified silver nanoparticles. Talanta 2023, 253, 123941. [Google Scholar] [CrossRef]
- Treffer, R.; Lin, X.; Bailo, E.; Deckert-Gaudig, T.; Deckert, V. Distinction of Nucleobases—A Tip-Enhanced Raman Approach. Beilstein J. Nanotechnol. 2011, 2, 628–637. [Google Scholar] [CrossRef]
- Sagar, D.M.; Korshoj, L.E.; Hanson, K.B.; Chowdhury, P.P.; Otoupal, P.B.; Chatterjee, A.; Nagpal, P. High-Throughput Block Optical DNA Sequence Identification. Small 2018, 14, 1703165. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, H.J.; Kim, J.H.; Chang, B.; Park, H.-K. Label-Free Detection for a DNA Methylation Assay Using Raman Spectroscopy. Chin. Med. J. 2017, 130, 1961–1967. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Chica, A.J. On the Interpretation of Raman Spectra of 1-Aminooxy-Spermine/DNA Complexes. Nucleic Acids Res. 2004, 32, 579–589. [Google Scholar] [CrossRef] [PubMed]
- AW, A.; JC, T. Double-Stranded DNA Damage Assessed with Raman Spectroscopy. Biochem. Anal. Biochem. 2016, 5, 3. [Google Scholar] [CrossRef]
- De Angelis, A.; Ferrara, M.A.; Di Caprio, G.; Managò, S.; Sirleto, L.; Coppola, G.; De Luca, A.C. Spermatozoa Quality Assessment: A Combined Holographic and Raman Microscopy Approach. In Proceedings of the SPIE 9529, 21–25 June 2015, Munich, Germany, Optical Methods for Inspection, Characterization, and Imaging of Biomaterials II; Ferraro, P., Grilli, S., Ritsch-Marte, M., Stifter, D., Eds.; SPIE: Bellingham, DC, USA, 2015; p. 952916. [Google Scholar] [CrossRef]
- Barhoumi, A.; Zhang, D.; Tam, F.; Halas, N.J. Surface-Enhanced Raman Spectroscopy of DNA. J. Am. Chem. Soc. 2008, 130, 5523–5529. [Google Scholar] [CrossRef]
- Otto, C.; van den Tweel, T.J.J.; de Mul, F.F.M.; Greve, J. Surface-enhanced Raman Spectroscopy of DNA Bases. J. Raman Spectrosc. 1986, 17, 289–298. [Google Scholar] [CrossRef]
- Pyrak, E.; Jaworska, A.; Kudelski, A. SERS Studies of Adsorption on Gold Surfaces of Mononucleotides with Attached Hexanethiol Moiety: Comparison with Selected Single-Stranded Thiolated DNA Fragments. Molecules 2019, 24, 3921. [Google Scholar] [CrossRef]
- Szekeres, G.P.; Kneipp, J. SERS Probing of Proteins in Gold Nanoparticle Agglomerates. Front. Chem. 2019, 7, 30. [Google Scholar] [CrossRef]
- Chan, T.-Y.; Liu, T.-Y.; Wang, K.-S.; Tsai, K.-T.; Chen, Z.-X.; Chang, Y.-C.; Tseng, Y.-Q.; Wang, C.-H.; Wang, J.-K.; Wang, Y.-L. SERS Detection of Biomolecules by Highly Sensitive and Reproducible Raman-Enhancing Nanoparticle Array. Nanoscale Res. Lett. 2017, 12, 344. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.; Lin, B.-Y. Silver SERS Adenine Sensors with a Very Low Detection Limit. Biosensors 2020, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Madzharova, F.; Heiner, Z.; Gühlke, M.; Kneipp, J. Surface-Enhanced Hyper-Raman Spectra of Adenine, Guanine, Cytosine, Thymine, and Uracil. J. Phys. Chem. C 2016, 120, 15415–15423. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Bi, L.; Rao, Y.; Tao, Q.; Dong, J.; Su, T.; Liu, F.; Qian, W. Fabrication of Large-Scale Gold Nanoplate Films as Highly Active SERS Substrates for Label-Free DNA Detection. Biosens. Bioelectron. 2013, 43, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.J.; Lei, Z.C.; Li, J.; Zong, C.; Yang, C.J.; Ren, B. Label-Free Surface-Enhanced Raman Spectroscopy Detection of DNA with Single-Base Sensitivity. J. Am. Chem. Soc. 2015, 137, 5149–5154. [Google Scholar] [CrossRef] [PubMed]
- Coluccio, M.L.; Gentile, F.; Das, G.; Perozziello, G.; Malara, N.; Alrasheed, S.; Candeloro, P.; Fabrizio, E.D. From Nucleotides to DNA Analysis by a SERS Substrate of a Self Similar Chain of Silver Nanospheres. J. Opt. 2015, 17, 114021. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Z.; Zong, S.; Zhang, R.; Zhu, D.; Xu, S.; Wang, C.; Cui, Y. SERS-Based DNA Detection in Aqueous Solutions Using Oligonucleotide- Modified Ag Nanoprisms and Gold Nanoparticles. Anal. Bioanal. Chem. 2013, 405, 6131–6136. [Google Scholar] [CrossRef]
- Huang, J.A.; Mousavi, M.Z.; Zhao, Y.; Hubarevich, A.; Omeis, F.; Giovannini, G.; Schütte, M.; Garoli, D.; De Angelis, F. SERS Discrimination of Single DNA Bases in Single Oligonucleotides by Electro-Plasmonic Trapping. Nat. Commun. 2019, 10, 5321. [Google Scholar] [CrossRef]
- Macdonald, D.; Smith, E.; Faulds, K.; Graham, D. DNA Detection by SERS: Hybridisation Parameters and the Potential for Asymmetric PCR. Analyst 2020, 145, 1871–1877. [Google Scholar] [CrossRef]
- Kamińska, A.; Sivanesan, A.; Witkowska, E.; Gołąb, J.; Winiarska, M.; Nowis, D.; Dzięcielewski, I.; Weyher, J.L.; Waluk, J. Detection of DNA Mutations Using Novel SERS (Surface-Enhanced Raman Spectroscopy) Diagnostic Platform. J. Chem. Chem. Eng. 2013, 7, 972–978. [Google Scholar]
- Eremina, O.E.; Zatsepin, T.S.; Farzan, V.M.; Veselova, I.A.; Zvereva, M.I. DNA Detection by Dye Labeled Oligonucleotides Using Surface Enhanced Raman Spectroscopy. Mendeleev Commun. 2020, 30, 18–21. [Google Scholar] [CrossRef]
- Lin, D.; Wu, Q.; Qiu, S.; Chen, G.; Feng, S.; Chen, R.; Zeng, H. Label-Free Liquid Biopsy Based on Blood Circulating DNA Detection Using SERS-Based Nanotechnology for Nasopharyngeal Cancer Screening. Nanomedicine 2019, 22, 102100. [Google Scholar] [CrossRef]
- Maruoka, K.; Sugano, K.; Isono, Y. SERS Detection and Analysis of a Single DNA Oligomer Using a Single Gold Nanoparticle Dimer. In Proceedings of the 2017 19th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS), Kaohsiung, Taiwan, 18–22 June 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 468–471. [Google Scholar]
- Caprara, D.; Ripanti, F.; Capocefalo, A.; Sarra, A.; Brasili, F.; Petrillo, C.; Fasolato, C.; Postorino, P. DNA-Functionalized Gold Nanoparticle Assemblies for Surface Enhanced Raman Scattering. Colloids Surf. A Physicochem. Eng. Asp. 2020, 589, 124399. [Google Scholar] [CrossRef]
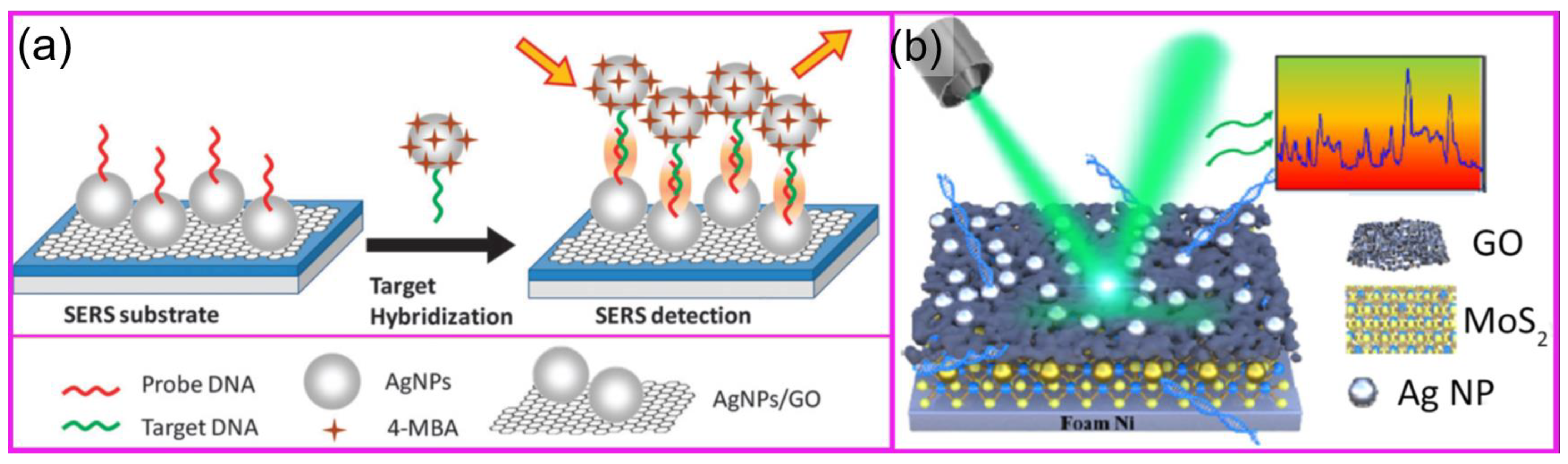
- Gao, X.; Zhang, H.; Fan, X.; Zhang, C.; Sun, Y.; Liu, C.; Li, Z.; Jiang, S.; Man, B.; Yang, C. Toward the Highly Sensitive SERS Detection of Bio-Molecules: The Formation of a 3D Self-Assembled Structure with a Uniform GO Mesh between Ag Nanoparticles and Au Nanoparticles. Opt. Express 2019, 27, 25091. [Google Scholar] [CrossRef]
- Fang, C.; Agarwal, A.; Buddharaju, K.D.; Khalid, N.M.; Salim, S.M.; Widjaja, E.; Garland, M.V.; Balasubramanian, N.; Kwong, D.L. DNA Detection Using Nanostructured SERS Substrates with Rhodamine B as Raman Label. Biosens. Bioelectron. 2008, 24, 216–221. [Google Scholar] [CrossRef]
- Keum, J.W.; Kim, M.; Park, J.M.; Yoo, C.; Huh, N.; Park, S.C. DNA-Directed Self-Assembly of Three-Dimensional Plasmonic Nanostructures for Detection by Surface-Enhanced Raman Scattering (SERS). Sens. Biosensing Res. 2014, 1, 21–25. [Google Scholar] [CrossRef]
- Frøhling, K.B.; Alstrøm, T.S.; Bache, M.; Schmidt, M.S.; Schmidt, M.N.; Larsen, J.; Jakobsen, M.H.; Boisen, A. Surface-Enhanced Raman Spectroscopic Study of DNA and 6-Mercapto-1-Hexanol Interactions Using Large Area Mapping. Vib. Spectrosc. 2016, 86, 331–336. [Google Scholar] [CrossRef]
- Le Ru, E.C.; Blackie, E.; Meyer, M.; Etchegoin, P.G. Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study. J. Phys. Chem. C 2007, 111, 13794–13803. [Google Scholar] [CrossRef]
- Xiong, Y.; Washio, I.; Chen, J.; Sadilek, M.; Xia, Y. Trimeric Clusters of Silver in Aqueous AgNO3 Solutions and Their Role as Nuclei in Forming Triangular Nanoplates of Silver. Angew. Chem.-Int. Ed. 2007, 46, 4917–4921. [Google Scholar] [CrossRef]
- Hoppe, C.E.; Lazzari, M.; Pardiñas-Blanco, I.; López-Quintela, M.A. One-Step Synthesis of Gold and Silver Hydrosols Using Poly(N-Vinyl-2- Pyrrolidone) as a Reducing Agent. Langmuir 2006, 22, 7027–7034. [Google Scholar] [CrossRef] [PubMed]
- Métraux, G.S.; Mirkin, C.A. Rapid Thermal Synthesis of Silver Nanoprisms with Chemically Tailorable Thickness. Adv. Mater. 2005, 17, 412–415. [Google Scholar] [CrossRef]
- Brennan, M.E.; Whelan, A.M.; Kelly, J.M.; Blau, W.J. Silver Nanoparticle Self-Organization into Dendritic Fractals. Synth. Met. 2005, 154, 205–208. [Google Scholar] [CrossRef]
- Sun, Y.; Mayers, B.; Xia, Y. Transformation of Silver Nanospheres into Nanobelts and Triangular Nanoplates through a Thermal Process. Nano Lett. 2003, 3, 675–679. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Salazar, J.R.; Camacho, S.A.; Constantino, C.J.L.; Oliveira Junior, O.N. New Trends in Plasmonic (Bio)Sensing. An. Acad. Bras. Cienc. 2018, 90, 779–801. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Jiang, S.; Zhang, C.; Yue, W.; Zou, Y.; Wang, G.; Liu, H.; Zhang, X.; Li, M.; Zhu, Z.; et al. Ultrasensitive Label-Free Detection of DNA Hybridization by Sapphire-Based Graphene Field-Effect Transistor Biosensor. Appl. Surf. Sci. 2018, 427, 1114–1119. [Google Scholar] [CrossRef]
- Chen, C.; Liu, W.; Tian, S.; Hong, T. Novel Surface-Enhanced Raman Spectroscopy Techniques for DNA, Protein and Drug Detection. Sensors 2019, 19, 1712. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Cao, W.; Su, S.; Weng, L.; Song, S.; Fan, C.; Wang, L. Nanostructure-Based Surface-Enhanced Raman Scattering Biosensors for Nucleic Acids and Proteins. J. Mater. Chem. B 2016, 4, 1757–1769. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Lee, J.U.; Sim, S.J. Plasmonic Coupling-Dependent SERS of Gold Nanoparticles Anchored on Methylated DNA and Detection of Global DNA Methylation in SERS-Based Platforms. J. Opt. 2015, 17, 114022. [Google Scholar] [CrossRef]
- Aherne, D.; Ledwith, D.M.; Gara, M.; Kelly, J.M. Optical Properties and Growth Aspects of Silver Nanoprisms Produced by a Highly Reproducible and Rapid Synthesis at Room Temperature. Adv. Funct. Mater. 2008, 18, 2005–2016. [Google Scholar] [CrossRef]
- Chen, S.; Fan, Z.; Carroll, D.L. Silver Nanodisks: Synthesis, Characterization, and Self-Assembly. J. Phys. Chem. B 2002, 106, 10777–10781. [Google Scholar] [CrossRef]
- Wang, H.; Kundu, J.; Halas, N.J. Plasmonic Nanoshell Arrays Combine Surface-Enhanced Vibrational Spectroscopies on a Single Substrate. Angew. Chem.-Int. Ed. 2007, 46, 9040–9044. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Guan, P.; Qin, D.; Golden, G.; Wallace, P.M. Inverted Size-Dependence of Surface-Enhanced Raman Scattering on Gold Nanohole and Nanodisk Arrays. Nano Lett. 2008, 8, 1923–1928. [Google Scholar] [CrossRef] [PubMed]
- Zangana, S.; Lednický, T.; Bonyár, A. Three Generations of Surface Nanocomposites Based on Hexagonally Ordered Gold Nanoparticle Layers and Their Application for Surface-Enhanced Raman Spectroscopy. Chemosensors 2023, 11, 235. [Google Scholar] [CrossRef]
- Ngo, H.T.; Gandra, N.; Fales, A.M.; Taylor, S.M.; Vo-Dinh, T. DNA Detection and Single Nucleotide Mutation Identification Using SERS for Molecular Diagnostics and Global Health. In Proceedings of the Advanced Biomedical and Clinical Diagnostic and Surgical Guidance Systems XV, San Diego, CA, UAS, 28 January–2 February 2017; SPIE: Bellingham, DC, USA, 2017; Volume 10054, p. 100540C. [Google Scholar]
- Gaidi, M.; Daoudi, K.; Tlili, A.; Columbus, S.; Leblanc-Lavoie, J.; Ramachandran, K.; Suleiman, B.; Alhazaa, A.N.; El Khakani, M.A. Fast, Highly Sensitive and Label Free Detection of Small Genetic Sequence Difference of DNA Using Novel Surface-Enhanced Raman Spectroscopy Nanostructured Sensor. Sens. Biosensing Res. 2021, 32, 100406. [Google Scholar] [CrossRef]
- Lin, T.W.; Wu, H.Y.; Tasi, T.T.; Lai, Y.H.; Shen, H.H. Surface-Enhanced Raman Spectroscopy for DNA Detection by the Self-Assembly of Ag Nanoparticles onto Ag Nanoparticle-Graphene Oxide Nanocomposites. Phys. Chem. Chem. Phys. 2015, 17, 18443–18448. [Google Scholar] [CrossRef] [PubMed]
- Bonyár, A.; Zangana, S.; Lednický, T.; Rigó, I.; Csarnovics, I.; Veres, M. Application of Gold Nanoparticles–Epoxy Surface Nanocomposites for Controlling Hotspot Density on a Large Surface Area for SERS Applications. Nano-Struct. Nano-Objects 2021, 28, 100787. [Google Scholar] [CrossRef]
- Ambia-Garrido, J.; Vainrub, A.; Pettitt, B.M. A Model for Structure and Thermodynamics of SsDNA and DsDNA near a Surface: A Coarse Grained Approach. Comput. Phys. Commun. 2010, 181, 2001–2007. [Google Scholar] [CrossRef]
- Han, J.; Kim, D.; Kim, J.; Kim, G.; Fischer, P.; Jeong, H. Plasmonic Nanostructure Engineering with Shadow Growth. Adv. Mater. 2023, 35, 2107917. [Google Scholar] [CrossRef]
- Kimling, J.; Maier, M.; Okenve, B.; Kotaidis, V.; Ballot, H.; Plech, A. Turkevich Method for Gold Nanoparticle Synthesis Revisited. J. Phys. Chem. B 2006, 110, 15700–15707. [Google Scholar] [CrossRef] [PubMed]
- Kattumuri, V.; Chandrasekhar, M.; Guha, S.; Raghuraman, K.; Katti, K.V.; Ghosh, K.; Patel, R.J. Agarose-Stabilized Gold Nanoparticles for Surface-Enhanced Raman Spectroscopic Detection of DNA Nucleosides. Appl. Phys. Lett. 2006, 88, 153114. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, D.; Hao, Y.; Liu, Y.; Si, M. Label-Free Detection of Wild Mushrooms DNA Based on Surface-Enhanced Raman Spectroscopy. J. Raman Spectrosc. 2020, 51, 46–54. [Google Scholar] [CrossRef]
- Ferreira, I.C.F.R.; Vaz, J.A.; Helena Vasconcelos, M.; Martins, A. Compounds from Wild Mushrooms with Antitumor Potential. Anticancer Agents Med. Chem. 2010, 10, 424–436. [Google Scholar] [CrossRef]
- Jilani, A.; Abdel-wahab, M.S.; Hammad, A.H. Advance Deposition Techniques for Thin Film and Coating. In Modern Technologies for Creating the Thin-Film Systems and Coatings; InTech: Houston, TX, USA, 2017. [Google Scholar]
- Jin, R.; Cao, Y.; Mirkin, C.A.; Kelly, K.L.; Schatz, G.C.; Zheng, J.G. Photoinduced Conversion of Silver Nanospheres to Nanoprisms. Science 2001, 294, 1901–1903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ge, J.; Pham, T.; Goebl, J.; Hu, Y.; Lu, Z.; Yin, Y. Reconstruction of Silver Nanoplates by UV Irradiation: Tailored Optical Properties and Enhanced Stability. Angew. Chem. 2009, 121, 3568–3571. [Google Scholar] [CrossRef]
- Xue, C.; Mirkin, C.A. PH-Switchable Silver Nanoprism Growth Pathways. Angew. Chem.-Int. Ed. 2007, 46, 2036–2038. [Google Scholar] [CrossRef]
- Callegari, A.; Tonti, D.; Chergui, M. Photochemically Grown Silver Nanoparticles with Wavelength-Controlled Size and Shape. Nano Lett. 2003, 3, 1565–1568. [Google Scholar] [CrossRef]
- Washio, I.; Xiong, Y.; Yin, Y.; Xia, Y. Reduction by the End Groups of Poly(Vinyl Pyrrolidone): A New and Versatile Route to the Kinetically Controlled Synthesis of Ag Triangular Nanoplates. Adv. Mater. 2006, 18, 1745–1749. [Google Scholar] [CrossRef]
- Thacker, V.V.; Herrmann, L.O.; Sigle, D.O.; Zhang, T.; Liedl, T.; Baumberg, J.J.; Keyser, U.F. DNA Origami Based Assembly of Gold Nanoparticle Dimers for Surface-Enhanced Raman Scattering. Nat. Commun. 2014, 5, 3448. [Google Scholar] [CrossRef]
- Li, L.; Wang, Z.; Lu, Y.; Zhu, K.; Zong, S.; Cui, Y. DNA-Assisted Synthesis of Ortho-NanoDimer with Sub-Nanoscale Controllable Gap for SERS Application. Biosens. Bioelectron. 2021, 172, 112769. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Bell, S.E.J. Label-Free Detection of Single-Base Mismatches in DNA by Surface-Enhanced Raman Spectroscopy. Angew. Chem. Int. Ed. 2011, 50, 9058–9061. [Google Scholar] [CrossRef] [PubMed]
- Bonyár, A.; Kovács, R. Towards Digital Twins of Plasmonic Sensors: Constructing the Complex Numerical Model of a Plasmonic Sensor Based on Hexagonally Arranged Gold Nanoparticles. Nanomaterials 2023, 13, 2044. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.B.; Halas, N.J. Surface-Enhanced Raman Scattering on Tunable Plasmonic Nanoparticle Substrates. Proc. Natl. Acad. Sci. USA 2004, 101, 17930–17935. [Google Scholar] [CrossRef]
- Levin, C.S.; Kundu, J.; Barhoumi, A.; Halas, N.J. Nanoshell-Based Substrates for Surface Enhanced Spectroscopic Detection of Biomolecules. Analyst 2009, 134, 1745–1750. [Google Scholar] [CrossRef] [PubMed]
- Weyher, J.L.; Kelly, J.J. Defect-Selective Etching of Semiconductors. In Springer Handbook of Crystal Growth; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Grand, J.; De La Chapelle, M.L.; Bijeon, J.L.; Adam, P.M.; Vial, A.; Royer, P. Role of Localized Surface Plasmons in Surface-Enhanced Raman Scattering of Shape-Controlled Metallic Particles in Regular Arrays. Phys. Rev. B Condens. Matter Mater. Phys. 2005, 72, 033407. [Google Scholar] [CrossRef]
- Greeneltch, N.G.; Blaber, M.G.; Schatz, G.C.; Van Duyne, R.P. Plasmon-Sampled Surface-Enhanced Raman Excitation Spectroscopy on Silver Immobilized Nanorod Assemblies and Optimization for near Infrared (Λex = 1064 Nm) Studies. J. Phys. Chem. C 2013, 117, 2554–2558. [Google Scholar] [CrossRef]
- Félidj, N.; Aubard, J.; Lévi, G.; Krenn, J.R.; Salerno, M.; Schider, G.; Lamprecht, B.; Leitner, A.; Aussenegg, F.R. Controlling the Optical Response of Regular Arrays of Gold Particles for Surface-Enhanced Raman Scattering. Phys. Rev. B Condens. Matter Mater. Phys. 2002, 65, 0754191–0754199. [Google Scholar] [CrossRef]
- McFarland, A.D.; Young, M.A.; Dieringer, J.A.; Van Duyne, R.P. Wavelength-Scanned Surface-Enhanced Raman Excitation Spectroscopy. J. Phys. Chem. B 2005, 109, 11279–11285. [Google Scholar] [CrossRef]
- Grillet, N.; Manchon, D.; Cottancin, E.; Bertorelle, F.; Bonnet, C.; Broyer, M.; Lermé, J.; Pellarin, M. Photo-Oxidation of Individual Silver Nanoparticles: A Real-Time Tracking of Optical and Morphological Changes. J. Phys. Chem. C 2013, 117, 2274–2282. [Google Scholar] [CrossRef]
- Hsieh, C.-C.; Balducci, A.; Doyle, P.S. Ionic Effects on the Equilibrium Dynamics of DNA Confined in Nanoslits. Nano Lett. 2008, 8, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, W.; Wang, D.; Gong, Z.; Fan, M. Evaluation of the Intrinsic PH Sensing Performance of Surface-Enhanced Raman Scattering PH Probes. Microchem. J. 2020, 154, 104565. [Google Scholar] [CrossRef]
- Nardo, V.M.; Renda, V.; Trusso, S.; Ponterio, R.C. Role of Ph on Nanostructured Sers Active Substrates for Detection of Organic Dyes. Molecules 2021, 26, 2360. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Chowdhury, J.; Dutta, S.; Pal, T. A PH Dependent Raman and Surface Enhanced Raman Spectroscopic Studies of Citrazinic Acid Aided by Theoretical Calculations. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 169, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Kazanci, M.; Schulte, J.P.; Douglas, C.; Fratzl, P.; Pink, D.; Smith-Palmer, T. Tuning the Surface-Enhanced Raman Scattering Effect to Different Molecular Groups by Switching the Silver Colloid Solution PH. Appl. Spectrosc. 2009, 63, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wong, J.H.; Liu, S.J. Fluorescence and Raman Study of PH Effect on the Adsorption Orientations of Methyl Red on Silver Colloids. Appl. Spectrosc. 2011, 65, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Chong, N.S.; Smith, K.A.; Setti, S.; Ooi, B.G. Application of Gold and Silver Colloidal Nanoparticles for the Surface-Enhanced Raman Spectrometric Analysis of Melamine and 4-Aminobiphenyl. Int. J. Environ. Technol. Manag. 2013, 16, 3–20. [Google Scholar] [CrossRef]
- Bi, L.; Wang, Y.; Yang, Y.; Li, Y.; Mo, S.; Zheng, Q.; Chen, L. Highly Sensitive and Reproducible SERS Sensor for Biological PH Detection Based on a Uniform Gold Nanorod Array Platform. ACS Appl. Mater. Interfaces 2018, 10, 15381–15387. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.; Neumann, O.; Janesko, B.G.; Zhang, D.; Lal, S.; Barhoumi, A.; Scuseria, G.E.; Halas, N.J. Adenine− and Adenosine Monophosphate (AMP)−Gold Binding Interactions Studied by Surface-Enhanced Raman and Infrared Spectroscopies. J. Phys. Chem. C 2009, 113, 14390–14397. [Google Scholar] [CrossRef]
- Paleček, E.; Jelen, F. Electrochemistry of Nucleic Acids. Perspect. Bioanal. 2005, 1, 73–173. [Google Scholar] [CrossRef]
- Tan, Z.J.; Chen, S.J. Nucleic Acid Helix Stability: Effects of Salt Concentration, Cation Valence and Size, and Chain Length. Biophys. J. 2006, 90, 1175–1190. [Google Scholar] [CrossRef] [PubMed]
- Bonyár, A.; Csarnovics, I.; Szántó, G. Simulation and Characterization of the Bulk Refractive Index Sensitivity of Coupled Plasmonic Nanostructures with the Enhancement Factor. Photonics Nanostruct. 2018, 31, 1–7. [Google Scholar] [CrossRef]
- Li, Y.; Gao, T.; Xu, G.; Xiang, X.; Zhao, B.; Han, X.X.; Guo, X. Direct Approach toward Label-Free DNA Detection by Surface-Enhanced Raman Spectroscopy: Discrimination of a Single-Base Mutation in 50 Base-Paired Double Helixes. Anal. Chem. 2019, 91, 7980–7984. [Google Scholar] [CrossRef] [PubMed]

| Peak Position (cm−1) | Assignment |
|---|---|
| 471 | T (ring stretching) |
| 574 | A (ring deformation) |
| 649, 659, 656 | G (ring breathing) |
| 667–669, 675 | G (ring breathing) |
| 685 | A (stretching), G (deformation (C-H)) |
| 707, 719 | A (scissoring, C–S) |
| 723–731 | A (ring breathing) |
| 745–754 | T (stretching in C5–CH3) |
| 750–758 | T (ring breathing) |
| 761 | A (ring breathing) |
| 783–787 | C (ring breathing) |
| 865 | G (ring stretching) |
| 828–835 | Phosphodiester O–P–O stretching, T (C4–C5 stretching) |
| 875 | Deoxyribose ring |
| 934, 936, 938, 939 | A/C/G (deoxyribose stretching) |
| 1005–1008 | C–O stretch in the deoxyribose |
| 1021–1034 | T (C–N–C bending) |
| 1061 | A (C–N stretching) |
| 1073–1080 | PO2 stretch in backbone |
| 1077 | PO2 stretch in backbone |
| 1124 | A (stretching of the deoxyribose phosphate backbone) |
| 1160 | A/G/T (stretching of the deoxyribose phosphate backbone) |
| 1190–1198 | C (C–N stretching, N-H bending) |
| 1203, 1207, 1208 | T (stretching of the deoxyribose phosphate backbone) |
| 1262 | A/T (C–C and C–N stretching) |
| 1269–1276 | C (ring stretching, C–N stretching) |
| 1290, 1299 | C (CH2 deformation) |
| 1333 | A/G (CH2 wagging mode) |
| 1341 | G (C–N stretching) |
| 1355, 1360, 1370 | A/C/G/T (C–N stretching) |
| 1398 | T (NH deformation/CH3 deformation) |
| 1461 | A (C–H deformation of deoxyribose) |
| 1507, 1514, 1518 | C (C–N stretching, NH2 deformation) |
| 1580–1590 | C/G/T (C–N stretching) |
| 1602 | A/G (C=C stretching) |
| 1643 | C/G/T (C=O stretching, C=C stretching) |
| 1670 | A (NH2 scissoring) |
| 1700 | T (C=O stretching) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zangana, S.; Veres, M.; Bonyár, A. Surface-Enhanced Raman Spectroscopy (SERS)-Based Sensors for Deoxyribonucleic Acid (DNA) Detection. Molecules 2024, 29, 3338. https://doi.org/10.3390/molecules29143338
Zangana S, Veres M, Bonyár A. Surface-Enhanced Raman Spectroscopy (SERS)-Based Sensors for Deoxyribonucleic Acid (DNA) Detection. Molecules. 2024; 29(14):3338. https://doi.org/10.3390/molecules29143338
Chicago/Turabian StyleZangana, Shireen, Miklós Veres, and Attila Bonyár. 2024. "Surface-Enhanced Raman Spectroscopy (SERS)-Based Sensors for Deoxyribonucleic Acid (DNA) Detection" Molecules 29, no. 14: 3338. https://doi.org/10.3390/molecules29143338




